Abstract
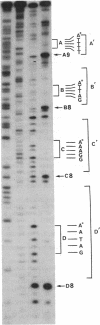
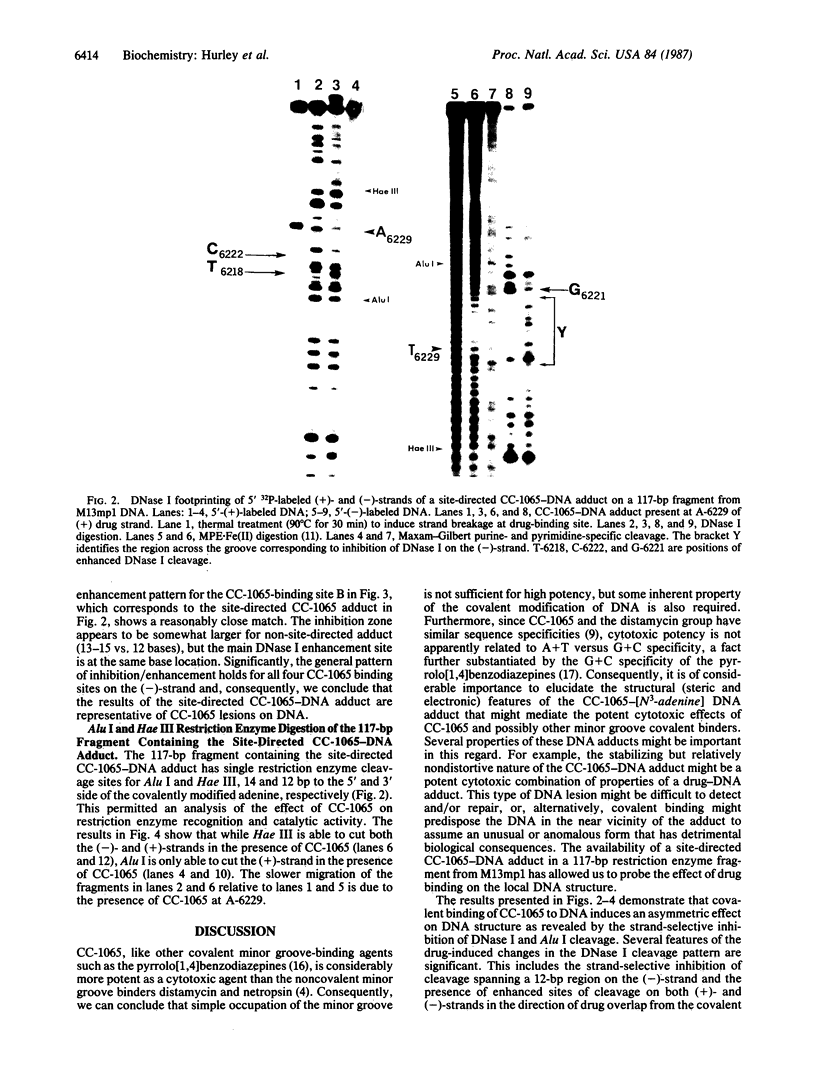
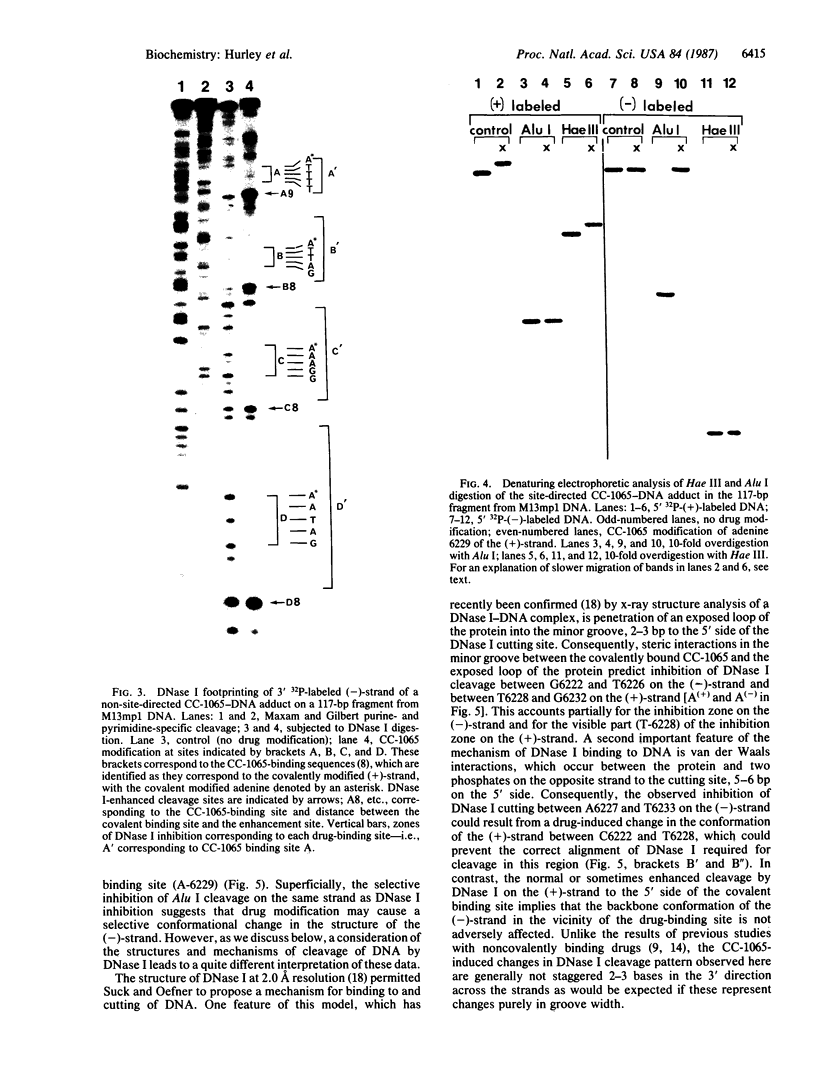
Using DNase I and Alu I endonuclease analysis of a site-directed CC-1065-[N3-adenine]DNA adduct in a 117-base-pair fragment from M13mp1 DNA, we have demonstrated that CC-1065 produces an asymmetric effect on DNA conformation that extends more than one helix turn to the 5' side of the covalently modified adenine. CC-1065 is a potent antitumor antibiotic produced by Streptomyces zelensis, which is believed to mediate its cytotoxic effects through covalent binding to DNA. Previous studies have demonstrated that CC-1065 binds covalently to N3 of adenine and lies within the minor groove of DNA spanning a 4-base-pair sequence to the 5' side of the modified adenine. DNase I footprinting of this site-directed CC-1065-DNA adduct on the noncovalently modified strand shows that inhibition of cleavage occurs over a 12-base region, which is bordered on the 3' side by a site of 2-fold enhancement of cleavage. On the covalently modified strand a much less pronounced inhibition/enhancement pattern of cleavage occurs as far as 11 bases to the 5' side of the covalently modified adenine. While Hae III is able to cleave the DNA on both strands on the 3' side of the covalently modified adenine, Alu I is only able to cleave the covalently modified strand on the 5' side of the adduct. By taking into account the recently published structure of DNase I, we are able to interpret these results and develop a model for the effect of CC-1065 on local DNA structure. In this model, we propose selective drug-induced distortion of the covalently modified strand as a consequence of the alkylation of adenine by CC-1065.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fox K. R., Howarth N. R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanka L. J., Dietz A., Gerpheide S. A., Kuentzel S. L., Martin D. G. CC-1065 (NSC-298223), a new antitumor antibiotic. Production, in vitro biological activity, microbiological assays and taxonomy of the producing microorganism. J Antibiot (Tokyo) 1978 Dec;31(12):1211–1217. doi: 10.7164/antibiotics.31.1211. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Hecht S. M., Reynolds V. L., Molineux I. J., Hurley L. H. DNA sequence specificity of the pyrrolo[1,4]benzodiazepine antitumor antibiotics. Methidiumpropyl-EDTA-iron(II) footprinting analysis of DNA binding sites for anthramycin and related drugs. Biochemistry. 1986 Mar 25;25(6):1249–1258. doi: 10.1021/bi00354a009. [DOI] [PubMed] [Google Scholar]
