Abstract
Many elderly individuals remain dementia-free throughout their life. However, some of these individuals exhibit Alzheimer disease neuropathology on autopsy, evidenced by neurofibrillary tangles (NFTs) in AD-specific brain regions. We conducted a genome-wide association study to identify genetic mechanisms that distinguish non-demented elderly with a heavy NFT burden from those with a low NFT burden. The study included 344 non-demented subjects with autopsy (201 subjects with low and 143 with high NFT levels). Both a genotype test, using logistic regression, and an allele test provided genome-wide significant evidence that variants in the RELNgene are associated with neuropathology in the context of cognitive health. Immunohistochemical data for reelin expression in AD-related brain regions added support for these findings. Reelin signaling pathways modulate phosphorylation of tau, the major component of NFTs, either directly or through β-amyloid pathways that influence tau phosphorylation. Our findings suggest that up-regulation of reelin may be a compensatory response to tau-related or beta-amyloid stress associated with AD even prior to the onset of dementia.
1. INTRODUCTION
Prevalence rates for Alzheimer disease (AD) climb steadily from ~1% in 65-year-olds to as high as 40% by the age of 85[Hebert, et al. 2003]. Despite these dire figures, some elderly individuals remain dementia-free throughout their life and, of these, a proportion exhibit substantial AD neuropathology, in the form of amyloid plaques and neurofibrillary tangles (NFTs),at autopsy [Erten-Lyons, et al. 2009], [Bennett, et al. 2005].Thus, individuals differ in their capacity to maintain normative cognitive function even within the context of neuropathological structural changes associated with AD.
There are a number of ways in which genetic mechanisms might promote cognitive health. On one hand, genetic variants may prevent development of the neuropathological substrate that underlies susceptibility to cognitive dysfunction. Alternatively, in the presence of neuropathology, genetic variants may mitigate the effects of AD-related lesions that otherwise result in cognitive decline. The first case provides greater guarantee of cognitive health. Thus, it is important to identify genetic variants associated with development of AD neuropathology within the context of cognitive resilience.
We conducted a genome-wide SNP association study (GWAS) to identify genetic mechanisms involved in healthy brain aging. This study was based on a sample of non-demented, deceased subjects with autopsy who were members of longitudinal healthy aging cohorts at 10 NIA-funded Alzheimer Disease Centers (ADC). These subjects comprised one group with little or no evidence of NFT formation, and one with substantial or severe levels of NFT formation in critical brain regions. We report results that implicate variants in the glycoprotein reelin (RELN)as key elements in the molecular basis of AD-related neuropathological processes. We provide additional support for these statistical findings with immunohistochemical data for reelin expression in AD-related brain regions in a subset of our study subjects. To our knowledge, this is the first GWAS that specifically addresses genetic mechanisms of AD neuropathology in non-demented elderly with post-mortem examinations.
2. Methods
2.1 Subjects
Subjects were recruited from aging research cohorts collected over the last two decades at 10 NIA-funded ADCs across the country. Eligibility criteria included the following: 1) ≥65 years old at enrollment; 2) deceased, with autopsy; 3) clinical diagnosis of “no dementia” at enrollment and death; 4) ≥1 clinical evaluation within the year before death; 5) DNA available; 6) Caucasian ancestry. A total of 412 subjects met initial criteria. All subjects had been previously genotyped for apolipoproteinE (APOE ) status and these data were provided by the respective ADC Data Cores. The study was approved by the IRB at OHSU.
2.2 Neuropathologic Diagnosis
One of the hallmarks of AD is the occurrence of NFTs in limbic and neocortical regions of the brain. The Braak score[Braak and Braak 1991] is a measure of the location and frequency of NFTs and is key to establishing the pathologic diagnosis of AD[1997]. Braak scores (BS) range from 0 (no NFTs) to 6 (NFTs in primary motor and/or sensory neocortex), and were obtained for all subjects from the ADC Clinical Data Cores.
We classified subjects on the basis of NFT burden to obtain subsets of individuals for analysis. Autopsy reports were reviewed for consistency with the Braak score provided in the data files from each ADC by a 3-member team including a neuropathologist (RW), a neurologist (JK) and a geneticist (PK). Subjects were classified into three groups: LOBraak (≤2), MEDBraak and HIBraak (≥4):
If the Braak score and information in the autopsy report were consistent with low NFT levels, the subject was classified as LOBraak (n=189).
If the Braak score and information in the autopsy were consistent with high NFT levels, the subject was classified as HIBraak (n=110).
If the Braak score and autopsy information were at odds with respect to classification of LO or HI NFT burden, preference was given to the autopsy (n=12).
If the Braak score and autopsy information agreed with a Braak score of 3, the subjects was classified as MEDBraak (n= 80); furthermore, if the BS was 2, 3 or 4, but the autopsy report lacked sufficient detail to confirm these scores, the subject was also classified as MEDBraak (n=21).In order to maximize phenotypic homogeneity between groups, and thus increase power, we excluded this group from the initial analysis.
The final sample consisted of 311 subjects (192 LOBraak, 119HIBraak).
2.3 Clinical Diagnosis
All study subjects were deceased and had been evaluated for cognitive decline and dementia within 12 months prior to death. Assessments for determining absence of dementia were consistent with standardized protocols in the DSM-III-R.
To minimize the extent of any cognitive impairment in the two groups, we obtained longitudinal clinical data for all subjects, consisting of Clinical Dementia Rating (CDR) scores[Morris 1993] and/or Mini-Mental State Exam (MMSE) scores[Folstein, et al. 1975], from the time of enrollment until death. The CDR is a dementia staging-tool in which a score of 0 represents no cognitive impairment, .5 may represent some cognitive impairment, and a score≥1 represents dementia; MMSE scores≥26are generally indicative of no dementia.
