Abstract
Polynuclear platinum compounds are more effective at killing glioblastoma cells than cisplatin, work by a different mechanism, and typically do not induce high levels of apoptosis at early time points after exposure. Here we tested the hypothesis that combining BBR3610, the most potent polynuclear platinum, with a PI3K inhibitor would promote apoptosis and enhance the impact on glioblastoma cells. The PI3K pathway is commonly activated in glioblastoma and promotes tumor cell survival, suggesting that its inhibition would make cells more sensitive to cytotoxic agents. We chose PX-866 as a PI3K inhibitor as it is a clinically promising agent being evaluated for brain tumor therapy. Combining BBR3610 and PX-866 resulted in synergistic killing of cultured glioma cells, and an extension of survival in an orthotopic xenograft animal model. Both agents alone induced autophagy, and this appeared to be saturated, because when they were combined no additional autophagy was observed. However, the combination of PX-866 and BBR3610 did induce statistically significant increases in the level of apoptosis, associated with a reduction in pAkt and pBad, as well as inhibition of transwell migration. We conclude that combining polynuclear platinums with PI3K inhibitors has translational potential and alters the cellular response to include early apoptosis.
Keywords: glioblastoma, platinum, PI3 Kinase, autophagy, apoptosis
Introduction
Currently primary emphasis in the research focused on developing novel treatments for glioma is being placed on promising new compounds that target signal transduction pathways. However, until such therapies result in clinical advances, the rationale for studying drugs whose action is not predicated on the presence of a particular molecular target or mutation 1, 2, such as platinum chemotherapeutics, remains strong. More effective yet may be combinations of broadly acting cytotoxic agents with a signal transduction inhibitor. In glioma conventional platinum compounds, such as cisplatin, often show limited therapeutic benefit, with responses restricted to rare sensitive patients and toxicities evident in proportion to high blood concentration 3. Promising, in this context, are the polynuclear platinum compounds, a class that is structurally distinct from cisplatin, and whose clinical profile and mechanism of action are different from those of the established platinum compounds 4–6. Cells with resistance to cisplatin show no cross-resistance to the polynuclear platinum BBR3464 5–8. The polynuclear platinum complexes are amphiphilic, due to their lipophilic alkanediamine chains and hydrophilic platinum-amine coordination spheres, a feature that is likely to enhance membrane permeability compared to cisplatin or oxaliplatin. When compared to conventional platinum chemotherapeutics, the polynuclear compounds show increased rates of cellular uptake and more rapid formation of DNA adducts 7–9. Furthermore, polynuclear platinum agents such as BBR3464 have been well tolerated in the clinic, with a toxicity profile distinct from that of cisplatin-based agents10.
We have previously examined “2nd generation” members of the polynuclear platinum family for pre-clinical potential, and found the dinuclear complex with a spermine-like linker, BBR3610, to be particularly effective against glioma cells 11. These polyamine-linked dinuclear compounds have essentially the same impact at the cellular and molecular levels as BBR3464, with DNA-binding profiles and cellular responses that are very similar. They also show cross-resistance with BBR3464, which suggests common elements in their mechanism of action 4, 7, 8, 12. The structural modifications made in “2nd generation” compounds enhances cytotoxicity but has no impact on their pharmacokinetic profile of the compound13, 14. BBR3610 had the greatest potency against glioma cells in culture and in xenograft models, and was more effective in the treatment of glioma in vivo than cisplatin or BBR3464 11. Interestingly, the polynuclear platinum agents induced predominantly G2/M arrest in glioma cells, an effect that was mediated by extracellular signal-regulated kinase (ERK) 3 activation, as was the induction of apoptosis by cisplatin 11.
When considering combination therapies between cytotoxic agents and signal transduction inhibitors, targeting the signaling pathways that support cell survival in cancer cells is particularly attractive. In the case of glioblastoma, the primary pathway that shows abnormally high activity in cancer and supports cell survival, is the phosphoinositide-3-kinase (PI3K) pathway 15. Overexpressed or mutated epidermal growth factor receptor (EGFR), deletion of the phosphatase and tensin homologue (PTEN) gene or both, lead to elevated levels of activity of the pro-survival Akt/protein kinase B molecule. Inhibition of PI3K has the potential of interrupting this signal, even in cells with overactive EGFR or PTEN loss, and so may potentiate the action of cytotoxic drugs. A PI3K inhibitor with significant promise is PX-866, for which it has been shown that PI3-K blocking concentrations can be achieved by oral administration and which is expected to have minimal toxicity 16–18.
Here we test the hypothesis that the combination of BBR3610 and PX-866 will enhance the killing of glioma cells when compared to the effect of either agent alone, and furthermore that suppression of the PI3K pathway may enhance the induction of apoptosis by BBR3610. Combining these agents showed synergistic killing of cells in culture, and an extension of survival time in an intracranial xenograft model of glioma. In addition, the combination of PX-866 and BBR3610 increased the level of apoptosis induced in the short term, when compared to BBR3610 alone, suggesting that this is an important mechanism of enhancement.
Materials and Methods
Cell Culture and Reagents
U87MG (p53 wild type, PTEN mutant), LNZ308 (both p53 and PTEN mutant) and LN229 (p53 mutant and PTEN wild type) cells were maintained in Dulbecco’s Modififed Eagle’s Medium (DMEM) supplemented with 50 units penicillin and streptomycin and 10%FBS at 37°C with 7% CO2. BBR3610 (> 99.9 % purity) was dissolved at 100 μM in distilled water and kept at −80°C until use. PX-866 was dissolved at 1.1 mM in ethanol and kept at −20°C as a stock solution. Cell lines were obtained from the stocks of the Ludwig Institute for Cancer Research, San Diego Branch, in 1997 and have been maintained in the Bogler laboratory. As of one year ago, they are routinely fingerprinted for identity using a PCR-based analysis (GenomeLab Human STR Primer set from Beckman Coulter) which interrogates a set of 12 short tandem repeats (Supplemental Table 1).
