Abstract
In a world of emerging and resurging infectious diseases, dominated by zoonoses, environmental monitoring plays a vital role in our understanding their dynamics and their spillover to humans. Here, we critically review the ecology, epidemiology and need for monitoring of a variety of directly transmitted (Sin Nombre virus, Avian Influenza) and vector-borne (Ross River virus, West Nile virus, Lyme disease, anaplasmosis and babesiosis) zoonoses. We focus on the valuable role that existing monitoring plays in the understanding of these zoonoses, the demands for new monitoring, and how improvements can be made to existing monitoring. We also identify the fruitful outcomes which would result from implementation of the monitoring demands we have highlighted. This review aims to promote improvements in our understanding of zoonoses, their management, and public health by encouraging discussion among researchers and public health officials.
Introduction and paper structure
A significant number of pathogens that cause human disease are maintained within and transmitted from wildlife and livestock.1–3 Spillover of these zoonotic pathogens to humans is related to contact of humans with disease reservoirs, either directly or indirectly, and is often the result of complex ecological interactions, influenced by changes in climate and the environment; both natural and anthropogenic.4–8 Monitoring of climatic conditions and of infection and abundance of hosts and vectors of zoonoses plays an important role in determining causes of pathogen dynamics and in predicting human incidence. For example, incidence of hantavirus pulmonary syndrome (HPS) has been associated with El-Niño events and enhanced human-deer mouse (reservoir) interactions in the Four Corners region of the US9,10 and increased summer rainfall in southwest Western Australia was correlated with increased abundance of vector mosquitoes and human infections of Ross River virus.11
Determining causes of emergence of zoonotic pathogens frequently requires combining experimentation and theoretical modeling.12 However, formulation of causal hypotheses and parameterization of particular variables (such as relationships between rainfall and increased mosquito abundance) often originate from monitoring approaches. Thus, given the general importance of monitoring, and the capacity of monitoring to complement experimental and theoretical approaches, critical evaluations of monitoring needs will contribute fruitful insights to enhance understanding of pathogen dynamics and determine remedies to complex public health problems. Here we focus on the role that environmental monitoring can play in understanding the ecology and epidemiology of zoonoses (Fig. 1). We examine an array of zoonoses, including those that are directly transmitted (Sin Nombre virus (SNV), Avian Influenza (AI)) and vector-borne (Ross River virus (RRV), West Nile virus (WNV), Lyme disease (LD), anaplasmosis and babesiosis). We emphasize how specific examples of existing or newly created environmental monitoring systems could be integrated with disease surveillance to enhance our understanding and deal with public health problems.
Fig. 1.
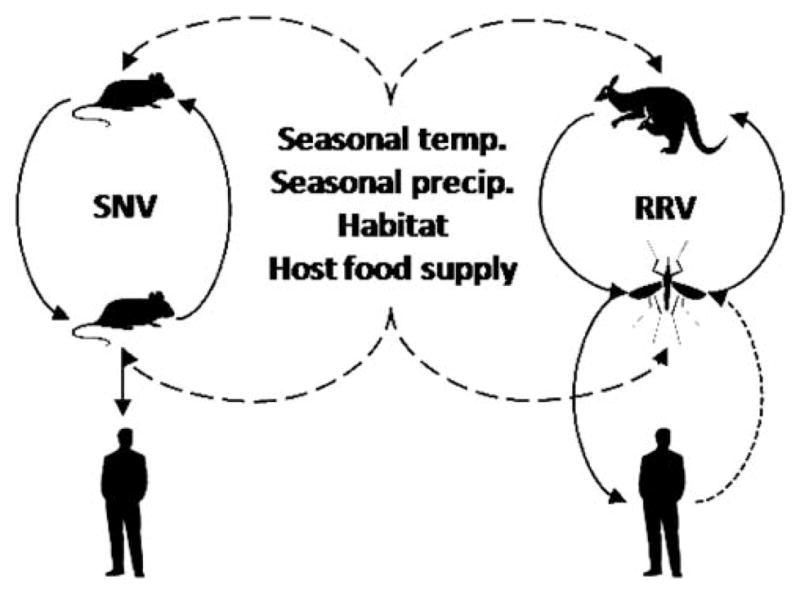
Conceptual diagram outlining aspects of the ecology and epidemiology of zoonoses that may be targeted for monitoring. Diagram represents one directly transmitted zoonosis (left, Sin Nombre virus, with rodent reservoir) and one vector-borne zoonosis (right, Ross River virus, with Macropod host and mosquito vector). Important candidate environmental variables identified that affect host and vector abundance (influence represented by long-dashed lines). Pathogen transmission represented by solid lines, except human to vector transmission (dotted line), which usually has a limited contribution to transmission.