Longitudinal CDR scores were available for 143 of the 311 subjects. Of these, 120 (84%) had no CDR scores>0; 23(16%) had one CDR of .5 at some point during participation. Among these 23, 15 were in the LO Braak and 8 in the HIBraak group. In the majority of these 23 subjects – 11 in the LO and 5 in the HI Braak group – the CDR of .5 occurred at some point prior to the last evaluation and was followed by CDRs of 0 in subsequent evaluations. Longitudinal MMSE scores were available for the remaining 168 subjects. All had MMSE ≥ 26 at death (mean = 28.3; sd = 1.3).
2.4 Genotyping
DNA was extracted from blood in 91 of the 311 subjects, using standard procedures. For the remaining 220 subjects, DNA was extracted from frozen brain tissue. Genomic DNA was extracted from 100 mg of brain tissue using the Wizard Genomic DNA Purification Kit according to the manufacturer (Promega,Inc.). DNA quality and concentration were determined using a Biomate-3 spectrophotometer.
Genotyping was performed by deCODE Genetics (Reykjavik, Iceland). Genotypes were obtained using the Illumina HumanCNV370v1_C array from 750 ng of genomic DNA following manufacturer's protocols (Illumina, Inc.). Illumina's BeadStudio3.1.14 genotyping module was used to cluster, call genotypes, and assign confidence scores.
2.5 Analysis
2.5.1Quality Control
We included only individuals with a missing genotype rate < 20%. Specifically, three individuals had missing rates of 19, 18 and 16%, and all others had rates < 2%. Since SNPs with low call rates may inflate association statistics [Plenge, et al. 2007], only autosomal SNPs (n=334,221) with call rates ≥ 99% across the entire sample were used (21387 SNPs excluded due to low call rates). Only SNPs with a minor allele frequency (MAF) > 0.05 in cases and controls were included (20,730 SNPs excluded for low MAF). We also required absence of strong deviation from Hardy-Weinberg equilibrium (HWE) in controls; specifically, likelihood ratio χ2< 50, corresponding to a nominal p-value of 1.54 × 10-12. This threshold was chosen so that SNPs with only the most extreme deviations from HWE would be deleted (30 SNPs excluded for deviation from HWE). As a result of these quality control measures, a total of 292,074 SNPs were used in this analysis.
2.5.2 Association Analysis
Two different methods were used to test for the association between SNPs and the phenotype --association at the genotype and at the allele level. The former was carried out via logistic regression analysis with covariates and the latter with the Set Association approach[Hoh, et al. 2001] without covariates. We tested the effect of putative non-genetic risk factors on the phenotype, and found that sex and age-at-death had a significant impact. Thus in the logistic regression approach, genotypes at a given SNP (a categorical variable), sex (a categorical variable) and age of death (a continuous variable) were included as independent variables, and the Braak score category, HI (“case”) vs. LO (“control”), as the dependent variable. The fitting of a general linear model (binomial family in this case) was conducted in R (http://cran.cnr.berkeley.edu/web/packages/qvalue/index.html). For the two degrees of freedom associated with the genotypes at a given SNP, logistic regression furnished two p-values. To obtain a single significance level for one variant, these two (independent) p-values were combined into one with the unweighted Z transform method[Whitlock 2005].
We chose agenotypic model because it is a general model, and is sensitive to different types of association. It is also ideally suited for logistic regression analysis, in which we allow for the total effects of genotypes and non-genetic factors, the latter of which act on individuals (i.e., genotypes) rather than alleles. Since the logistic regression result is most robust when all three possible genotypes are represented in both comparison groups, we only included SNPs in which this was the case; of the 292,074 SNPs which passed quality control measures described above, 12,163 were excluded because there were < 3 genotype classes in the HI Braak and/or LO Braak groups. Thus, the total number of SNPs in the logistic regression analysis was 279,911. To assess the association of SNP alleles with our phenotype, we initially carried out 1 df chi-square tests in 2 × 2 tables of alleles for each of the 279,911 SNPs and constructed a Q-Q plot for the resulting chi-square values using the Systat 12 statistics package (http://www.systat.com/).In Set Association analysis[Hoh, et al. 2001], we evaluated the combined effects of multiple SNPs. The combined association of a given number of SNPs (wherever they are in the genome) was measured by the sum of their corresponding test statistics (1 df chi-squares). We focused on the most significant m SNPs, with m ranging successively from 1 through 15. Significance levels for each such sum were evaluated with permutation analysis, and the smallest p-value for these sums was taken as the genome-wide test statistic, whose significance level was again evaluated in randomization samples. This approach represents an approximation to multivariate analysis[Manly 2007] and, thus, assesses the joint effects of the most significant SNPs on disease association. In this analysis, the sumstat program was used with 20,000 randomization samples per run (http://linkage.rockefeller.edu/ott/sumstat.html).
2.5.3 Population Stratification
We used the agglomerative clustering method based on identity by state (IBS), as implemented in PLINK v1.05[Purcell, et al. 2007] (http://pngu.mgh.harvard.edu/~purcell/plink/) to test for population stratification in our data.
2.5.4 Post-hoc Analysis
For particular SNPs of interest, we conducted a post-hoc analysis in which allelic association to the phenotype was tested with a likelihood ratio(LR) chi-square test, and the odds ratio(OR) was calculated with a 95% confidence interval (CI).