Growth Inhibition Assay
Growth inhibition in culture was measured by two assays. In the first, cell viability was measured by clonogenicity. Cells were plated at clonal density in 6-well plates and treated with drugs as indicated in the results section the next day. Cells were then allowed to form colonies for two weeks, with regular media changes but no additional drug treatments, before fixation followed by Eosin and Azure staining (DiffQuick from IMEB Inc, San Marcos, CA). A cluster of 50 cells or more was scored as a colony and the surviving fraction was calculated in relation to the control, untreated wells. In the second, cell viability was analyzed by water-soluble tetrazolium (WST) assay. Cells were seeded in 96-well plates at 750 cells/well and allowed to attach overnight, before treatment with drugs. Five to seven days after exposure to drug, 10 μl of WST reagent solution (Roche, Indianapolis, IN) was added to each well, and incubated for an additional 4 hours at 37°C. The WST dye absorption was measured at 450nm.
To differentiate synergism from additive or antagonistic action, we used CalcuSyn software (version 2.0, Biosoft, Cambridge, UK) to calculate the Combination Index (CI) according to published methods 19. A CI of less than 1.0 was considered indicative of synergism and this was further classified as strong synergism (CI < 0.3), synergism (CI of 0.3–0.7) and slight to moderate synergism (CI of 0.7 – 0.9).
Cell Cycle Analysis
Cells were harvested at the indicated time after drug treatment, including the collection of non-adherent cells, and fixed in 70% ethanol. Fixed cells were incubated at 1 × 106 cells per ml with of propidium iodide (50 μg/ml in PBS) containing 100 μM RNase (Qiagen, Valencia, CA) at 37°C for 1 hour. DNA content was analyzed on a fluorescence-activated cell sorter (FACSCalibur, BD Biosciences, San Diego, CA) and data collected and analyzed using CellQuest software (BD Biosciences).
Analysis of Apoptosis with Annexin-V FITC
Following treatment with drugs as indicated, cells were trypsinized, washed twice with PBS, and then resuspended at a concentration of 1 × 106 cells/ml in binding buffer (10 mM HEPES/NaOH pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). Annexin V-FITC (PharMingen, San Diego, CA) and propidium iodide (PI) were added, and the cells incubated at room temperature for 15 minutes in the dark. The stained cells were analyzed by fluorescence-activated cell sorter (FACSCalibur, BD Biosciences, San Diego, CA) and data collected and analyzed using CellQuest software (BD Biosciences).
Autophagy Measurement Using GFP-LC3
Cells were transfected with a green fluorescent protein-microtubule-associated protein 1 light chain 3 (GFP-LC3) expression plasmid (gift from Dr. Seiji Kondo; 20) using Fugene (Invitrogen Life Technologies, NE, USA). After 24h, cells were treated with various drug combinations as indicated and observed the fluorescence of GFP-LC3 under a fluorescence microscope and counted LC3 punctate spots two days later. Cells were scored as undergoing autophagy if they had 6 or more LC3-GFP spots. The percentage of cells undergoing autophagy was calculated from the ratio of autophagic cells to normal cells bearing GFP-LC3 fluorescence.
Western Blotting
Cells were lysed in a lysis buffer (20mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1 mM EDTA, 1 mM EGTA, 2 mM sodium orthovanadate, 2 mM NaF, and Complete™ Protease Inhibitor Mix (Roche Molecular Biochemicals, Germany)) for 20 min on ice, cleared by centrifugation at 12,000 × g at 4°C. Proteins were resolved by 10% SDS-PAGE, transferred onto nitrocellulose membrane, blocked with 5% nonfat dry milk in TBST (10mM Tris-HCl pH 7.5, 100 mM NaCl, and 0.05% Tween 20) followed by primary antibody: anti- Poly (ADP-ribose) polymerase (PARP): N-19, Santa Cruz Biotechnology, San Diego, CA: anti-LC3B antibody gift of Dr. Seiji Kondo 21; total and anti-phospho-AKT (Ser473), total and phospho-S6K (Thr389), and total and phospho-Bad (Ser136) antibodies were from Cell Signaling Technology (Danvers, MA); anti-β-actin antibody was from Sigma-Aldrich (St. Louis, MO).. Blots were washed and incubated with horse radish peroxidase- conjugated secondary antibody. Antibody complexes were visualized by enhanced chemiluminescence-Western blotting detection system (Pierce Biotechnology, Rockford, IL).
Transwell Migration Assay
The assay was performed by the modified method of procedure previously described 22. Transwell inserts (8 μm pore size; Corning, NY) were coated with a final concentration of 0.78 mg/ml of Matrigel. After rinsing with PBS, 100 μl of cell suspension (1×107 cells/ml) was added to the upper chamber. Serum free media containing 0.1mg/ml BSA (Sigma, St Louis, MO) was added to the lower compartment. BBR3610 and PX-866 at various concentrations were added to the upper chamber. After incubation for 16 h, cells that had migrated to the lower surface of the matrigel were stained with the Diff-Quick staining kit (IMEB Inc., San Marcos, CA). The average number of cells in 10 randomly chosen microscopic fields (20x magnification) was determined and final values for each condition calculated as the average from the three invasion chambers.
