Abstract
A unnatural base pair that is replicated and transcribed with good efficiency would lay the foundation for the long term goal of creating a semi-synthetic organism, but also would have immediate in vitro applications, such as the enzymatic synthesis of site-specifically modified DNA and/or RNA. One of the most promising of the unnatural base pairs that we have identified is that formed between d5SICS and dMMO2. The ortho substituents of these nucleotides are included to facilitate unnatural base pair extension, presumably by forming a hydrogen-bond with the polymerase, but the synthesis of the unnatural base pair still requires optimization. Recently, we have shown that meta and/or para substituents within the dMMO2 scaffold can facilitate unnatural base pair synthesis, although the mechanism remains unclear. To explore this issue, we synthesized and evaluated several dMMO2 derivatives with meta-chlorine, -bromine, -iodine, -methyl, or -propinyl substituents. Complete characterization of unnatural base pair and mispair synthesis and extension reveal that the modifications have large effects only on the efficiency of unnatural base pair synthesis and that the effects likely result from a combination of changes in steric interactions, polarity, and polarizability. The results also suggest that functionalized versions of the propinyl moiety of d5PrM should serve as suitable linkers to site-specifically incorporate other chemical functionalities into DNA. Similar modifications of d5SICS should allow labeling of DNA with two different functionalities, and the previously demonstrated efficient transcription of the unnatural base pair suggests that derivatives might similarly enable site-specific labeling of RNA.
Keywords: DNA replication, halides, hydrophobic effect, nucleobases, synthetic biology
Introduction
The four letter genetic alphabet is conserved throughout nature and is based on the two Watson-Crick base pairs formed via the complementary hydrogen-bonding (H-bonding) patterns of adenine with thymine and guanine with cytosine. However, significant and increasing effort has been directed toward re-engineering the alphabet to include a third, unnatural base pair.[1-14] Such unnatural base pairs might eventually be used as part of an expanded genetic code in vivo, but are likely to find more immediate in vitro applications, such as the site-specific labeling of enzymatically synthesized DNA or RNA. The earliest attempts toward expanding the genetic alphabet, pioneered by Benner and co-workers,[1] relied on designing nucleobases with alternate H-bonding patterns. While several stable pairs were identified, this approach is complicated by facile tautomerization,[15] although recent efforts have made progress in the identification of substituents that reduce the problem.[2, 15] Recent efforts have also drawn on results from Kool and co-workers demonstrating that H-bonding is not an absolute requirement for polymerase-mediated replication,[16] and work from this[4, 5, 17-19] and other labs[12-14, 20-25] has clearly shown that hydrophobic and packing forces are sufficient not only for selective pairing in duplex DNA, but also for enzymatic DNA synthesis, and in some cases even RNA synthesis.[6, 12, 26]
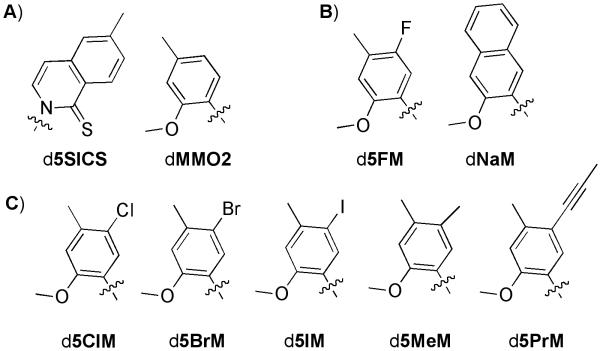
Among the most promising unnatural base pairs that we have identified are those based on small, predominantly hydrophobic aromatic nucleobases that are derivatized with an H-bond acceptor at the position ortho to the glycosidic bond.[4-6, 19] After synthesis of the unnatural base pair (by insertion of the unnatural triphosphate opposite its cognate nucleotide in the template), these ortho substituents are required for efficient extension (by insertion of the next correct dNTP).[2, 7, 19, 23, 27-29] This likely results from the nucleotides adopting a syn orientation about the glycosidic bonds (in analogy to the natural nucleotides[30]), which would position the H-bond acceptors in the developing minor groove where they participate in a functionally critical interaction with H-bond donors of A family polymerases.[29, 31, 32] For example, the nucleotides d5SICS and dMMO2 (Scheme 1A) form a particularly promising unnatural base pair whose relatively efficient replication is critically dependent upon the ortho sulfur and O-methoxy substituents.[3]
Scheme 1.
A) The dMMO2:d5SICS unnatural base pair. B) dMMO2 analogues d5FM and dNaM. C) dMMO2 analogues synthesized and evaluated in this study. Only nucleobase moieties are shown; sugar and phosphate backbone are omitted for clarity.