For each zoonosis, or group of zoonoses, discussed in this review we briefly outline the natural history of that system and existing monitoring that contributes to our comprehension of transmission and illness to humans. We then identify specific improvements in monitoring that would contribute to understanding pathogen dynamics and spillover to humans. Finally, we make recommendations for how identified demands in monitoring could be implemented. A summary of these zoonotic case studies are outlined in Table 1. A common theme among examples outlined in this review is the need for implementation of new monitoring programs, improved integration between researchers and governmental organizations, and better knowledge and access to data spanning multiple disciplines.
Table 1.
Summary of zoonotic disease systems and their hosts and vectors. Identified demand for targeted monitoring, suggested means of implementation and benefits of improved monitoring.
| System | Principle reservoir(s) | Principle vector(s) | Monitoring demand | Implementation | Benefits |
|---|---|---|---|---|---|
| Directly transmitted pathogens | |||||
| Sin Nombre virus | Deer mouse (Peromyscus maniculatus) | N/A | Linkage of sylvan population monitoring with peridomestic monitoring | Addition of peridomestic monitoring to existing sylvan deer mouse population monitoring Development of monitoring of deer mouse excreta and SNV in particulate matter in peridomestic environments |
Improved understanding of linkage between environment and HPS incidence Improved prediction of HPS risk |
| Avian Influenza viruses | Poultry, Swine, Waterbirds (Anseriformes, Charadriformes) | N/A | Transmission intensity, viral diversity and viral recombination | Standardized monitoring system for wild birds and swine Sharing of poultry surveillance and virus characterization by all parties |
Improved understanding of spillover of viruses from wild birds Improved understanding of outbreaks of influenza in poultry Improved understanding of recombination between viruses of wild birds, swine, poultry and humans |
| Vector-borne pathogens | |||||
| Ross River virus | Macropods and other marsupials | Cx. annulirostris, Ae. camptorhynchus, Ae. vigilax, | Host population dynamics and immunity in and around epidemic zones | Coordination of RRV surveillance with expertise of marsupial mammalogists | Improved epidemic forecasting |
| West Nile virus | Birds | Culex mosquitoes | Abundance of WNV-infected mosquitoes for selected species | Support for regional monitoring and testing, data sharing and integration | Improved regional risk assessment and early warning systems Better allocation of resources |
| Lyme disease, babesiosis and anaplasmosis | White-footed mice (Peromyscus leucopus), other mammals and, to a lesser extent, birds and reptiles | I. ricinus, I. scapularis, I. pacificus | Reliable prediction of abundance of infected ticks and human incidence | Monitoring of infected tick abundance Identification of climatic variables that consistently predict tick-borne disease risk Population monitoring of white-footed mice and other hosts permissive to tick feeding Monitoring local acorn production |
Forecasting of human exposure risk and disease incidence |
Examples of directly transmitted pathogens
Sin Nombre virus
SNV was first recognized in 1993, following an outbreak of acute respiratory illness (later known as hantavirus pulmonary syndrome) with high mortality rates in the Four Corners region of the US.13,14 The reservoir for this pathogen was later determined to be the deer mouse, Peromyscus maniculatus, with apparent spillover to other rodent species and, under certain conditions, to humans.15 Human outbreaks of HPS in the Four Corners region appeared to follow El-Niño events, which were favorable for deer mouse reproduction, and human-deer mouse interactions.16 Since 1993, SNV has been identified across the US (coinciding with the distribution of deer mice) and HPS incidence rates are greatest in western states,17,18 although the relationship between climate and HPS cases outside of the Four Corners regions is less well understood. Human exposure to SNV is primarily associated with rural peridomestic environments (houses, barns, sheds and other types of out-buildings) and is believed to occur through inhalation of SNV contaminated deer mouse excreta while undertaking activities that generate particulate matter, such as sweeping.19,20
Monitoring of sylvan small mammal populations and SNV dynamics was undertaken from 1994–2004 among states with high rural HPS incidence rates (New Mexico, Colorado, Utah, Arizona and Montana),18,21–23 and this monitoring is ongoing in Montana.24,25 This monitoring has yielded substantial insight into the ecological dynamics of SNV and its reservoir. For example, prevalence of SNV antibodies (a marker of chronic infection) is highest among older male deer mice,17,26 transmission appears to be due to intraspecific aggression,22,26 antibody prevalence increases after a delay in deer mouse density,17,27 and host community structure influences antibody prevalence of deer mice.3,28 While much has been learned about dynamics of SNV in sylvan environments, there remain significant gaps in knowledge about small mammal SNV dynamics in peridomestic settings, human-deer mouse interactions, and human SNV exposure.