Intracranial Xenograft Analysis
The intracranial tumor model was made as described previously 23. Briefly, male nude mice (Experimental Radiation Oncology at M. D. Anderson Cancer Center) were anesthetized with intraperitoneal injection of ketamine/xylazine solution (ketamine 10 mg/ml and xylazine 1 mg/ml in saline) at a dosage of 0.1 ml/10 g (ketamine 100 mg/kg and xylazine 10 mg/kg) body weight. A 0.9-mm burr hole was then drilled into the skulls of the mice 1.0 mm anterior and 2.5 mm lateral to the bregma to expose the dura. A screw guide with a 0.5 mm central hole and a stylet were placed into the burr hole. Two weeks after the screw guide was set in the skull, a 10 μL syringe (Hamilton Co., Reno, NV) fitted with a 26-gauge needle was connected to a microinfusion pump (Harvard Apparatus Co., Cambridge, MA). Through the screw guide, at a depth of 3.5 mm from the skull (corresponding to the region of the caudate nucleus), 5 μL aliquots of 5 × 105 U87 cells in serum-free DMEM were inoculated at a rate of 1.0 μL/min. Mice were randomized into four groups (10 mice for each group) and treatment was initiated one week after tumor implantation. BBR3610 was dissolved in PBS and administered at a dose of 0.1 mg/kg through injection of the tail vein once a week for a total of three treatments in both BBR3610 treatment group and combination treatment group. PX-866 was dissolved in PBS and administered at a dose of 2 mg/kg orally three times a week (Monday, Wednesday, and Friday) for a total of twelve treatments in both PX-866 treatment group and combination treatment group. Mice of control group were treated with PBS. All mouse studies were performed in the veterinary facilities of the University of Texas M. D. Anderson Cancer Center in accordance with institutional, state, federal, and ethical regulations for experimental animal care.
Statistics
Quantitative data were expressed as mean ± standard deviation or ± standard error of the mean, as indicated. All measurements for each condition were performed in triplicate. IC50 values were obtained automatically using Prism software (version 3.0, San Diego, CA). Statistical significance of differences was assessed using a two-tailed unpaired Student’s t-test and one-way analysis of variance (ANOVA; SPSS version 11.0, Chicago, IL). Statistical significance was considered to have been reached if the p value was less than 0.05. For the animal experiment pairs of Kaplan-Meier survival curves were compared in Prism 5 for Macintosh software using the log-rank Mantel-Cox test.
Results
BBR3610 and PX-866 are synergistic in the inhibition of glioma cell growth in culture
To determine whether the suppression of PI3K signaling by PX-866 altered the sensitivity of glioma cells to BBR3610 we combined the two agents, and performed both colony formation and WST-1 assays. U87 cells are too motile to form discrete colonies, and so only LNZ308 and LN229 cells were measured with the colony formation assay. Both cell lines showed a greater suppression of colony formation by BBR3610 when PX-866 was also present (Table 1; Supplemental Fig. 1). The concentrations of PX-866 used in these studies (400 nM and 800 nM) induced little growth suppression in our glioma cells in the absence of BBR3610 (Supplemental Fig. 1; see points on ordinate), as expected from prior studies which showed maximum suppression of PI3K activity at 800 nM PX-866 in the absence of cytotoxicity 16, 17. Direct analysis of synergism by the calculation of a Combination Index (CI; 19) revealed synergism when PX-866 and BBR3610 were combined, as measured by both colony formation (Table 1) and WST-1 assay (Supplemental Table 2). All observed combinations of the two drugs at different concentrations yielded a combination index below 1 indicating synergism. Combinations of PX-866 with 10 nM BBR3610, which is close to its IC90 (8 nM as measured by clonogenic assay 11) gave CIs at or below 0.3 in all but one case when measured by WST-1 assay, indicating strong synergism (Supplemental Table 2).
PX-866 increases the level of apoptosis induced by BBR3610
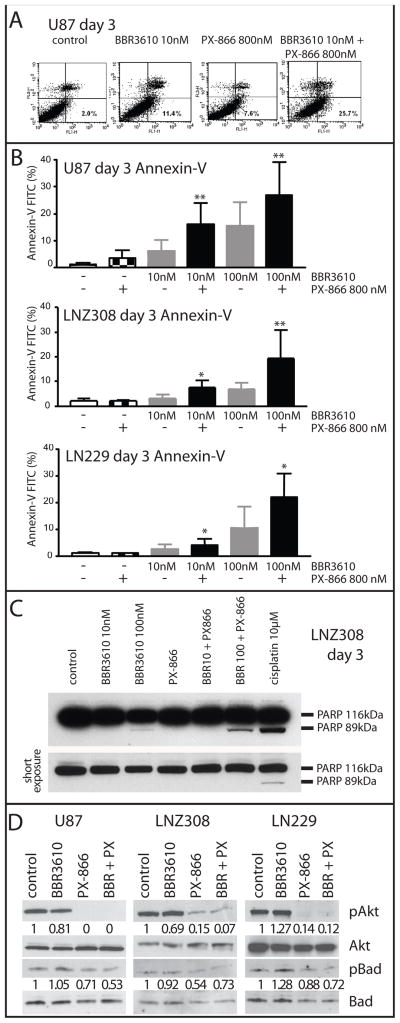
To explore whether the enhancement of BBR3610’s effect by PX-866 involved an alteration of the nature of the cellular response to the platinum agent, we analyzed apoptosis. The early response of glioma cells to BBR3610 is G2/M arrest with little apoptosis 11, while apoptosis is commonly triggered by conventional platinums such as cisplatin. At the same time it is well recognized that an active PI3K pathway is important for cellular survival in glioma, leading to the hypothesis that suppression of this signal may increase the level of apoptosis induced by BBR3610. To test this, we analyzed the proportion of cells stained by Annexin-V, which measures the appearance of phosphatidyl serine on the plasma membrane, an early event in apoptosis. We observed an increase in the percentage of Annexin-V positive cells when BBR3610 and PX-866 were combined, as shown in a representative experiment where the levels increased from 11.4% to 25.7% three