- Hurley L. H., Reynolds V. L., Swenson D. H., Petzold G. L., Scahill T. A. Reaction of the antitumor antibiotic CC-1065 with DNA: structure of a DNA adduct with DNA sequence specificity. Science. 1984 Nov 16;226(4676):843–844. doi: 10.1126/science.6494915. [DOI] [PubMed] [Google Scholar]
- Jacobson M. K., Twehous D., Hurley L. H. Depletion of nicotinamide adenine dinucleotide in normal and xeroderma pigmentosum fibroblast cells by the antitumor drug CC-1065. Biochemistry. 1986 Oct 7;25(20):5929–5932. doi: 10.1021/bi00368a014. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L., Lesser D., Kurpiewski M. The enfolding arms of EcoRI endonuclease: role in DNA binding and cleavage. Cell. 1986 May 23;45(4):619–629. doi: 10.1016/0092-8674(86)90294-1. [DOI] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Needham-VanDevanter D. R., Hurley L. H. Construction and characterization of a site-directed CC-1065-N3-adenine adduct within a 117 base pair DNA restriction fragment. Biochemistry. 1986 Dec 30;25(26):8430–8436. doi: 10.1021/bi00374a016. [DOI] [PubMed] [Google Scholar]
- Petrusek R. L., Anderson G. L., Garner T. F., Fannin Q. L., Kaplan D. J., Zimmer S. G., Hurley L. H. Pyrrol[1,4]benzodiazepine antibiotics. Proposed structures and characteristics of the in vitro deoxyribonucleic acid adducts of anthramycin, tomaymycin, sibiromycin, and neothramycins A and B. Biochemistry. 1981 Mar 3;20(5):1111–1119. doi: 10.1021/bi00508a011. [DOI] [PubMed] [Google Scholar]
- Reynolds V. L., McGovren J. P., Hurley L. H. The chemistry, mechanism of action and biological properties of CC-1065, a potent antitumor antibiotic. J Antibiot (Tokyo) 1986 Mar;39(3):319–334. doi: 10.7164/antibiotics.39.319. [DOI] [PubMed] [Google Scholar]
- Reynolds V. L., Molineux I. J., Kaplan D. J., Swenson D. H., Hurley L. H. Reaction of the antitumor antibiotic CC-1065 with DNA. Location of the site of thermally induced strand breakage and analysis of DNA sequence specificity. Biochemistry. 1985 Oct 22;24(22):6228–6237. doi: 10.1021/bi00343a029. [DOI] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska K., Pullman B. A theoretical investigation of the sequence specificity in the binding of the antitumor drug anthramycin to DNA. J Biomol Struct Dyn. 1986 Aug;4(1):127–136. doi: 10.1080/07391102.1986.10507650. [DOI] [PubMed] [Google Scholar]