Sin Nombre virus antibody prevalence among deer mice is higher within and around peridomestic settings than sylvan settings.29–32 This may be due to smaller home ranges of peri-domestic deer mice, leading to increased contact and transmission events between individuals.33 In terms of human-deer mouse interactions in peridomestic settings, Childs et al.34 found deer mouse abundance (but surprisingly not infection prevalence) was greater in HPS case houses than non-case houses. This suggests that human risk of infection with SNV may be a function of the number of infected deer mice using peridomestic environments. Additionally, it has been hypothesized that human risk of infection is greater in peridomestic settings than sylvan because SNV may persist in deer mouse excreta longer, due to less exposure to ultraviolet radiation,33 which can inactivate many types of viruses.35
Paired monitoring of sylvan and peridomestic deer mouse populations and SNV dynamics among these two environments is needed to quantify linkages, and how this relates to human incidence of HPS. This type of monitoring is currently being conducted near Cascade and Anaconda Montana,26,31 but detailed analysis of linkage between these two environments has yet to be published. Additional long-term studies of peridomestic sites in high HPS incidence areas, such as the Four Corners region, are also needed. To achieve this, peridomestic surveys of deer mice could be locally matched with existing sylvan monitoring that may be ongoing, thus not requiring much additional survey effort. Understanding human exposure risk to SNV would also be enhanced by the development of monitoring of particulate matter (detecting the amount of deer mouse excreta and SNV in dust particles or suspended droplets) in peridomestic settings. Such a type of monitoring and its relative feasibility has yet to be described, but is in development (K. S. Richardson, personal communication) and, if feasible, could be undertaken in concordance with peridomestic deer mouse monitoring. These additional monitoring practices would enable better linkage between environmental data, sylvan and peri-domestic deer mouse and SNV dyanamics, and forecasting of conditions that lead to outbreaks of HPS.
Avian influenza viruses
Waterbirds are generally recognized as the original reservoir for most influenza viruses, including those that contribute genetic material to human influenza strains such as the recently emerged H1N1,36,37 as well as highly pathogenic poultry strains such as H5N1. A large diversity of viral subtypes (characterized by two surface proteins, hemagglutinin and neuraminidase) is transmitted in each the two groups of wild birds thought to be most important in transmission; Anseriformes (including ducks, geese and swans), and Charadriiformes (including gulls, plovers, and sandpipers).38 Although some subtypes of AI can be found in both taxa, some viral subtypes are found exclusively in one group,39 and the spillover of individual viral strains between the two groups is poorly known, despite its importance for viral spread.40,41
The emergence of novel strains of AI in both poultry and humans is thought to be linked to viral circulation in wild birds. Nonetheless, transmission dynamics outside large scale poultry farms are poorly known. For example, the H1N1 virus that caused the recent pandemic in humans was separated from the most recently sampled ancestor by more than a decade, demonstrating the paucity of monitoring of influenza in swine.36 In contrast, the re-emergence and nearly pandemic spread of H5N1 in 2003–2006 (with as-of-yet limited human infection), was anticipated, but not prevented, by prospective surveillance and monitoring of influenza in poultry markets following the initial emergence of H5N1 in China in 1996–1997.42 Monitoring and characterization of AI viruses thus has the potential to inform public health, and veterinary officials of the presence and/ or emergence of novel and potentially pathogenic strains.