Despite the relatively efficient recognition of the unnatural base pair, it is still not replicated as efficiently as a natural base pair and it is most limited by the rate of insertion of dMMO2 opposite d5SICS in the template.[3, 5] Given the essentiality of the ortho substituents of dMMO2, our continuing efforts toward its optimization have focused on meta and para derivatizations.[5] We found that d5FMTP and dNaMTP (Scheme 1B) are each inserted opposite d5SICS with greater efficiency and fidelity than dMMO2TP, suggesting that, similar to ortho substituents, meta and para substituents can also facilitate polymerase recognition. However, unlike the ortho substituents, the mechanism by which the meta and para substituents affect replication remains unclear. Meta derivatization of dMMO2 is also interesting because such substituents are expected to positioned into the developing major groove, where, in analogy to the C5 substituents of natural pyrimidines,[33] they might be used to attach functional groups, such as fluorophores or other moieties with interesting chemical or physical properties. Thus, understanding how different meta substituents affect replication is not only expected to help further optimize the unnatural base pair, but it should also help develop methodologies to site-specifically label oligonucleotides. The unnatural pairs formed between d5SICS and either dMMO2 or dNaM are also efficiently transcribed in both directions (i.e. each analogue efficiently directs the incorporation of the other in to RNA)[6] and thus these studies are also expected to help develop methodologies to site-specifically label RNA with two different functional groups, each attached to one of the unnatural nucleotides.
To systematically explore the effect of meta substituents on DNA polymerase recognition, we synthesized and evaluated five dMMO2 derivatives (Scheme 1C). These include derivatives bearing meta-chlorine, -bromine, -iodine, -methyl, or -propinyl substituents. Unnatural base pair synthesis and extension are fully characterized, as is the synthesis and extension of all possible mispairs. The data allow a thorough assessment of the contribution of major groove substituent size and electrostatics to unnatural base pair replication. We find that the modifications only have large effects on the rates at which the unnatural triphosphates are inserted into the growing primer terminus, which appear to result from a combination of steric effects, polarity, and polarizability.
Results
Nucleotide analogue design, synthesis, and evaluation
The unnatural nucleotides, d5ClM, d5BrM, d5IM, d5MeM, and d5PrM are dMMO2 derivatives that were designed as potential partners for d5SICS (Scheme 1C). When evaluated with the previously characterized nucleotides d5FM and dNaM (Scheme 1B), d5ClM, d5BrM, d5IM provide a systematic variation of the size and electrostatics of the major groove substituent. In addition, d5MeM and d5PrM were designed to characterize the effects of size and polarizability in the absence of large changes in dipole moment, and to also begin to explore the possibility of using meta attached linkers to append other functional groups.
The synthesis of each analogue is described in the Supporting Information. Briefly, all derivatives were synthesized via Heck coupling using the appropriate 2-methoxy benzene derivatives and 2′-deoxyribose glycal. In the case of d5BrM and d5IM, synthesis commenced with the dMMO2 nucleobase moiety, which was first coupled to tert-butyl-[[3-(tert-butyl-dimethylsilyloxy)-2,3-dihydrofuran-2-yl]methoxy]dimethylsilane to give the corresponding nucleoside and then halogenated. For d5ClM and d5MeM, 4-chloro-3-methylanisole and 3,4-dimethylanisol, respectively, were iodinated and then coupled to the same protected sugar. In each case, the major coupling product was the desired β-anomer, which was separated from the minor α-anomer by silica gel column chromatography. d5PrM was synthesized from d5IM via Sonogashira coupling.
Free nucleosides were converted to triphosphates or phosphoramidites using standard procedures, and phosphoramidites were used to synthesize DNA containing the unnatural nucleotides at a single position via standard procedures. In all cases, the effect of the modification on polymerase recognition was assessed by determining the steady-state efficiency (i.e., the second order rate constant kcat/KM) with which the Klenow fragment of E. coli DNA polymerase I (Kf) synthesizes the unnatural base pair, by insertion of the unnatural triphosphate opposite an analogue in the template, and extends the resulting unnatural primer terminus, by insertion of the next correct natural triphosphate. The corresponding rates of synthesis and extension for mispairs with natural nucleotides were also measured to determine fidelity.
Polymerase-mediated synthesis of the unnatural base pairs
Polymerase recognition of dMMO2TP derivatives
To begin to characterize how the different substituents impact unnatural base pair replication, we first determined the steady-state rates of insertion of dMMO2 derivatives opposite d5SICS in the template (Table 1). For reference, with an otherwise identical primer-template, Kf inserts dATP opposite dT with a second-order rate constant of 3.2 × 108 M−1 min−1 and dMMO2, d5FM, and dNaM opposite d5SICS with second order rate constants of 3.6 × 105, 3.6 × 106, and 5.0 × 106 M−1 min−1, respectively.[5]
Table 1.