days after drug treatment (Fig. 1A). Combining the data from three experiments showed that 800 nM PX-866 increased the proportion of Annexin-V-positive cells in U87, LNZ308 and LN229 cells treated with either 10 nM or 100 nM BBR3610 (Fig. 1B). Similar results were obtained from monitoring PARP cleavage, a measure of early caspase activation, by western blot (Fig. 1C). While detectable cleavage was observed with 100 nM BBR3610 alone, a significant increase was observed when PX-866 was added in combination. However, the level of PARP cleavage induced by 10 μM cisplatin was considerably higher, suggesting that the combination of BBR3610 and PX-866 does not induce maximal apoptosis in these cells. Phosphorylation of Bad at S136 inhibits its pro-apoptotic activity by preventing interaction with the anti-apoptotic Bcl-2 and Bcl-XL, and is catalyzed by Akt, among other signals24, 25. Therefore, we examined whether a reduction in pBad was induced by treatment of glioblastoma cells with PX-866. As expected we saw a reduction in pAkt in response to PX-866 in all three cell lines (Fig. 1D). A reduction in pBad was also observed, although it was not as profound as the reduction in pAkt. However, the degree of reduction in pBad seen in response to PX-866 alone and the combination of PX-866 and BBR3610 was similar (Fig. 1D) while the level of apoptosis was significantly higher when both drugs were present (Fig. 1A–C). This suggests that a reduction in pBad induced by PX-866 in glioblastoma cells is not sufficient to induce an increase in apoptosis alone, but may be an important in promoting an apoptotic response to BBR3610. To determine whether the induction of apoptosis resulted in a compensatory upregulation of anti-apoptotic regulators we examined levels of Bcl-2, a key regulator of glioblastoma apoptosis which is regulated both at the mRNA and protein levels26. Examination of mRNA by quantitative RT-PCR did not reveal a consistent pattern of change in response to BBR3610, PX-866 or both, in three glioblastoma cell lines (Suppl. Fig. 2).
Figure 1. Induction of Apoptosis in glioma cells by BBR3610 and PX-866.
Glioma cells were treated with BBR3610 and PX-866 as indicated, stained with AnnexinV-FITC three days later and the proportion of positive cells determined by FACS. (A) A typical experiment with U87 cells is shown, and the percentages of Annexin-V positive cells are indicated in the lower right quadrant. (B) Data from 3 independent experiments were combined and are presented as mean and standard deviation. Statistical analysis showed that the combination of BBR3610 + PX-866 significantly increased Annexin-V positive cells when compared to BBR3610 alone at the same concentration (*; p < 0.05, **; p < 0.01). (C) Western blot analysis of PARP cleavage shows intact PARP (116 KDa) and cleaved product (89 KDa). The combination of BBR3610 + PX-866 showed increased PARP cleavage compared to BBR3610 alone. A lower exposure is shown to demonstrate similar loading between lanes. (D) Western blot of lysates from cells left untreated, or treated with 100 nM BBR3610, 800 nM PX-866 or both for 60 minutes. Blots were probed with antibodies against total Akt or pAkt (Ser 473), total Bad or pBad (Ser136) as indicated. Quantification of phospho bands relative to total bands is indicated. One representative experiment is shown.
PX-866 does not abrogate the G2/M arrest induced by BBR3610
To determine whether the increase in apoptosis induced by combining PX-866 with BBR3610 is accompanied by a suppression of the G2/M arrest usually induced by the latter, we analyzed the DNA content of cells treated with these agents (Fig. 2). One day after treatment the G2/M arrest induced by BBR3610 was readily detectable in a single, representative experiment, and this was not eliminated by the combination with PX-866 (Fig. 2A). Statistical analysis of data from three experiments showed no significant differences in any of the cell-cycle compartments in any of the three cell lines analyzed (data for U87 cells shown in Fig. 2B; LNZ308 and LN229 data not shown). These data are consistent with the Annexin-V data described above, as DNA fragmentation is a later event in apoptosis than phosphatidyl-serine display, and so detecting 10 to 20% Annexin-V positive cells (Fig. 1) at the same time point as no significant increase in sub-G1 cells is detected (Fig. 2) is consistent. Furthermore, they suggest that the initial G2/M arrest induced by BBR3610 is not abrogated by PX-866.
Figure 2. Cell-cycle analysis of glioma cells treated with BBR3610 and PX-866.
Glioma cells were treated with drugs as indicated, stained with propidium iodide and analyzed by FACS. (A) A representative set of FACS traces from U87 cells one day after drug exposure is shown. (B) Data from three experiments in which U87 cells were analyzed three days after drug treatment were combined, and are presented as mean and standard deviation. No statistical differences between the profiles obtained from cells treated with BBR3610 and BBR3610 + PX-866 at the same concentration was detected.