2.5.6 Immunohistochemistry
Immunohistochemistry for reelin expression was performed on cases with autopsy performed at OHSU and for which the postmortem interval was less than 24 hours. Brains were fixed in neutral-buffered formaldehyde solution for at least 2 weeks and fixed hippocampal sections were processed in paraffin 7-μm sections, deparaffinized, subjected to standard antigen retrieval techniques (5 minute treatment at room temperature with 95% formic acid, followed by incubation at 85-90 °C in citrate buffer, pH 6.0 for 30 min) and stained with antibody to reelin (Santa Cruz Biotechnology, Santa Cruz, CA, antibody H221 used at 1:200 dilution). Sections were developed using the Vectastain ABC technique (Vector Laboratories, Burlingame, CA). Reelin staining of pyramidal neurons was evaluated in CA1-4 regions of the hippocampus and in subiculum by a neuropathologist (RLW) who was blinded to case status. A semiquantitative score was assigned to reelin positivity in pyramidal neurons in each hippocampal sector (0=no detectable reelin staining, 1=cytoplasmic blush staining detectable at 40x magnification, 2=clear cytoplasmic staining identifiable at 10x magnification, 3=pyramidal neurons identified as strong band of cytoplasmic staining visible at 4x magnification. Total reelin staining was calculated as the sum of the score in each region of hippocampus.
3. Results
3.1 Sample Characteristics
Sample characteristics of the two groups are provided in Table 1. The proportion of females is increased in the HIBraak group, and the difference is significant (p=0.019). Subjects in the HIBraak category were significantly older at death than those in the LOBraak group (p<.0001); thus, we incorporated both gender and age-at-death as covariates. APOE allele frequencies(based on pre-determined APOE genotypes) did not differ significantly between groups (p=.556), nor did education level (p=.544).
Table 1.
Sample Characteristics
| LO Braak (n=192) # (%) | HI Braak (n=119) # (%) | p-value | ||
|---|---|---|---|---|
| Sex: | male | 98 (51%) | 44 (37%) | .019 |
| female | 94 (49%) | 75 (63%) | Fisher's exact, 2-tailed | |
| Age at Death: (years) | mean (sd) | 83.83 (7.62) | 87.77 (6.12) | < .0001 |
| range | 65-104 | 72-106 | t-test | |
| Education: (years) | mean(sd) | 17.08 (3.88) | 17.30 (3.80) | .544 |
| range | t-test | |||
| APOE allele frequency n (%) | ε2 | 38 (10%) | 23 (10%) | .556 |
| ε3 | 299(80%) | 180 (77%) | χ2 = 1.17,2 df | |
| ε4 | 39 (10%) | 31(13%) | ||
| Neuritic plaque density: | frequent | 9 (5%) | 31(26%) | < .001 |
| moderate | 44 (23%) | 50 (42%) | χ2 = 58.1, 3df | |
| sparse | 40 (21%) | 16 (14%) | ||
| none | 97 (51%) | 21 (18%) | ||
The distribution of neuritic plaque density was different between groups (p<0.0001). Only 28% of subjects in the LOBraak category had “frequent” or “moderate” plaques, compared to 68% in the HI category. Hence, although Braak score was used to stratify groups by AD-associated neuropathological features, stratification by neuritic plaque burden also occurred.
3.2 Analysis
Results of the test for population stratification indicated that all subjects were assigned to the same cluster, and there was no significant difference between pair-wise IBS distances in the LO and HI Braak groups. Thus, we detected no evidence for population stratification.
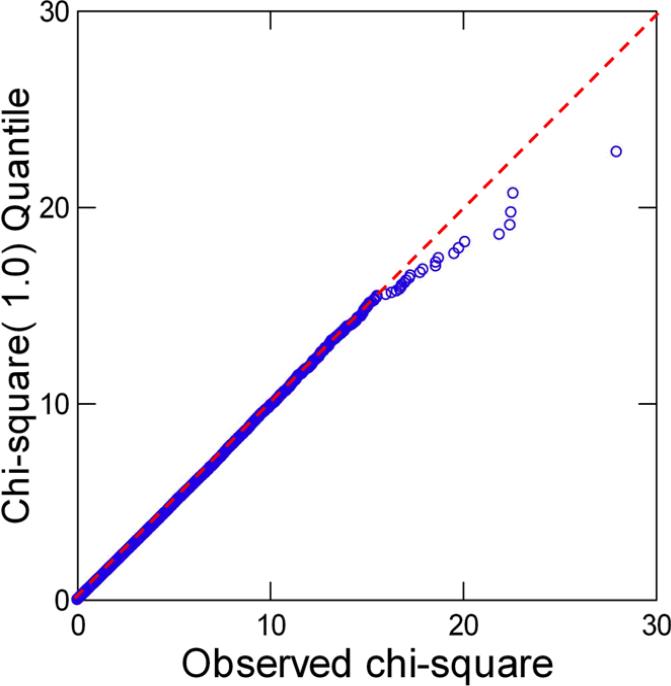
Allelic association for one SNP at a time resulted in one SNP, rs4298437 on chromosome 7, showing a Bonferroni-corrected significance level of p = 0.0341. Results for the ten SNPs with lowest p-values are shown in Table 2. The Q-Q plot in Figure 1 for all 279,911 SNPs demonstrates that most of the chi-square values perfectly follow theoretically expected values, thus confirming the conclusion of absence of population stratification. The plot also shows that at least ten SNPs have elevated test statistics.
Table 2.