Kinetic data for insertion of dMMO2TP derivatives opposite d5SICS in the template.[a]
| 5′–d (TAATACGACTCACTATAGGGAGA) | |||
|---|---|---|---|
| 3′–d (ATTATGCTGAGTGATATCCCTCT5SICSGCTAGGTTACGGCAGGATCGC) | |||
| Triphosphate | kcat (min−1) | kM (μM) | kcat/kM (M−1 min−1) |
| dMMO2TP[b] | 2.9 ± 0.7 | 6.0 ± 1.9 | 3.6 × 105 |
| d5FMTP[b] | 8.2 ± 1.8 | 2.3 ± 0.3 | 3.6 × 106 |
| d5ClMTP | 6.9 ± 1.4 | 7.9 ± 0.8 | 8.7 × 105 |
| d5BrMTP | 0.08 ± 0.02 | 3.4 ± 1.7 | 2.4 × 104 |
| d5IMTP | 0.80 ± 0.4 | 2.9 ±1.5 | 2.6 × 105 |
| d5MeMTP | 7.9 ± 0.5 | 57 ± 16 | 1.4 × 105 |
| d5PrMTP | 3.2 ± 0.4 | 9.6 ± 1.5 | 3.4 × 105 |
| d5SICSTP[b] | 1.7 ± 0.5 | 63 ± 4 | 2.7 × 104 |
see Experimental section for details.
Reference 5.
Replacing the fluoro substituent of d5FM with a chloro, bromo, or iodo substituent results in reduced efficiencies of insertion for the unnatural triphosphates opposite d5SICS; however, the magnitude of the reduction is highly variable. For the chloro substituent, efficiency is reduced only 4-fold, while for the bromo substituent, it is reduced 150-fold. The pronounced effect with bromine substitution is not simply due to size, as the iodo substituted derivative d5IMTP is inserted with an efficiency that is intermediate between the chloro and bromo analogues. In fact, the efficiency of d5IMTP insertion is similar to that for insertion of the parent analogue dMMO2TP, which lacks a major groove substituent. This suggests that efficient insertion is disfavored by large major groove substituents, but favored by large dipole moments (i.e. d5FMTP) and polarizability (i.e. d5IMTP).
To further examine the effects of major groove derivatization in the absence of large changes in dipole moment, we examined d5MeMTP and d5PrMTP, in which a methyl and a propinyl group, respectively, are positioned in the developing major groove. Insertion efficiencies opposite d5SICS in the template are similar for the two analogues and nearly identical to that of dMMO2TP. This data suggests that simple hydrophobicity is not the dominant factor differentiating the recognition of the nucleotides. In addition, while substituent size within the halide series appears to be important, the increased size associated with the propinyl group relative to the methyl group has little affect on insertion efficiency. This may be due to a different spatial distribution, or to the offset of a small disfavorable interaction by a favorable increase in polarizability.
Polymerase recognition of dMMO2 derivatives in the template
To examine how the modifications affect unnatural base pair synthesis when the dMMO2 derivative is in the template, we measured the rates of d5SICSTP incorporation (Table 2). In contrast to the large effects observed with triphosphate derivatization, modifications to the templating nucleobase had relatively little effect on the efficiency of unnatural base pair synthesis. Thus, we conclude that Kf is not sensitive to the major groove substituent of the dMMO2 derivatives in the template.
Table 2.
Kinetics data for insertion of triphosphates opposite dMMO2 derivatives (X) in the template.[a]
| 5′–d (TAATACGACTCACTATAGGGAGA) | ||||
|---|---|---|---|---|
| 3′–d (ATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC) | ||||
| X | Triphosphate | kcat (min−1) | KM (μM) | kcat/KM (M−1 min−1) |
| dMMO2 | d5SICSTP[b] | 11 ± 2.4 | 0.16 ± 0.04 | 4.7 × 107 |
| dMMO2TP[b] | 15 ± 3.4 | 8.3 ± 3.8 | 1.2 × 105 | |
| dATP[b] | 17 ± 4.1 | 10.2 ± 3.3 | 1.0 × 105 | |
| dGTP[b] | 0.16 ± 0.001 | 20.0 ± 5.0 | <1.0 × 103 | |
| dCTP[b] | 0.65 ± 0.04 | 220 ± 46 | <1.0 × 103 | |
| dTTP[c][b] | 1.2 ± 0.06 | 230 ± 123 | 2.4 × 103 | |
| d5FM | d5SICS | 3.3 ± 1.0 | 0.23 ± 0.03 | 1.4 × 107 |
| d5FMTP | 10.8 ± 2.4 | 12 ± 3.9 | 8.9 × 105 | |