The combination of PX-866 and BBR3610 does not induce additional autophagy
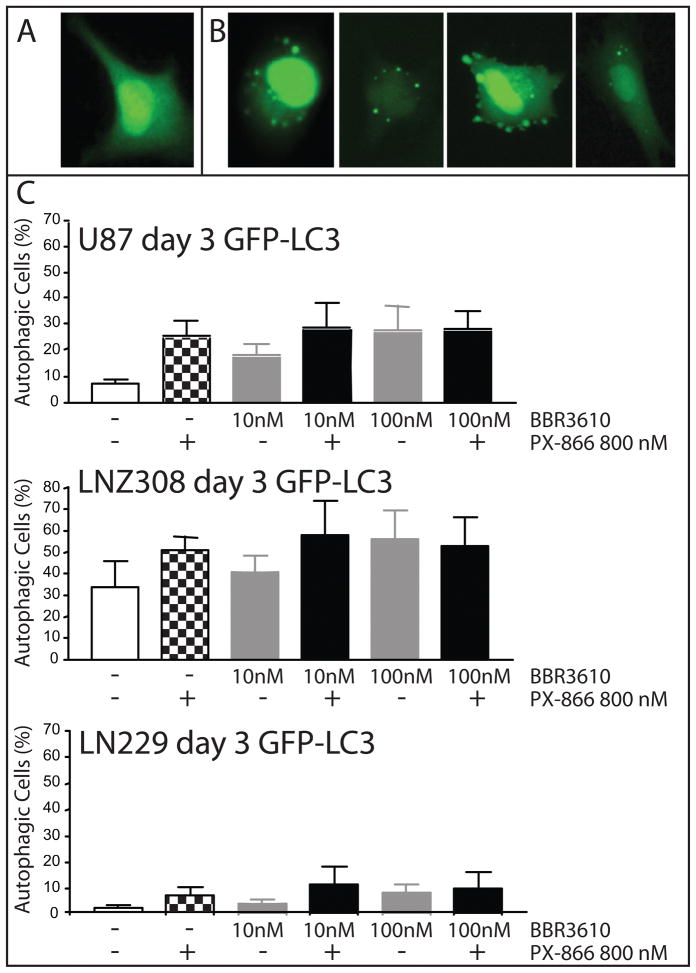
Both PX-866 27 and BBR3610 (Shingu et al., in preparation) induce autophagy. Therefore we investigated whether the synergy on growth suppression of glioma cells was also reflected in this mode of cell death. We transfected glioma cells with GFP-LC3 prior to drug exposure, and counted the proportion of GFP-positive cells that showed a punctate staining pattern, defined here as having more than 6 visible granules (Fig. 3A and B). While the basal level of autophagy varied between the cell lines used (LNZ308: 33.5 ± 12.1%; U87: 8.3 ± 3.1%; and LN229: 1.4 ± 1.0%), no statistical increase was observed when cells were treated with both agents in combination (Fig. 3C). We confirmed these finding by LC3-B Western blot, where either PX-866 alone or BBR3610 alone induced a stronger, lower LC3-B2 band as compared to untreated control, but we could not detect an increase when the agents were combined (data not shown).
Figure 3. Analysis of Autophagy in glioma cells treated with BBR3610 and PX-866.
Glioma cells were transiently transfected with GFP-LC3 and the formation of “LC3 dots” monitored using a fluorescent microscope. Examples of transfected cells that were scored as (A) not undergoing autophagy as they showed no LC3 dots and (B) those scored as undergoing autophagy as they showed six or more GFP-LC3 dots are shown. (C) Quantification of LC3 dot assay, presented as percentages of autophagic cells three days after the indicated drug treatments in U87, LLNZ308 and LN229 as indicated. Data from three independent experiments are presented as mean with standard deviation. The combination groups showed no significant increase in autophagy compared to single drug treated groups.
BBR3610 enhances the suppression of cell migration by PX-866
Cell migration is an important component of invasion, itself a key hallmark of glioma malignancy, prompting us to investigate the impact of our combination of agents on cell movement in culture. PX-866 inhibits the invasion of glioma cells thorough Matrigel-coated membranes at concentrations at which it inhibits PI3K activity 27, 28, prompting us to investigate whether the synergy observed with BBR3610 extends to this activity. As expected 800 nM PX-866 inhibited migration of U87 and LN229 cells significantly, within the first 16 hours after exposure (Fig. 4; p < 0.01). In contrast BBR3610 alone, at either 10 nM or 100 nM failed to show any impact on this behavior. However, when the two agents were combined, the suppression of transwell migration was greater than that observed with PX-866 alone in both U87 (p < 0.01) and LN229 (p < 0.05; Fig. 4).
Figure 4. BBR3610 increases the suppression of cell migration by PX-866.
Glioma cell migration was analyzed using Matrigel-coated transwells, in which the number of cells that migrated through the matrigel-coated membrane was estimated by counting 10 high power fields (20x objective). The mean number of cells per field was calculated and is shown with standard deviation. PX-866 inhibited migration of U87 and LN229 cells significantly at 800 nM when compared to untreated control (p < 0.001; not indicated), while BBR3610 alone did not at either 10 nM or 100 nM. The combination of 100 nM BBR3610 + PX-866 significantly reduced migration when compared to PX-866 alone in both cell lines (*; p < 0.05, **; p < 0.01).
The combination of PX-866 and BBR3610 is more effective in vivo than either agent alone
Lastly, we tested the impact of BBR3610 and PX-866 together on the growth of intracranial U87 xenografts. Nude mice bearing intracranial tumor were treated with PBS, BBR3610 alone, PX-866 alone, or combination of BBR3610 and PX-866, and survival curves were analyzed. Treatment was initiated one week after tumor cell implantation. BBR3610 was given three times, one week apart at a dose of 0.1mg/kg, concluding on day 21. PX-866 was given orally, three times a week for four weeks, at a dose of 2mg/kg and concluded on day 32. Animals were observed for signs of morbidity, sacrificed when necessary and survival curves calculated (Fig. 5). While untreated animals had a median survival of 24.5 days, and treatment with BBR3610 or PX-866 alone caused a median survival of 39 and 35.5 days, respectively, the combination extended this to 42.5 days. The combination of BBR3610 and PX-866 was a statistically significant improvement over BBR3610 (p=0.0066) and PX-866 (P<0.0001) alone.