Results of the allele association tests, showing the ten best SNPs, listed in order of significance
| Rank | SNP | Chr | Position | Chi-square | pNom1 | pBon2 | pRand3 | pSum4 | Gene5 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs4298437 * | 7 | 103413113 | 27.9567 | 1.241E-07 | 0.0341 | 0.0354 | 0.0354 | RELN |
| 2 | rs11782819 * | 8 | 10372191 | 22.5979 | 1.997E-06 | 0.4282 | 0.4086 | 0.0538 | UNQ9391 |
| 3 | rs6943822 * | 7 | 103385907 | 22.4898 | 2.113E-06 | 0.4464 | 0.4263 | 0.0463 | RELN |
| 4 | rs3791523 | 2 | 239753412 | 22.4401 | 2.168E-06 | 0.4549 | 0.4337 | 0.0355 | HDAC4 |
| 5 | rs1481650 | 8 | 26829428 | 21.8844 | 2.896E-06 | 0.5554 | 0.5347 | 0.0288 | ADRA1A |
| 6 | rs6734151 | 2 | 5387284 | 20.0985 | 7.355E-06 | 0.8724 | 0.8510 | 0.0284 | unknown |
| 7 | rs10956170 | 8 | 125381841 | 19.7785 | 8.695E-06 | 0.9123 | 0.8897 | 0.0280 | TMEM65 |
| 8 | rs894520 | 9 | 38179527 | 19.5413 | 9.845E-06 | 0.9364 | 0.9185 | 0.0273 | unknown |
| 9 | rs9530579 | 13 | 76123603 | 18.7333 | 1.503E-05 | 0.9851 | 0.9768 | 0.0284 | unknown |
| 10 | rs4790069 | 17 | 74561296 | 18.593 | 1.618E-05 | 0.9892 | 0.9825 | 0.0285 | C1QTNF1 |
gene (in which SNP is located) appears also in the list of significant genes obtained in the logistic regression analysis (Table 3): RELN (rs4298437 and rs6943822) and UNQ9391 (rs11782819)
pNom = nominal p-value
pBon = Bonferroni-corrected p-value (n = 279,911 SNPs)
pRand = p-value computed in 20,000 randomization samples
pSum = p-value associated with chi-square values summed up to given rank
data from NCBI MapViewer Buuild 36.3 (http://www.ncbi.nlm.nih.gov/mapview/)
Figure 1.
Q-Q plot based on nominal p-values resulting from the allelic association test
In the Set Association analysis, the sum of chi-squares for SNPs (Table 2; pSum) ranked 1-8 exhibited the smallest significance level, pmin = 0.0273. This analysis indicated that approximately eight SNPs jointly contribute to AD neuropathology although this conclusion is only marginally significant (the significance level associated with pmin, corrected for testing 15 sums, was p = 0.0577). Nonetheless, this result confirms an analogous conclusion based on the QQ plot. For the ten largest chi-square values, Table 2 also shows p-values based on permutation tests (pRand) but these values are close to those obtained by Bonferroni correction for multiple testing, which indicates that test results for the 279,911 SNPs are largely independent.
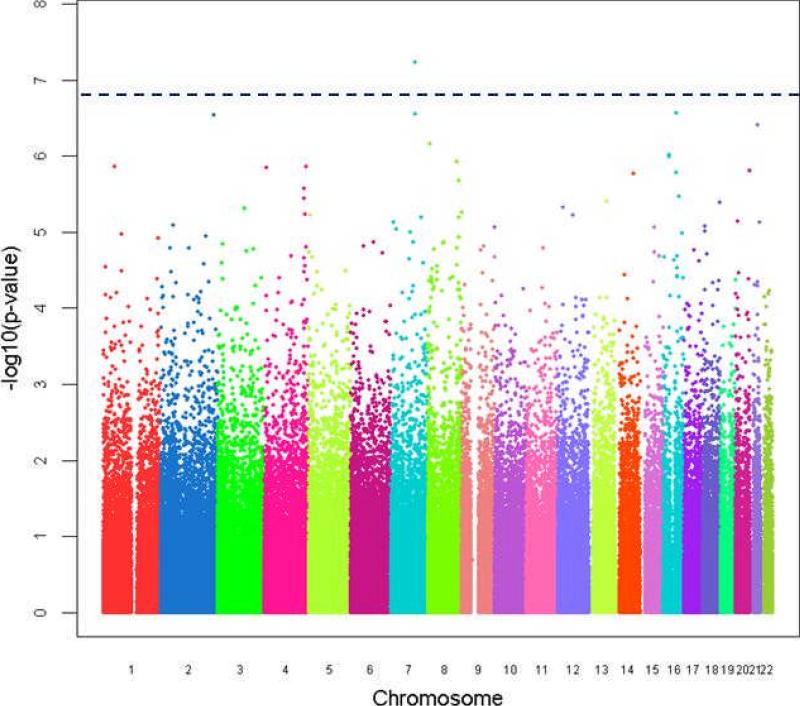
For the logistic regression controlling for age of death and sex, the genome-wide distribution of all SNPs and their regression p-values are presented in Figure 2. The ten SNPs with the lowest p-values for an association with the AD neuropathology phenotype are listed in Table 3.Two of the three most significant SNPs, rs4298437 and rs6943822, are in reelin (RELN); the p-value for rs4298437 reached genome-wide significance with the conservative Bonferroni correction (p = .0162). Both reelin SNPs are located in the 5’end of the gene.
Figure 2.
Manhattan plot showing the distribution of analyzed SNPs and their corresponding genotypic association p-values from logistic regression. The SNPs are ordered by their positions on the chromosomes, and the chromosomes (number 1 to 22 from left to right) are alternately colored. The y axis is –log10(p-value).
Table 3.