| dATP | 6.1 ± 1.0 | 20 ± 2 | 3.0 × 105 | |
| dGTP | nd[c] | nd[c] | <1.0 × 103 | |
| dCTP | nd[c] | nd[c] | <1.0 × 103 | |
| dTTP | 0.31 ± 0.1 | 130 ± 15 | 2.4 × 103 | |
| d5ClM | d5SICSTP | 3.5 ± 0.9 | 0.11 ± 0.04 | 3.2 × 107 |
| d5ClMTP | 9.1± 1.2 | 17.0 ± 0.7 | 5.2 × 105 | |
| dATP | 9.1 ± 2.2 | 22.0 ± 2.5 | 4.2× 105 | |
| dGTP | nd[a] | nd[a] | <1.0 × 103 | |
| dCTP | nd[c] | nd[c] | <1.0 × 103 | |
| dTTP | 0.52 ± 0.1 | 120 ± 44 | 4.2 × 103 | |
| d5BrM | d5SICSTP | 2.9 ± 0.6 | 0.12 ± 0.02 | 2.3 × 107 |
| d5BrMTP | 0.11 ± 0.04 | 8.7 ± 3.9 | 1.3 × 104 | |
| dATP | 8.5 ± 1.6 | 15 ± 2.7 | 5.6 × 105 | |
| dGTP | nd[c] | nd[c] | <1.0 × 103 | |
| dCTP | 0.52 ± 0.01 | 210 ± 11 | 2.5 × 103 | |
| dTTP | 0.65 ± 0.11 | 150 ± 15 | 4.4 × 103 | |
| d5IM | d5SICSTP | 8.1 ± 1.9 | 0.34 ± 0.15 | 2.4 × 107 |
| d5IMTP | 0.50 ± 0.2 | 4.1 ± 1.0 | 1.2 × 105 | |
| dATP | 7.6 ± 1.4 | 17 ± 2.4 | 4.5 × 105 | |
| dGTP | nd[c] | nd[c] | <1.0 × 103 | |
| dCTP | 0.45 ± 0.2 | 220 ± 106 | 2.0 × 103 | |
| dTTP | 0.55 ± 0.1 | 120 ± 22 | 4.6 × 103 | |
| d5MeM | d5SICSTP | 4.7 ± 0.6 | 0.24 ± 0.08 | 1.9 × 107 |
| d5MeMTP | 8.7 ± 0.7 | 28 ± 2 | 2.5 × 105 | |
| dATP | 6.7 ± 0.3 | 12 ± 2.0 | 5.4 × 105 | |
| dGTP | nd[c] | nd[c] | <1.0 × 103 | |
| dCTP | nd[c] | nd[c] | <1.0 × 103 | |
| dTTP | 0.56 ± 0.1 | 75 ± 1.6 | 7.4× 103 | |
| d5PrM | d5SICSTP | 3.6 ± 2.07 | 0.17 ± 0.03 | 2.1 × 107 |
| d5PrMTP | 4.0 ± 0.2 | 6.5 ± 1.0 | 6.2 × 105 | |
| dATP | 10.1 ± 3.1 | 14 ± 2.9 | 7.4 × 105 | |
| dGTP | nd[c] | nd[c] | <1.0 × 103 | |
| dCTP | nd[c] | nd[c] | <1.0 × 103 | |
| dTTP | 0.69 ± 0.1 | 150 ± 4.1 | 4.3 × 103 |
see Experimental section for details.
Reference 5.
Reaction was too inefficient for kcat and KM to be determined independently.
We next examined whether modification of the dMMO2 derivatives in the template affect the rates of mispair synthesis. We first measured the rate of insertion of each unnatural nucleotide opposite itself in the template (i.e. ‘self pair’ synthesis; note that with self pairs, modifications are made by definition to both the incoming and templating nucleotide) (Table 2). We observed the same substituent dependence for the efficiency of self pair synthesis as we did for insertion of the dMMO2 analogues opposite d5SICS in the template: the d5FMTP self pair was synthesized most efficiently, followed by the d5ClM and d5IM self pairs. The efficiency of self pair synthesis with d5MeM and d5PrM were again similar to each other, as well as to dMMO2, with rates ranging only from 1.2 × 105 to 6.2 × 105 M−1 min−1. Given these observations, we conclude that like unnatural base pair synthesis, the efficiency of self pair synthesis is more influenced by the nature of the triphosphate than the nature of the templating nucleobase.
Characterization of natural dNTP insertion opposite a dMMO2 derivative (Table 2) revealed that neither dGTP nor dCTP insertion is detectable (kcat/KM < 103 M−1 min−1) and that dTTP insertion is only barely detectable (kcat/KM = 2.4 – 7.3 × 103 M−1 min−1). In contrast, dATP is generally inserted more efficiently (1.0 × 105 M−1 min−1 – 7.4 × 105 M−1 min−1). As has been suggested with other predominantly hydrophobic nucleobases in the template,[34] this likely results from a combination of adenine’s hydrophobicity, its packing ability, and an interstrand intercalation mode of pairing as discussed in detail previously.[5] While mispair synthesis is highly dependent on which natural dNTP is inserted, the major groove substituent of the dMMO2 derivative in the template was again less important. Thus, the data support the hypothesis that the nature of the natural or unnatural triphosphate is more important for efficient synthesis than the nature of the templating unnatural nucleotide.