Figure 5. The combination of BBR3610 plus PX-866 significantly extends survival in an intracranial animal model.
U87 cells were implanted intracranially in nude mice (n = 10 in each group), and treatment was commenced one week later. BBR3610 (0.1 mg/kg) was administered intravenously once a week for a total of three treatments in both the BBR3610 treatment group and the combination treatment group. PX-866 (2 mg/kg) was administered orally three times a week for a total of twelve treatments in both the PX-866 treatment group and the combination treatment group. The timing of treatments is indicated. The control group mice were treated with PBS. Mice were sacrificed at morbidity, and survival curves were compared by using Prism 5 software. The combination of BBR3610 and PX-866 was a statistically significant improvement over BBR3610 (p=0.0066) and PX-866 (P<0.0001) alone, as determined by pair wise log-rank Mantel-Cox test of the Kaplan-Meier survival curves.
Discussion
Polynuclear platinum compounds were developed with the goal of treating cancers that were resistant to established platinums 4, 29. They have shown improved cellular uptake and DNA adduct formation, and a lack of cross-resistance with conventional platinums, suggesting the potential for a better cellular pharmacology and implying a different mechanism of action 4. In an analysis of three polynuclear platinums in glioma, BBR3610 showed the greatest efficacy, with an IC90 in culture of 8 nM, 250 times lower than that of cisplatin 11, and prompting further investigation of this compound in the context of glioma. Interestingly, the main effect of BBR3610 was to induce G2/M arrest in the absence of apoptosis, while cisplatin induced apoptosis 11. This was consistent with an earlier observation that BBR3464 provoked a modest induction of p53 with consequent upregulation of the cell cycle inhibitor p21 but not the proapoptotic Bax, whereas cisplatin gave a robust induction of p53 as well as increased expression of Bax and consequent apoptosis 6. The lack of apoptosis seen in response to polynuclear platinums may also be due to counteraction of their effects by strong pro-survival signals. In gliomas the most well recognized pathway responsible for supporting cell survival is the PI3K pathway, and so here we tested the hypothesis that combining an inhibitor of PI3K with BBR3610 would improve glioma cell killing, and increase the proportion of cells that underwent apoptosis.
Our data clearly support a synergy between BBR3610 and PX-866 in culture with three different cell lines studied by two assays (Table 1 and Supplemental Table 2; Supplemental Fig. 1). Furthermore, the combination of the two agents was more effective than either agent alone in prolonging survival in an orthotopic xenograft model (Fig. 5). This suggests that this combination has translational potential. Existing clinical data on cytotoxic drugs and signal transduction inhibitors in glioma firmly establish that no single compound suffices to achieve control of the disease. PI3K inhibitors may be a good example of this, as the activity of the PI3K pathway is responsive to cell stress, and so combination with a cytotoxic agent may be required for the full effects of inhibition of the PI3K to become apparent 27, 28. PX-866 is a superior PI3K inhibitor, in contrast to its parent compound wortmannin, by being more selective for the p110α isoform resulting in a more prolonged inhibition of phosphorylation of Akt and less toxicity 16, 17. Furthermore, recent work has shown that the hyperglycemia induced by PX-866 can be controlled by the peroxisome proliferator-activated receptor gamma agonist pioglitazone without affecting antitumor activity 18. BBR3610 and analogues are viable 2nd-generation analogues of BBR3464 and are in pre-clinical development (http://www.celltherapeutics.com/novel_platinum_complexes). The similar DNA binding profile to BBR3464 but reduced reactivity with deactivating thiols are desirable properties for such 2nd-generation drugs 30. The data presented here further strengthen the case for the viability of BBR3610 as a clinical candidate, and that a pre-clinical investigation of the combination of PX-866 and BBR3610 is warranted.
Investigation of the mechanism underlying the enhanced effect of the combination suggests that augmented apoptosis in response to BBR3610, when PX-866 is present, is a measurable component. In each of the three cell lines analyzed the addition of PX-866 significantly increased the level of Annexin-V staining and PARP cleavage (Fig. 1). This was less noticeable in our analysis of sub-G1 cells (Fig. 2), perhaps because this represents a later phase of apoptosis, and we measured the impact on day 3. From these data we conclude that suppression of PI3K signaling promotes apoptosis in response to BBR3610, and that this contributes to the combination effect observed. A reduction of pBad levels are likely a component of this response, as we observe a measurable decrease in pBad in the three cell lines studied here. However, the levels of apoptosis achieved even by the combination of 800 nM PX-866 and 100 nM BBR3610 was not as high as that seen in response to cisplatin at isodose concentrations (see for example Fig. 1C), suggesting that either the combination of PI3K inhibitor and polynuclear platinum is less effective at triggering this pathway, or does so with slower kinetics.