Results of the genotype association test using logistic regression analysis with covariates, showing the 10 SNPs with the lowest p-values, in order of significance
| SNP | Chr | Position | pNom1 | pBon2 | Gene3 | Location |
|---|---|---|---|---|---|---|
| rs4298437 * | 7 | 103413113 | 5.86E-08 | 0.0162 | RELN | Intron |
| rs4784670 | 16 | 55056012 | 2.73E-07 | 0.0764 | OGFOD1 | Intron |
| rs6943822 * | 7 | 103385907 | 2.80E-07 | 0.0783 | RELN | Intron |
| rs10498214 | 2 | 227772424 | 2.90E-07 | 0.0812 | COL4A3 | Intron |
| rs2833546 | 21 | 32170575 | 3.94E-07 | 0.1103 | HUNK | Intron |
| rs11782819 * | 8 | 10372191 | 6.84E-07 | 0.1914 | UNQ9391 | 45 kb upstream |
| rs11649538 | 16 | 24415618 | 9.54E-07 | 0.267 | RBBP6 | 50 kb upstream |
| rs152745 | 16 | 23273923 | 1.02E-06 | 0.2855 | SCNN1B | Intron |
| rs4457311 | 8 | 125353540 | 1.20E-06 | 0.3359 | LOC442396 | Intron |
| rs11205449 | 1 | 48419711 | 1.38E-06 | 0.3863 | SLC5A9 | 40 kb upstream |
gene (in which SNP is located) appears also in the list of significant genes obtained in the allele association analysis (Table 2)
pNom = nominal p-value
pBon = Bonferroni-corrected p-value (n = 279,911 SNPs)
data from NCBI MapViewer Buuild 36.3 (http://www.ncbi.nlm.nih.gov/mapview/)
In the post-hoc analysis both reelin SNPs showed highly significant allelic association with the phenotype, as shown in Table 4. Alleles associated with the HIBraak group are the A allele of rs4298437 (OR=2.51), and the A allele of rs6943822 (OR=2.23).
Table 4.
Results of post-hoc allelic association analysis of SNPs in RELN
| SNP | Likelihood χ2 p-value | Freq Allele G | Freq Allele G | Freq Allele A | Freq Allele B | Odds Ratio | 95% CI |
|---|---|---|---|---|---|---|---|
| rs6943822 | 0.000 | 0.33 | 0.52 | 0.67 | 0.48 | 2.23 | 1.59 - 3.12 |
| rs4298437 | 0.000 | 0.54 | 0.74 | 0.46 | 0.26 | 2.51 | 1.77 - 3.53 |
The overlap between the results of the allelic association test and the logistic regression analysis was minimal; three of the most significant SNPs in the former were included among the most significant SNPs in the latter. We attribute two reasons to this apparent discrepancy. First, we incorporated age-at-death and sex as covariates in the logistic regression analysis, whereas the allele test was carried out without inclusion of covariates. Also, the allele test may be less powerful than the genotype test. In spite of these differences, the prominent feature of this analysis is that the RELN SNP rs4298437 is the most significant SNP in both the allele and the genotype (with covariates) tests, with genome-wide significant Bonferroni-corrected p-values of .0341 and .0162, respectively; furthermore, in both tests, a second RELN SNP, rs6943822, is the third most significant SNP.
At the outset of this study, we excluded 101 subjects with mid-range (MED) Braak scores in order to maximize phenotypic differences between groups and avoid potential confounding effects of including subjects in this transitional category. Following the analysis, we examined the frequency of the RELN genotypes in rs4298437 and rs6943822 in this group of subjects. As indicated in Table 5, frequencies for both SNPs in the MED Braak subjects were intermediate between those of the LO and HI Braak subjects. This substantiates our approach of using extreme phenotypes rather than introducing the potential confounding effect of choosing an arbitrary midrange value for dichotomizing groups.
Table 5.
Frequencies(n, %) of RELNSNP genotypes in Braak score categories
| Genotype | rs4298437 | ||
|---|---|---|---|
| LO Braak | MED Braak | HI Braak | |
| AA | 10 (.05) | 10 (.10) | 25 (.18) |
| AG | 81 (.41) | 49 (.48) | 74 (.52) |
| GG | 105 (.54) | 42 (.42) | 42 (.30) |
| Total | 196 | 101 | 141 |
| rs6943822 | |||
| AA | 47 (.24) | 29 (.28) | 54 (.38) |
| AG | 95 (.48) | 48 (.48) | 73 (.52) |
| GG | 54 (.28) | 24 (.24) | 14 (.10) |
| Total | 196 | 101 | 141 |
Despite differences in AD neuropathology, there was no significant difference between HI and LO Braak subjects with respect to the frequency of the APOEε4 allele (Table 1). This lack of association between the ε4 allele and high levels of AD neuropathology may be related to the advanced age of these subjects (mean age at death in the HI Braak group = 87.8 years; Table 1). This would be consistent with a number of studies that suggest that the positive association between APOEε4 and AD declines after the age of 85 years [Juva, et al. 2000]. In order to further validate this finding, we examined the frequency of APOEε4 carriers (at least one ε4 allele) vs. non-carriers among the three genotypes for both RELN SNPs. As shown in Table 6, there were no significant differences in the frequency of ε4 carriers and non-carriers across SNP genotypes for rs6943822 (p = .50) or rs4298437 (p = .13).
Table 6.