Polymerase-mediated extension of the unnatural base pairs
Extension of primers terminating with dMMO2 derivatives paired opposite d5SICS
To characterize how the different major groove substituents affect the extension of the unnatural base pair, we synthesized primers that terminate at their 3′ end with one of the dMMO2 derivatives paired opposite d5SICS in the template. The next correct base in the template is dG, and the steady-state rate at which Kf inserts dCTP was measured (Table 3). For comparison, the unnatural pairs with dMMO2, d5FM, and dNaM, paired opposite d5SICS are extended with efficiencies of 1.9 × 106, 5.5 × 106, and 1.2 × 106 M−1 min−1, respectively.[5] While the extension efficiencies of d5ClM:d5SICS (primer:template) and d5BrM:d5SICS are similar (2.0 × 106, and 2.6 × 106 M−1 min−1), extension of d5IM:d5SICS is slightly less efficient (7-fold). The contributions of sterics and electrostatics were deconvoluted by characterizing extension of the dMMO2:d5SICS, d5MeM:d5SICS, and d5PrM:d5SICS unnatural base pairs. While the meta methyl substituent has little effect on extension, while the propinyl group has a slightly deleterious effect (7-fold decreased). Thus, while substituents with increased size at the primer terminus (i.e. d5IM and dPrM) reduce the efficiency of extension, the effects are generally small and the data suggest that the modifications have less of an effect on replication once incorporated into the growing primer terminus than they do during unnatural triphosphate insertion.
Table 3.
Kinetic data for unnatural base pair extension via dCTP incorporation with dMMO2 derivatives in the primer.[a]
| 5′–d(TAATACGACTCACTATAGGGAGAY) | |||
|---|---|---|---|
| 3′–d(ATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC) | |||
| X=d5SICS | |||
| Y | kcat (min−1) | KM (μM) | kcat/KM (M−1 min−1) |
| dMMO2[b] | 6.4 ± 1.1 | 3.4 ± 0.3 | 1.9 × 106 |
| d5FM | 3.5 ± 0.11 | 0.63 ± 0.1 | 5.5 × 106 |
| d5ClM | 4.2 ± 0.8 | 2.0 ± 0.06 | 2.0 × 106 |
| d5BrM | 5.3 ± 0.9 | 2.1 ± 0.07 | 2.6 × 106 |
| d5IM | 4.3 ± 1.3 | 15 ± 0.3 | 2.9 × 105 |
| d5MeM | 2.4 ± 0.06 | 1.7 ± 1.18 | 1.4 × 106 |
| d5PrM | 2.6 ± 0.52 | 9.6 ± 1.78 | 2.7 × 105 |
| d5SICS[b] | nd[c] | nd[c] | <1.0 × 103 |
see Experimental section for details.
Reference 5.
Reaction was too inefficient for kcat and KM to be determined independently.
Extension of primers terminating with d5SICS paired opposite dMMO2 derivatives
Finally, we characterized extension efficiency and fidelity of primers terminating with d5SICS paired opposite each dMMO2 derivative in the template (Table 4). For comparison, the d5SICS:dMMO2, d5SICS:d5FM, and d5SICS:d5NaM pairs are extended with efficiencies of 6.9 × 105, 2.3 × 106, and 2.7 × 106 M−1 min−1.[5] As with unnatural base pair synthesis, the extension efficiency of the modified unnatural base pairs is largely independent of template derivatization, with efficiencies ranging only from 2.2 × 106 to 5.3 × 106 M−1 min−1.
Table 4.
