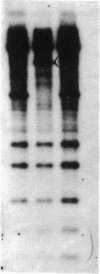
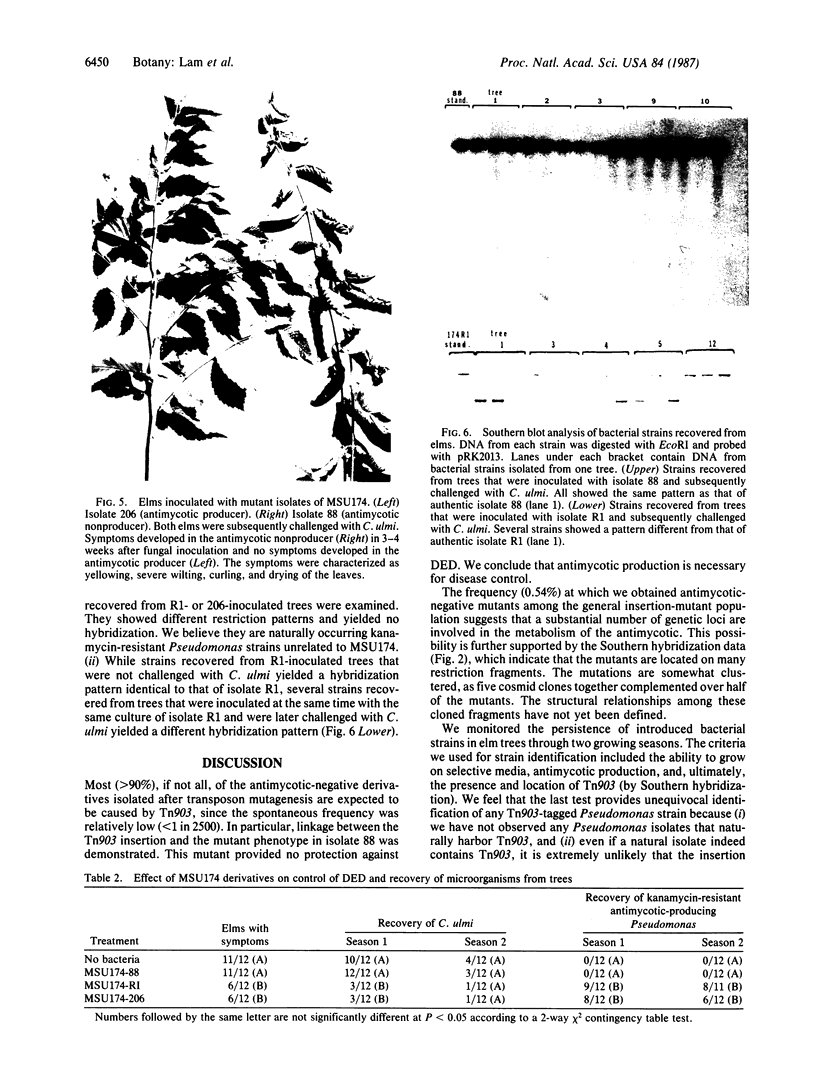
Abstract
Antimycotic-producing strains of Pseudomonas syringae are being tested as Dutch elm disease control agents. We examined the role of antimycotic production in disease control. Transposon Tn903 was used to mutagenize the antimycotic-producing strain MSU174. Eighty-one mutants that did not inhibit fungal growth were identified among 15,000 Tn903-containing derivatives. Linkages between Tn903 insertions and defects in antimycotic metabolism were established. Three Tn903-containing strains (two antimycotic producers and one nonproducer) were individually introduced into American elm seedlings. The seedlings were subsequently challenged with Ceratocystis ulmi, the causal agent of Dutch elm disease. Protection of the elms was observed with the two antimycotic-producing strains but not with the nonproducing strain. The introduced strains could be readily recovered from the seedlings after two growing seasons. They were unequivocally identified by the Tn903 insertions they contain.
Keywords: cosmid library, functional complementation, biological control, strain identification by DNA hybridization
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Lam S. T., Lam B. S., Strobel G. A vehicle for the introduction of transposons into plant-associated pseudomonads. Plasmid. 1985 May;13(3):200–204. doi: 10.1016/0147-619x(85)90043-5. [DOI] [PubMed] [Google Scholar]
- Nomura N., Yamagishi H., Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene. Gene. 1978 Feb;3(1):39–51. doi: 10.1016/0378-1119(78)90006-9. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]