Relationship between APOE ε4 frequency and RELN genotypes
| SNP rs6943822 genotype* | # subjects w/ ≥ 1 ε4 allele | # subjects w/ no ε4 allele | SNP rs4298437 genotype** | # subjects w/ ≥ 1 ε4 allele | # subjects w/ no ε4 allele |
|---|---|---|---|---|---|
| GG | 10 (.15) | 51 (.21) | GG | 23 (.35) | 110 (.46) |
| AG | 35 (.52) | 114 (.48) | AG | 33 (.49) | 105 (.44) |
| AA | 22 (.33) | 73 (.31) | AA | 11 (.16) | 23 (.10) |
| total | 67 | 238 | total | 67 | 238 |
p = .50
p = .13
3.3 Reelin Immunohistochemistry
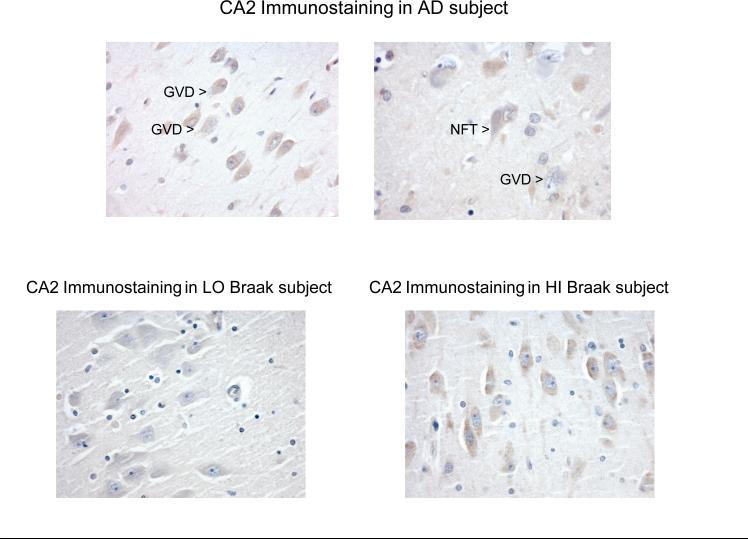
Reelin immunohistochemistry was performed on hippocampal sections from 19 (10 from the HI and 9 from the LO Braak group) subjects. Immunohistochemical staining for reelin was also performed on hippocampi from four subjects with clinical and neuropathological diagnoses of AD (Braak stage VI with moderate to frequent neuritic plaques). In general, reelin expression was confined to pyramidal neurons where it was present as variably intense cytoplasmic staining with highest expression in theCA2/3 sector of the hippocampus as previously described[Martinez-Cerdeno, et al. 2002]. Reelin expression tended to be very pronounced in AD with strong staining encountered especially in regions with many neurons undergoing granulovacuolar degeneration (GVD), a cytoplasmic feature associated with AD (Figure 3, top). Neurons with mild to moderate GVD in particular tended to have strong cytoplasmic reelin expression; granules and vacuoles themselves did not contain reelin and neurons with marked GVD tended to express less reelin due to extensive degeneration in general and to replacement of cytoplasmic with vacuolar lesions in particular. Reelin staining of neurofibrillary tangles and neuritic plaques was not conspicuous.
Figure 3.
Reelin expression in hippocampal pyramidal neurons. Hippocampal immunohistochemistry was performed on paraffin sections of hippocampus and representative pyramidal neurons from the CA2 region are depicted in AD (upper panel) and in non-demented subjects (lower panel). Strong cytoplasmic staining was noted in pyramidal neurons in AD, especially in regions in which significant granulovacuolar degeneration (GVD) was noted. Neuritic plaques and neurofibrillary tangles (NFTs) did not stain strongly for reelin. Cytoplasmic expression of reelin in pyramidal neurons was also present in non-demented subjects with HI Braak stages as defined in the text (lower right panel), similar to that observed in AD. Expression of reelin in LO Braak stages was less conspicuous (lower left panel).
In contrast, cognitively intact subjects in the LO Braak group tended to have lower reelin expression in the hippocampus , whereas reelin expression in the HI Braak group was similar to that observed in AD hippocampi. Hippocampal pyramidal neurons in both the LO and HI Braak groups displayed less GVD than in AD, but the tendency for increased reelin expression in the HI Braak group was still notable in neurons in which GVD was not present or was present to only a very limited extent (Figure 3, bottom).
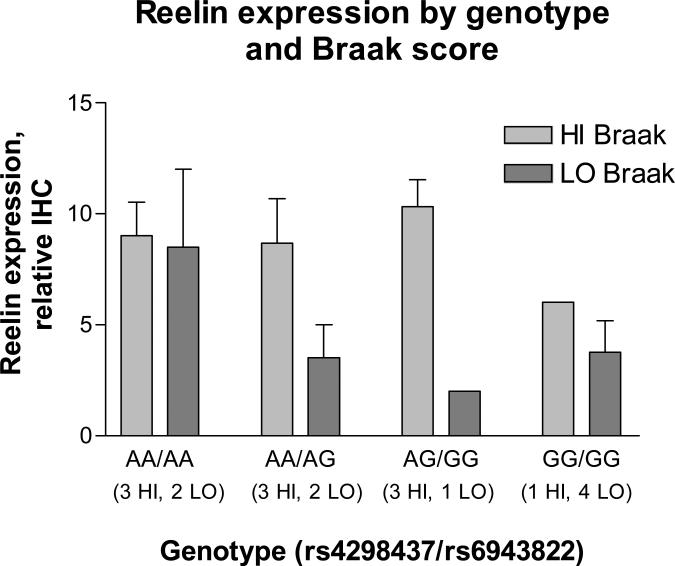
Total reelin expression was quantified as described in Methods. In both HI and LO Braak categories, subjects were stratified by two-SNP genotypes for rs4298437 and rs6943822. Total hippocampal reelin expression was significantly reduced in the LO Braak group (p = 0.025; Figure 4). In addition, when the LO and HI Braak groups were combined, there was a suggestion of an association between GG homozygotes (for either or both reelin SNPs) and reduced reelin expression, although this trend did not reach statistical significance (p = 0.096).
Figure 4.