Kinetic data for unnatural base pair and mispair extension via dCTP incorporation with dMMO2 derivatives in the template.[a]
| 5′-d(TAATACGACTCACTATAGGGAGAY | ||||
|---|---|---|---|---|
| 3′-d(ATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC | ||||
| X | Y | kcat (min−1) | KM (μM) | kcat/KM (M−1 min−1) |
| dMMO2 | d5SICS[b] | 3.8 ± 0.3 | 5.7 ± 1.2 | 6.7 × 105 |
| dMMO2[b] | 0.87 ± 0.28 | 160 ± 47 | 5.3 × 103 | |
| dA[b] | 8.7 ± 0.9 | 190 ± 17 | 4.6 × 104 | |
| dG[b] | nd[c] | nd[c] | <1.0 × 103 | |
| dC[b] | 3.9 ± 1.32 | 3.2 ± 0.23 | 1.2 × 106 | |
| dT[b] | 6.2 ± 0.8 | 9.4 ± 1.0 | 6.6 × 105 | |
| d5FM | d5SICS | 3.2 ± 0.7 | 1.4 ± 0.3 | 2.3 × 106 |
| d5FM | 2.6 ± 0.39 | 99 ± 17.9 | 2.6 × 104 | |
| dA | 5.5 ± 0.08 | 17 ± 1.6 | 3.2 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 2.4 ± 0.23 | 1.2 ± 0.10 | 2.0 × 106 | |
| dT | 4.1 ± 0.19 | 2.1 ± 0.17 | 2.0 × 106 | |
| d5ClM | d5SICS | 7.8 ± 1.47 | 1.5 ± 0.37 | 5.3 × 106 |
| d5ClM | 2.7 ± 0.19 | 80 ± 14.5 | 3.4 × 104 | |
| dA | 7.0 ± 0.19 | 7.3 ± 3.1 | 9.6 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 2.8 ± 0.34 | 0.54 ± 0.14 | 5.2 × 106 | |
| dT | 4.2 ± 0.79 | 0.95 ± 0.10 | 4.4 × 106 | |
| d5BrM | d5SICS | 6.8 ± 2.0 | 2.0 ± 0.18 | 3.4 × 106 |
| d5BrM | 1.5 ± 0.1 | 46 ± 8.3 | 3.2 × 104 | |
| dA | 5.6 ± 0.2 | 8.5 ± 1.1 | 6.5 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 2.5 ± 0.07 | 2.4 ± 0.02 | 5.9 × 106 | |
| dT | 3.6 ± 0.1 | 0.85 ± 0.07 | 4.3 × 106 | |
| d5IM | d5SICS | 4.1 ± 1.6 | 1.1 ± 0.3 | 3.6 × 106 |
| d5IM | 0.91 ± 0.18 | 120 ± 13 | 7.4 × 103 | |
| dA | 5.9 ± 0.9 | 12 ± 0.3 | 4.9 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 1.8 ± 0.02 | 0.30 ± 0.03 | 5.9 × 106 | |
| dT | 3.1 ± 0.4 | 0.58 ± 0.05 | 5.2 × 106 | |
| d5MeM | d5SICS | 4.6 ± 0.5 | 2.1 ± 0.05 | 2.2 × 106 |
| d5MeM | 3.9 ± 0.12 | 59 ± 11 | 6.5 × 104 | |
| dA | 3.8 ± 1.0 | 18 ± 5.7 | 2.2 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 2.4 ± 0.08 | 0.80 ± 0.07 | 3.0 × 106 | |
| dT | 5.2 ± 1.4 | 4.8 ± 1.9 | 1.1 × 106 | |
| d5PrM | d5SICS | 7.5 ± 2.81 | 5.8 ± 2.5 | 1.3 × 106 |
| d5PrM | 0.98 ± 0.17 | 230 ±150 | 4.2 × 103 | |
| dA | 5.2 ± 0.09 | 21 ± 6.6 | 2.4 × 105 | |
| dG | nd[c] | nd[c] | <1.0 × 103 | |
| dC | 2.9 ± 0.25 | 0.67 ± 0.03 | 4.3 × 106 | |
| dT | 2.7 ± 0.37 | 1.8 ± 0.8 | 1.2 × 106 | |
see Experimental section for details.
Reference 5.
Reaction was too inefficient for kcat and KM to be determined independently.
Extension of the dMMO2, d5FM, and dNaM self pairs is inefficient, with a kcat/KM of only 5.3 × 103 M−1 min−1 M to 2.6 × 104.[5]. The derivatized self pairs are extended with similar or slightly reduced efficiencies, varying between 4.2 × 103 and 3.4 × 104 M−1 min−1 (Table 4). The d5MeM and d5PrM self pairs are extended with efficiencies of 6.5 × 104 and 4.2 × 103 M−1 min−1, respectively. Interestingly, we observed the same dependence on extension efficiency for the self pairs as we did for derivative triphosphate insertion opposite d5SICS, although the effects were smaller (10-fold versus 150-fold). As with dMMO2,[19] extension of mispairs with dT or dC at the primer terminus is surprisingly efficient (6.6 × 105 - 5.6 × 106 M−1 min−1), while extension of mispairs with dA is slightly less efficient (2.2 × 105 – 9.6 × 105 M−1 min−1), and extension of mispairs with dG is not detectable. Generally, the nature of the templating dMMO2 derivative had little effect on mispair extension.