A predictive approach to understanding and controlling the spread of influenza viruses and the emergence of novel strains would require a global monitoring program to complement human surveillance targeted at the three groups of animals thought to be important in influenza dynamics: wild birds, poultry, and swine. In order to maximize the informational content gained from monitoring, the sampling scheme should aim to estimate the intensity of viral transmission in a standardized and comparable way. This means that the age and species of wild birds, the farm type (poultry or swine at industrial or backyard scale), breed of poultry or swine, and livestock age must be consistent in repeated samples from the same locations (stratified by geography, and farm type for livestock) using identical methodology and diagnostics. In order to be cost-effective, this monitoring program would need to take advantage of testing already done at the local scale, and scale up the results through data integration at the national and international levels. An independent program at the national or global scale would be logistically and financially prohibitive, but in order to provide adequate information, the data integration must occur at the global level.
This contrasts sharply with current schemes that are highly opportunistic, unstandardized, and poorly integrated. In addition, viruses from surveillance need to be characterized in terms of infectivity to and virulence in wild birds, poultry, swine, and humans. The techniques for high-throughput processing of AI virus samples are currently being developed,43 and should facilitate improved monitoring programs needed to address this important threat to human and livestock health and wildlife. A greater challenge is coordinating and implementing a global standardized surveillance plan for wild birds, and integrating the results from all three animal monitoring programs.
Examples of vector-borne pathogens
Ross river virus
Epidemiologically RRV is Australia’s most significant (causing the most human illness) mosquito-borne pathogen, resulting in approximately 5000 human notifications of polyarthritis, the clinical manifestation of RRV infection, annually.44 This arbo-virus naturally cycles between mosquito vectors and mammalian reservoir hosts. Human cases often occur in or near areas with abundant vector and host populations, where epizootics likely lead to spillover and epidemics, e.g. ref. 11. There are three principle vectors of RRV across Australia (Culex annulirostris, Aedes camptorhynchus and Aedes vigilax), but other species can also be involved.44,45 Culex annulirostris breeds in freshwater and contributes to transmission across the continent, Ae. camptorhynchus is halotolerant and is a dominant vector in southern Australia, and Ae. vigilax is also halotolerant replacing Ae. camptorhynchus north of its distribution.44,45 Across the continent macropod marsupials are generally considered the principle reservoir hosts, although other marsupials also have a role in transmission.44–48
Much research has been conducted linking prevailing environmental conditions, such as temperature and precipitation, to vector mosquitoes and human RRV infections in frequent epidemic zones across all Australian states and territories, e.g. ref. 49, 50–53. In temperate southern Australia RRV activity exhibits spring-summer seasonality, associated with seasonal climatic conditions and vector populations.11,54–58 In tropical northern Australia seasonality of RRV is less pronounced, reflecting more stable environmental conditions for breeding of vectors.44,45,59,60
Overall, research on reservoir hosts has been significantly underrepresented in efforts to understand the ecology and epidemiology of RRV in Australia, and advances in this area will likely have rewarding outcomes.61 Host population dynamics, and consequent changing herd immunity, likely contribute to instigation of epizootics and consequent spillover to humans and epidemic activity. This is evidenced by epidemics of RRV in southwest Western Australia which exhibit four year cycles. In the mid-late 1990s RRV epidemics in this region were thought to be due to high summer rainfall, but this was called into question when later epidemics occurred without this high summer rainfall.11,55,62 Subsequent monitoring of antibodies to RRV among western gray kangaroos (Macropus fuliginosus, the probable RRV reservoir in this region) found 25% were antibody positive to RRV prior to the 2003–2004 epidemic, but this rose to 75% by the time the epidemic was two thirds to completion,55 (C. J. Gordon, unpublished data). This important antibody survey indicates epizootic activity preceding, and possibly instigating, epidemic activity.