Analysis of reelin expression in hippocampus. Immunohistochemical (IHC) expression of reelin was scored in the hippocampus of OHSU cases with postmortem interval less than 24 hours. Scores are the sum of semiquantitative assessments of cytoplasmic reelin positivity of pyramidal neurons in each sector as described in the text. Cases were stratified by Braak grouping and by genotype. Two-way ANOVA showed a significant association of reelin expression with HI vs. LO Braak stage (P=0.025) but no association with genotype.
4. Discussion
We identified variants in the RELN gene that are significantly associated with AD neuropathology in cognitively healthy elderly individuals. Analysis of allelic association in two SNPs in RELN generated significant ORs, ranging from 2.23-2.51, for the alleles associated with the HIBraak category. A significant association with SNPs in RELN was also identified in a study of female AD patients [Seripa, et al. 2008]. The region of RELN we found associated with NFT pathology and the region found in Seripa et al (2008) are both near promoter elements, which have been postulated to play a role in hypermethylation and therefore modification in expression of this gene [Chin, et al. 2007; Dong, et al. 2005].
4.1 Reelin
Reelin is an extracellular matrix glycoprotein, encoded by the RELN gene, which is secreted throughout the CNS and is important in neuronal development [Bar, et al. 2000; D'Arcangelo 2006].It is a ligand for at least three classes of cell membrane receptors involved in actin cytoskeleton regulation/reorganization and in tau phosphorylation. Tau is a microtubule-associated protein abundant in neurons and the primary component of NFTs. Normal reelin levels are necessary to prevent abnormal phosphorylation of tau [Hiesberger, et al. 1999; Ohkubo, et al. 2003; Trommsdorff, et al. 1999]. Reelin is also associated with plaques in β-amyloid precursor protein/presenilin-1 double-transgenic mouse brains [Wirths, et al. 2001]. A recent report [Durakoglugil, et al. 2009]indicates a role for reelin in protecting against beta-amyloid-induced suppression of long-term potentiation and NMDA receptors. Thus, reelin may play key roles in AD pathology both extracellularly with its involvement in plaques and intracellularly via inhibition of tau phosphorylation. Since tau phosphorylation is a key step in the formation of NFTs, our finding of an association between RELN polymorphisms and high levels of AD neuropathology is compelling. We note that the two RELN SNPs found to be associated with high levels of AD pathology in this study are probably not the actual functional SNPs in this regard, but instead are in strong linkage disequilibrium with other causative variants yet to be identified.
Evidence for direct involvement in pathways related to AD neuropathology is less apparent for other genes listed in Tables 2 and 3, with the exception of the histone deacetylase (HDAC) gene HDAC4. HDAC activities have been linked to neurodegenerative disorders [Timmermann, et al. 2001] and recently been shown to restore cognition in AD transgenic mice [Green, et al. 2008]. Possible roles for a sodium/glucose transporter, SLC5A9, or a transmembrane protein, TMEM65, are also conceivable. Variants in these genes, or others represented in the tables, may function in as yet unknown ways to either prevent or promote the formation of NFTs and/or amyloid plaques.
Our discovery of increased expression of reelin in pyramidal neurons of the hippocampus in AD and in cognitively intact controls with AD-associated pathology suggests that up-regulation of reelin may be a compensatory response to β-amyloid or tau-related stress associated with AD even prior to the onset of dementia. If so, polymorphisms in RELN may influence the capacity for up-regulation in affected neuronal populations and thereby exert effects on cognitive reserve in the context of AD-associated stressors and pathologic lesions.
The purpose of this study was to identify genetic variants that distinguish cognitively healthy, elderly individuals with a heavy NFT burden from those with minimal NFT burden. The former group also had a significantly greater amyloid plaque burden than the latter. There are three broad mechanisms whereby genetic variants could mediate NFT burden in cognitively healthy individuals. They could act directly through pathways related to tau, indirectly through pathways related to β-amyloid pathology (which, in turn influences tau), or through other mechanisms that affect brain reserve (e.g., neuronal or synaptic plasticity). One interpretation of this study is that genetic variants in RELN result in perturbations associated with the development of NFTs (and amyloid plaques). Since these subjects were cognitively healthy, the implication is that other genetic variants and/or environmental factors are required for the development of cognitive dysfunction.
This is the first GWAS that specifically addresses genetic differences related to AD neuropathology in clinically non-demented elderly. This is important because these individuals are traditionally used as control subjects in case-control studies of AD, under the assumption that, as a group, they are free of genetic factors related to AD pathophysiology. If, as this analysis suggests, a proportion of clinically non-demented individuals harbor genetic variants that predispose them to the underlying neuropathology that characterizes AD, including them in a control group will likely confound the results of genetic case-control studies.
These findings require replication in larger datasets with comparable phenotypes. Well-characterized, non-demented elderly with autopsy are difficult to obtain. The NIH is sponsoring the AD Genetics Consortium to assemble and analyze a comprehensive collection of AD cases and controls representing the collective resources of the AD research community. This collection will provide an outstanding opportunity for confirmation and expansion of the results reported here.
ACKNOWLEDGEMENTS
The authors would like to thank the following individuals for their generous and varied contributions to the production of this report, including clinical and neuropathological data verification, DNA and brain tissue procurement and transportation, and data organization and transmission: Charles DeCarli MD, at the University of California at Davis; Marilyn Albert MD, at Johns Hopkins University; Christine Hulette MD, James Burke MD, Jeffrey Browndyke PhD, John Ervin, Michelle McCart and Mari Szmanski RN at Duke University Medical Center; Alison Goate PhD, at the Washington University School of Medicine; Joseph Parisi MD and Kris Johnson RN, at Mayo Clinic, Rochester, and Dennis Dixon at Mayo Clinic, Jacksonville; Sonya Anderson at the University of Kentucky; Mary Sundsmo at the University of California at San Diego; Eszter Gombosi at the University of Michigan; Jamie Laut at Oregon Health & Science University. We thank Dr. Chungsheng He, Rockefeller University, for writing an R program to create the Manhattan plot.This work was supported by the National Institute on Aging (grants R01-AG026916, P30-AG028377, P50-AG005146, P30-AG028383, P50-AG16574, U01-AG06786, P30-AG008017, P30-AG10161, R01-AG17917, P30-AG10129, P50-AG05131, P50-AG08671, P50-AG05681, P01-AG03991, U01-AG016976) of the National Institutes of Health, and by the Natural Science Foundation of China NSFC, project number 30730057 (to JO).