Discussion and Conclusion
The unnatural base pair formed between d5SICS and dMMO2 is reasonably well replicated by Kf and was originally identified from the optimization of the most promising unnatural base pair identified from a screen of 3600 candidates.[3] The substituents that are ortho to the glycosidic bond, and presumably oriented into the developing minor groove during DNA synthesis, appear to be essential. However, other modifications, for example, the increased aromatic surface area of dNaM or the halide of dFM result in even more efficient replication,[5] suggesting that meta and para substituents may also be important. Thus, we examined meta derivatization of dMMO2 through halide substitution to alter substituent size and dipole moment, as well as through methyl and propinyl substitution to specifically examine the effects of substituent size and polarizability. In addition to elucidating the physical forces mediating unnatural base pair recognition, these studies were expected to aid development of a methodology to use the unnatural base pairs for site-specific modification of DNA and RNA.
Meta substitution had little effect on unnatural base pair or mispair synthesis with the dMMO2 derivatives in the template, and relatively small effects on extension with the derivatives present at either the primer terminus or in the template. Thus, the meta substituents do not significantly facilitate or interfere with these steps of replication. In contrast, the modifications examined had large effects on the efficiency of Kf-mediated triphosphate insertion, with the second-order rate constants varying 150-fold. The fluoro-modified triphosphate is inserted by Kf opposite d5SICS the most efficiently, while the chloro derivative is inserted only 4-fold less efficiently. While the bromo derivative is inserted 150-fold less efficiently than the fluoro derivative, the iodo derivative is inserted only 10-fold less efficiently. These trends do not simply parallel nucleobase hydrophobicity, dipole moment of the aryl-halide bond, or the van der Waals radii of the halide. Considering that significant packing interactions are likely to be introduced during nucleotide insertion, it is reasonable to assume that modifications that favor packing will facilitate insertion. However, because the van der Waals radii of bromine and iodine are the largest of the substituents tested (1.85 Å and 1.98 Å, respectively), and significantly larger than that expected to be easily accommodated between the nucleobases (which are separated by 3.34 Å in native B-form DNA), their presence is likely to be at least marginally destabilizing. Thus, it seems that increased dipole moment (5FM) or polarizability (5IM) favors unnatural triphosphate insertion, while the inclusion of substituents that are too large (5BrM and 5IM) disfavor it. The intermediate efficiency with which d5IMTP is inserted likely reflects opposed and somewhat compensating effects of favorable polarizability and disfavorable sterics. The similar efficiency with which d5PrMTP is inserted opposite d5SICS, relative to dMeMTP, may reflect similarly compensating effects of polarizability and sterics.
The results are particularly interesting from the perspective of the effort to expand the genetic alphabet. While relatively well replicated compared to other analogues, the replication of the unnatural base pair formed between d5SICS and dMMO2 is limited by the relatively slow insertion of dMMO2TP opposite d5SICS in the template. However, dMMO2 is better recognized by the polymerase during the other steps of replication (i.e. when in the template or during extension when present in either the primer or the template). Importantly, the data suggests that derivatizations at the meta position of dMMO2 might be used to specifically optimize the insertion of triphosphate, without interfering with the other steps of replication. The search for such meta modifications is currently underway.
For a site-specific labeling strategy that is compatible with polymerase amplification, the data suggest that propinyl linkers, such as those used already with the natural nucleotides,[33] other unnatural base pairs,[12, 14] and present in d5PrM, are promising. These linkers are comprised of propargyl amines, with the amine serving as a reactive site to attach other functionalities. The data suggest that such linkers are also likely to be well tolerated within the dMMO2 scaffold. In this manner, virtually any functionality might be attached to the dMMO2 derivative and site-specifically incorporated into DNA during enzymatic synthesis. Moreover, it seems likely that d5SICS derivatives will allow analogous site-specific modification, such that a single duplex could be site-specifically modified with two different functional groups. The efficient transcription of the unnatural base pair would also allow site-specific labeling of RNA with two different moieties. Such DNA and RNA should find different academic and biotechnological applications, and efforts toward these applications are currently underway.
Experimental Section
General Methods
All reactions were carried out in oven-dried glassware under inert atmosphere, and all solvents were dried over 4 Å molecular sieves with the exceptions of dichloromethane, which was distilled from CaH2, and tetrahydrofuran, which was distilled from sodium and potassium metal. All other reagents were purchased from Aldrich. All unnatural nucleosides and nucleotides used in this study were synthesized as described in Supporting Information. 1H, 13C, and 31P NMR spectra were recorded on Bruker DRX-500, Varian Inova-400, or Mercury 300 spectrometers. High resolution mass spectroscopic data were obtained from the core facility at The Scripps Research Institute. T4 polynucleotide kinase was purchased from New England Biolabs, Kf from GE Healthcare, and [γ-32P]-ATP was purchased from MP Biomedicals.