Another form of cell death, autophagy, was also found to occur in cells treated with BBR3610. We are the first to report that BBR3610 induces autophagy at nanomolar concentrations, which are close the to the IC90 as measured by colony formation 11. We observed this both via autophagic granule formation (Fig. 3) and LC3-B Western blot (Shingu et al, in preparation). In our study, PX-866 alone also induced autophagy at levels similar to that seen with BBR3610 (Fig. 3). Interestingly the combination of the two agents did not enhance the level of autophagy measured. Many studies have shown that tumor cells could undergo both apoptosis and autophagy in response to noxious stimuli such as irradiation or chemotherapy, and that there can be cross-talk between these two forms of cell death 31. When apoptosis was inhibited, for example by knock-out of the proapoptotic proteins BAX and BAK in mice, increased levels of autophagy were observed in mouse embryonic fibroblasts 32. Conversely, inhibition of autophagy by the H+-ATPase inhibitor bafilomycin A1 increased apoptotic cell death in various cancer cells treated by chemotherapy 33. What has been studied less intensely is the result of combining two agents that induce autophagy. In our experiments we combined maximal combinations of two agents: BBR3610 at 100 nM is at ten-times the IC90 concentration required to suppress colony formation and PX-866 at 800 nM is maximal for inhibition of PI3K, and both agents alone induced similar levels of autophagy in three different cell lines, alone or in combination (Fig. 3). In all three lines 10 nM BBR3610 induced sub-maximal levels of autophagy. This suggests that autophagy is saturated by 100 nM BBR3610 and 800 nM PX-866, and that once saturation is reached, addition of another agent that induces autophagy did not increase its level. Interestingly, the actual levels of maximal autophagy varied between cell lines: about 60% in LNZ308, 30% in U87 and 10% in LN229 (Fig. 3), implying that the threshold may be cell-type specific.
Furthermore, while these combinations failed to increase autophagy, they did result in more apoptosis. The molecular basis of additional triggering of apoptosis will require further investigation. We included cell lines representing both wild-type and mutant PTEN and p53 to allow detection of a strong influence of either of these genes on the response to treatment, but saw no clear cut differences. While this is consistent with PTEN positive and p53 wild-type cells also being impacted by our treatment, it must be borne in mind that the majority of glioblastomas have inactivating mutations somewhere in these two pathways 34. Complete understanding of the molecular correlate of responsiveness to these agents will require a greater understanding particularly of the molecular basis of polynuclear platinum response. Extensive analyses have shown a lack of shared mechanism with the established, and fairly well understood platinums 4, but the exact pathways that BBR3610 triggers in glioma cells are not yet well defined 11. Understanding whether the apoptosis observed here is triggered as a consequence of saturating the autophagy response, or independently by activation of other pathways, will lead to important insights on how our findings can be best translated to the clinic.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by a Grant from the Center for Targeted Therapy of The University of Texas M. D. Anderson Cancer Center (OB) as well as by funding from the National Cancer Institute/National Institutes of Health (RO1CA052995 to GP; RO1CA128991 to OB and NPF; 1P50CA127001 to WKAY, GP and OB and R21CA1108499 to OB). RD was supported in part by a research fellowship from the American Brain Tumor Association in honor of the Segal Family.
Abbreviations
- ANOVA
analysis of variance
- CI
Combination Index
- DMEM
Dulbecco’s Modififed Eagle’s Medium
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinase
- GFP
green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- PARP
Poly (ADP-ribose) polymerase
- PI
propidium iodide
- PI3K
phosphoinositide-3-kinase
- PTEN
phosphatase and tensin homologue
- WST
water-soluble tetrazolium
Footnotes
Conflict of Interest: Dr. Powis is an inventor of PX-866 and holds stock in Oncothyreon. Dr. Farrell is a named inventor on patents relating to BBR3610, which are licensed for possible clinical development.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, Riggs BL, Horvath S, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 3.Gwak HS, Youn SM, Kwon AH, Lee SH, Kim JH, Rhee CH. ACNU-cisplatin continuous infusion chemotherapy as salvage therapy for recurrent glioblastomas: phase II study. J Neurooncol. 2005;75:173–80. doi: 10.1007/s11060-005-1858-8. [DOI] [PubMed] [Google Scholar]
- 4.Farrell N. Polynuclear platinum drugs. Met Ions Biol Syst. 2004;42:251–96. [PubMed] [Google Scholar]
- 5.Perego P, Gatti L, Caserini C, Supino R, Colangelo D, Leone R, Spinelli S, Farrell N, Zunino F. The cellular basis of the efficacy of the trinuclear platinum complex BBR 3464 against cisplatin-resistant cells. J Inorg Biochem. 1999;77:59–64. doi: 10.1016/s0162-0134(99)00142-7. [DOI] [PubMed] [Google Scholar]
- 6.Servidei T, Ferlini C, Riccardi A, Meco D, Scambia G, Segni G, Manzotti C, Riccardi R. The novel trinuclear platinum complex BBR3464 induces a cellular response different from cisplatin. Eur J Cancer. 2001;37:930–8. doi: 10.1016/s0959-8049(01)00061-2. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JD, Peroutka J, Beggiolin G, Manzotti C, Piazzoni L, Farrell N. Comparison of cytotoxicity and cellular accumulation of polynuclear platinum complexes in L1210 murine leukemia cell lines. J Inorg Biochem. 1999;77:47–50. doi: 10.1016/s0162-0134(99)00137-3. [DOI] [PubMed] [Google Scholar]