Footnotes
DISCLOSURE STATEMENT
Dr. Ronald Petersen serves as Chair of the Safety Monitoring Committee and consultant for Elan Pharmaceuticals, Chair of Safety Monitoring for Wyeth Pharmaceuticals and consultant for GE Healthcare.Dr. John Morris has participated or is currently participating in clinical trials of antidementia drugs sponsored by Elan Pharmaceuticals, Eli Lilly and Wyeth Pharmaceuticals. Dr. Morris has served as a consultant or has received speaking honoraria from AstraZeneca, Bristol-Myers Squibb, Genentech, Lilly, Merck, Novartis, Pfizer, Schering Plough, Wyeth, and Elan. Appropriate approval and procedures were used concerning human subjects.
References
- Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18(4 Suppl):S1–2. [PubMed] [Google Scholar]
- Bar I, Lambert de Rouvroit C, Goffinet AM. The evolution of cortical development. An hypothesis based on the role of the Reelin signaling pathway. Trends Neurosci. 2000;23(12):633–8. doi: 10.1016/s0166-2236(00)01675-1. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Bienias JL, Evans DA, Wilson RS. Mild cognitive impairment is related to Alzheimer disease pathology and cerebral infarctions. Neurology. 2005;64(5):834–41. doi: 10.1212/01.WNL.0000152982.47274.9E. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Chin J, Massaro CM, Palop JJ, Thwin MT, Yu GQ, Bien-Ly N, Bender A, Mucke L. Reelin depletion in the entorhinal cortex of human amyloid precursor protein transgenic mice and humans with Alzheimer's disease. J Neurosci. 2007;27(11):2727–33. doi: 10.1523/JNEUROSCI.3758-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G. Reelin mouse mutants as models of cortical development disorders. Epilepsy Behav. 2006;8(1):81–90. doi: 10.1016/j.yebeh.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci U S A. 2005;102(35):12578–83. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durakoglugil MS, Chen Y, White CL, Kavalali ET, Herz J. Reelin signaling antagonizes beta-amyloid at the synapse. Proc Natl Acad Sci U S A. 2009;106(37):15938–43. doi: 10.1073/pnas.0908176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erten-Lyons D, Woltjer RL, Dodge H, Nixon R, Vorobik R, Calvert JF, Leahy M, Montine T, Kaye J. Factors associated with resistance to dementia despite high Alzheimer disease pathology. Neurology. 2009;72(4):354–60. doi: 10.1212/01.wnl.0000341273.18141.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28(45):11500–10. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60(8):1119–22. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24(2):481–9. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Hoh J, Wille A, Ott J. Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res. 2001;11(12):2115–9. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juva K, Verkkoniemi A, Viramo P, Polvikoski T, Kainulainen K, Kontula K, Sulkava R. APOE epsilon4 does not predict mortality, cognitive decline, or dementia in the oldest old. Neurology. 2000;54(2):412–5. doi: 10.1212/wnl.54.2.412. [DOI] [PubMed] [Google Scholar]
- Manly BFJ. Randomization, bootstrap, and Monte Carlo methods in biology. Chapman & Hall/ CRC; Boca Raton, FL: 2007. [Google Scholar]
- Martinez-Cerdeno V, Galazo MJ, Cavada C, Clasca F. Reelin immunoreactivity in the adult primate brain: intracellular localization in projecting and local circuit neurons of the cerebral cortex, hippocampus and subcortical regions. Cereb Cortex. 2002;12(12):1298–311. doi: 10.1093/cercor/12.12.1298. [DOI] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–4. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Ohkubo N, Lee YD, Morishima A, Terashima T, Kikkawa S, Tohyama M, Sakanaka M, Tanaka J, Maeda N, Vitek MP. Apolipoprotein E and Reelin ligands modulate tau phosphorylation through an apolipoprotein E receptor/disabled-1/glycogen synthase kinase-3beta cascade. FASEB J. 2003;17(2):295–7. doi: 10.1096/fj.02-0434fje. others. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe'er I, Burtt NP, Blumenstiel B, DeFelice M. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–82. doi: 10.1038/ng.2007.27. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seripa D, Matera MG, Franceschi M, Daniele A, Bizzarro A, Rinaldi M, Panza F, Fazio VM, Gravina C, D'Onofrio G. The RELN locus in Alzheimer's disease. J Alzheimers Dis. 2008;14(3):335–44. doi: 10.3233/jad-2008-14308. others. [DOI] [PubMed] [Google Scholar]
- Timmermann S, Lehrmann H, Polesskaya A, Harel-Bellan A. Histone acetylation and disease. Cell Mol Life Sci. 2001;58(5-6):728–36. doi: 10.1007/PL00000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97(6):689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18(5):1368–73. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Wirths O, Multhaup G, Czech C, Blanchard V, Tremp G, Pradier L, Beyreuther K, Bayer TA. Reelin in plaques of beta-amyloid precursor protein and presenilin-1 double-transgenic mice. Neurosci Lett. 2001;316(3):145–8. doi: 10.1016/s0304-3940(01)02399-0. [DOI] [PubMed] [Google Scholar]