Oligonucleotide Synthesis
Oligonucleotides were prepared by the β-cyanoethylphosphoramidite method on controlled pore glass supports (1 μmol) by using an Applied Biosystems Inc. 392 DNA/RNA synthesizer as standard method. After automated synthesis, the oligonucleotides were cleaved from the support and deprotected by heating at 55 °C for 12 h. The crude product was further purified by polyacrylamide gel electrophoresis, followed by electroelution. The resulting purified oligonucleotides were precipitated with 80% ethanol and dried overnight. Oligonucleotide concentration was determined by UV absorption. DNA primers with unnatural nucleotides at their 3′-termini were synthesized using 3′-phosphate CPG (Glen Research) and then purified as described above, followed by treatment for 1 hour at 37 °C with CIP (0.5 U) (New England Biolabs) to produce free 3′-OH groups.
Gel-Based Kinetic Assay
Primer oligonucleotides were 5′ radiolabeled with [γ-32P]-ATP and T4 polynucleotide kinase. Templates were annealed to primers in the reaction buffer by heating to 90 °C followed by slow cooling to ambient temperature. Assay conditions included 40 nM primer/template, 0.1-1.3 nM Kf, 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 50 μg/mL acetylated BSA. The reactions were carried out by combining the DNA-enzyme mixture with an equal volume (5 μL) of 2× dNTP stock solution, incubating at 25 °C for 1-10 min, and quenching by the addition of loading dye (95% formamide, 20 mM EDTA, and sufficient amounts of bromophenol blue and xylene cyanol; 20 μL). The reaction mixture was then analyzed by 15% polyacrylamide/8 M urea denaturing gel electrophoresis. Radioactivity was quantified using a Phosphorimager and the ImageQuant program (Molecular Dynamics) with overnight exposures. The kobs values were plotted against triphosphate concentration and the data was fit to the Michaelis-Menten equation (Kaleidagraph, Synergy Software) to determine kcat and KM. The data presented are the average of three independent determinations.
Supplementary Material
Acknowledgements
Funding was provided by the National Institutes of Health (GM60005 to F.E.R.) and Korea Research Foundation (KRF-2006-352-C00047 to Y.J.S.).
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- [1].Piccirilli JA, Krauch T, Moroney SE, Benner SA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- [2].Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hwang GT, Romesberg FE. J. Am. Chem. Soc. 2008;130:14872–14882. doi: 10.1021/ja803833h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seo YJ, Matsuda S, Romesberg FE. J. Am. Chem. Soc. 2009;131:5046–5047. doi: 10.1021/ja9006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Leconte AM, Romesberg FE. In: Protein Engineering. Köhrer C, RajBhandary UL, editors. Springer-Verlag; Berlin: 2009. pp. 291–314. [Google Scholar]
- [8].Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, Kodama K, Yokogawa T, Nishikawa K, Yokoyama S. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- [9].Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- [10].Mitsui T, Kimoto M, Harada Y, Yokoyama S, Hirao I. J. Am. Chem. Soc. 2005;127:8652–8658. doi: 10.1021/ja0425280. [DOI] [PubMed] [Google Scholar]
- [11].Hirao I. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- [12].Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- [13].Hirao I, Mitsui T, Kimoto M, Yokoyama S. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- [14].Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sismour AM, Benner SA. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moran S, Ren RX-F, Rumney SI, Kool ET. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- [18].Ogawa AK, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- [19].Leconte AM, Matsuda S, Romesberg FE. J. Am. Chem. Soc. 2006;128:6780–6781. doi: 10.1021/ja060853c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- [21].Kincaid K, Beckman J, Zivkovik A, Halcomb RL, Engels JW, Kuchta RD. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matray TJ, Kool ET. Nature. 1999;399:704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
- [23].Morales JC, Kool ET. J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang X, Lee I, Berdis AJ. Biochemistry. 2005;44:13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- [25].Zhang X, Lee I, Zhou X, Berdis AJ. J. Am. Chem. Soc. 2006;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- [26].Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I. Nucleic Acids Res. 2007;35:5360–5369. doi: 10.1093/nar/gkm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leconte AM, Matsuda S, Hwang GT, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:4326–4329. doi: 10.1002/anie.200601272. [DOI] [PubMed] [Google Scholar]
- [28].Kim Y, Leconte AM, Hari Y, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:7809–7812. doi: 10.1002/anie.200602579. [DOI] [PubMed] [Google Scholar]
- [29].Matsuda S, Leconte AM, Romesberg FE. J. Am. Chem. Soc. 2007;129:5551–5558. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- [31].Spratt TE. Biochemistry. 2001;40:2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- [32].Guo MJ, Hildbrand S, Leumann CJ, McLaughlin LW, Waring MJ. Nucleic Acids Res. 1998;26:1863–1869. doi: 10.1093/nar/26.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hocek M, Fojta M. Org. Biomol. Chem. 2008;6:2233–2241. doi: 10.1039/b803664k. [DOI] [PubMed] [Google Scholar]
- [34].Henry AA, Olsen AG, Matsuda S, Yu C, Geierstanger BH, Romesberg FE. J. Am. Chem. Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.