- 8.Roberts JD, Peroutka J, Farrell N. Cellular pharmacology of polynuclear platinum anti-cancer agents. J Inorg Biochem. 1999;77:51–7. doi: 10.1016/s0162-0134(99)00147-6. [DOI] [PubMed] [Google Scholar]
- 9.Harris AL, Yang X, Hegmans A, Povirk L, Ryan JJ, Kelland L, Farrell NP. Synthesis, characterization, and cytotoxicity of a novel highly charged trinuclear platinum compound. Enhancement of cellular uptake with charge. Inorg Chem. 2005;44:9598–600. doi: 10.1021/ic051390z. [DOI] [PubMed] [Google Scholar]
- 10.Sessa C, Capri G, Gianni L, Peccatori F, Grasselli G, Bauer J, Zucchetti M, Vigano L, Gatti A, Minoia C, Liati P, Van den Bosch S, et al. Clinical and pharmacological phase I study with accelerated titration design of a daily times five schedule of BBR3464, a novel cationic triplatinum complex. Ann Oncol. 2000;11:977–83. doi: 10.1023/a:1008302309734. [DOI] [PubMed] [Google Scholar]
- 11.Billecke C, Finniss S, Tahash L, Miller C, Mikkelsen T, Farrell NP, Bögler O. Polynuclear platinum anticancer drugs are more potent than cisplatin and induce cell cycle arrest in glioma. Neuro-Oncology. 2006;8:215–26. doi: 10.1215/15228517-2006-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGregor TD, Hegmans A, Kasparkova J, Neplechova K, Novakova O, Penazova H, Vrana O, Brabec V, Farrell N. A comparison of DNA binding profiles of dinuclear platinum compounds with polyamine linkers and the trinuclear platinum phase II clinical agent BBR3464. J Biol Inorg Chem. 2002;7:397–404. doi: 10.1007/s00775-001-0312-4. [DOI] [PubMed] [Google Scholar]
- 13.Gatti L, Perego P, Leone R, Apostoli P, Carenini N, Corna E, Allievi C, Bastrup U, De Munari S, Di Giovine S, Nicoli P, Grugni M, et al. Novel bis-platinum complexes endowed with an improved pharmacological profile. Mol Pharm. 7:207–16. doi: 10.1021/mp900211j. [DOI] [PubMed] [Google Scholar]
- 14.Manzotti C, Nicoli P, Torriani D, Menta E, Farrell N, Pezzoni G. Polynuclear Platinum Complexes. A New Class of Anticancer Agents 6th World Congress on Advances in Oncology; 2001. [Google Scholar]
- 15.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes & Development. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 16.Ihle NT, Paine-Murrieta G, Berggren MI, Baker A, Tate WR, Wipf P, Abraham RT, Kirkpatrick DL, Powis G. The phosphatidylinositol-3-kinase inhibitor PX-866 overcomes resistance to the epidermal growth factor receptor inhibitor gefitinib in A-549 human non-small cell lung cancer xenografts. Mol Cancer Ther. 2005;4:1349–57. doi: 10.1158/1535-7163.MCT-05-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihle NT, Williams R, Chow S, Chew W, Berggren MI, Paine-Murrieta G, Minion DJ, Halter RJ, Wipf P, Abraham R, Kirkpatrick L, Powis G. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther. 2004;3:763–72. [PubMed] [Google Scholar]
- 18.Ihle NT, Lemos R, Schwartz D, Oh J, Halter RJ, Wipf P, Kirkpatrick L, Powis G. Peroxisome proliferator-activated receptor gamma agonist pioglitazone prevents the hyperglycemia caused by phosphatidylinositol 3-kinase pathway inhibition by PX-866 without affecting antitumor activity. Mol Cancer Ther. 2009;8:94–100. doi: 10.1158/1535-7163.MCT-08-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Aoki H, Kuhnel F, Kondo Y, Kubicka S, Wirth T, Iwado E, Iwamaru A, Fujiwara K, Hess KR, Lang FF, Sawaya R, et al. Autophagic cell death of malignant glioma cells induced by a conditionally replicating adenovirus. J Natl Cancer Inst. 2006;98:625–36. doi: 10.1093/jnci/djj161. [DOI] [PubMed] [Google Scholar]
- 21.Aoki H, Kondo Y, Aldape K, Yamamoto A, Iwado E, Yokoyama T, Hollingsworth EF, Kobayashi R, Hess K, Shinojima N, Shingu T, Tamada Y, et al. Monitoring autophagy in glioblastoma with antibody against isoform B of human microtubule-associated protein 1 light chain 3. Autophagy. 2008;4:467–75. doi: 10.4161/auto.5668. [DOI] [PubMed] [Google Scholar]
- 22.Yoon SO, Kim MM, Chung AS. Inhibitory effect of selenite on invasion of HT1080 tumor cells. J Biol Chem. 2001;276:20085–92. doi: 10.1074/jbc.M101143200. [DOI] [PubMed] [Google Scholar]
- 23.Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. Journal of Neurosurgery. 2000;92:326–33. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 24.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–41. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 25.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–91. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 26.Nagane M, Coufal F, Lin H, Bögler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–86. [PubMed] [Google Scholar]
- 27.Powis G, Ihle NT, Yung WK. Inhibiting PI-3-K for glioma therapy. Cell Cycle. 2009;8:335. doi: 10.4161/cc.8.3.7884. [DOI] [PubMed] [Google Scholar]
- 28.Powis G, Ihle N, Kirkpatrick DL. Practicalities of drugging the phosphatidylinositol-3-kinase/Akt cell survival signaling pathway. Clin Cancer Res. 2006;12:2964–6. doi: 10.1158/1078-0432.CCR-06-0617. [DOI] [PubMed] [Google Scholar]
- 29.Pratesi G, Perego P, Polizzi D, Righetti SC, Supino R, Caserini C, Manzotti C, Giuliani FC, Pezzoni G, Tognella S, Spinelli S, Farrell N, et al. A novel charged trinuclear platinum complex effective against cisplatin- resistant tumours: hypersensitivity of p53-mutant human tumour xenografts. Br J Cancer. 1999;80:1912–9. doi: 10.1038/sj.bjc.6690620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerzankova L, Suchankova T, Vrana O, Farrell NP, Brabec V, Kasparkova J. Conformation and recognition of DNA modified by a new antitumor dinuclear Pt(II) complex resistant to decomposition by sulfur nucleophiles. Biochem Pharmacol. 2009 doi: 10.1016/j.bcp.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–8. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 33.Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–91. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 34.Network TCGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8.s. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.