Reproduction of kangaroos in southern Australia is seasonal and emergence of joeys from the pouch, which constitutes an influx of susceptible hosts into the population, coincides with the timing of RRV epidemics.11,54–58,63–66 Epidemics of RRV may occur when a sufficient number of susceptible hosts have been recruited into the population, and vector populations are adequate to support an epizootic. Thus, prediction of interannual epidemics of RRV, in southern Australia in particular, may be significantly improved by establishment of monitoring programs which follow population dynamics and herd immunity of macropods. Despite fundamental involvement of marsupials in the transmission of RRV see reviews by44,45,61 and references therein and many marsupial mammalogists across the continent, to date there has been little incorporation of their expertise into existing surveillance strategies across Australia. Efforts to integrate survey and trapping expertise from mammalogists in areas in and around RRV epidemic zones would be a valuable addition (albeit one that would require additional investment in surveillance) to the high quality surveillance of vectors and human incidence that already exists. In particular, basic information on the population dynamics of marsupials in and around RRV epidemic zones will enable the development of more precise epidemic forecast models.
West Nile virus
WNV is a single stranded RNA flavivirus transmitted on five continents primarily by Culex mosquitoes, and is amplified by competent avian hosts.67–69 It occasionally spills over to humans and other mammals, often from an enzootic vector that also feeds on mammals.70,71 Recent work has quantified the relationships in the laboratory between temperature, simulated rainfall, and several aspects of mosquito and virus ecology, including vector competence, and survival of larval mosquitoes.72–74
The introduction of WNV into the Western Hemisphere in 1999 was followed by rapid spread and enormous variability in the intensity of local transmission on several spatial scales and between years.75 However, the cause of year to year variation in transmission is unknown, despite its importance for public health and the allocation of limited control resources. Predicting the intensity of local and regional transmission would require uncovering the links between transmission and climate, vectors, and host populations far enough in advance to allow for control efforts to decrease the epidemic potential of the virus.
Following the introduction of WNV into New York in 1999, the Centers for Disease Control and Prevention partnered with local health departments to track the weekly incidence of WNV in mosquitoes, dead birds, humans, and horses through a surveillance data integration system called ArboNET. The strengths of this system include a broad geographic scope (over half of counties in the lower 48 states in 2003–2005, but decreasing recently), and information on both enzootic (bird to bird) transmission through WNV-infected dead birds and mosquitoes, and transmission to humans and horses. Key pieces of data that have not been successfully incorporated into this monitoring system include reliable data on the number of specimens collected and tested (for humans, horses, mosquitoes or birds), surveillance effort (e.g. number of mosquito traps and trapnight run), or the abundance of mosquitoes, which is required to estimate entomological risk or the force of infection. This is especially unfortunate, because two studies have linked the abundance or incidence of WNV-infected mosquitoes with the number of human cases.76,77
Although surveillance for WNV has surpassed most other pathogens in terms of both geographic scope, and taxonomic richness (birds, mosquitoes, humans, and horses), monitoring efforts by state and local health departments have decreased enormously in the past few years and promise to fall much further because funds for surveillance have been drastically reduced. The most valuable use of scarce resources at present would be a geographically stratified standardized monitoring network that captures the variability in transmission ecologies across North America, including different vectors, hosts, and climate regimes. The most useful tool in this system would be a set of standardized mosquito traps, such as Centers for Disease Control and Prevention light traps baited with CO2, run weekly at the same locations throughout each transmission season, and WNV testing of the most important mosquito species from these traps.70 These data, in addition to human samples collected through passive surveillance, should be integrated into a national network like ArboNET while also capturing the additional data outlined above (surveillance effort, mosquito abundance by species (mosquitoes per trap night), and numbers of samples tested (humans and mosquitoes)). This would enable estimation of the entomological risk or force of infection for humans, which can then be linked to the wealth of real time climate data currently being collected. When refined by our ever-growing understanding of the climatic and other drivers of year to year variation in WNV transmission this monitoring system should enable unprecedented opportunities and efficiency for targeted control.78,79
Lyme disease and other tick-borne diseases
Ticks in the Ixodes ricinus group, including I. scapularis in most of North America, transmit a series of zoonotic pathogens that cause serious illness. These illnesses include, most prominently, LD but also human babesiosis and anaplasmosis. Risk of human exposure is a function of the abundance of infected ticks in an environment and human behavior that influences contacts with ticks. Owing to the sensitivity of these ticks to temperature extremes and low humidity when held in the laboratory, reviewed by80 researchers interested in monitoring temporal changes in tick abundance and distribution have focused on climatic variables. Several short-term studies have found support for season-specific temperature or precipitation variables as predictors of I. scapularis abundance or LD risk.81–83 Others have identified specific bioclimatic variables that appear to be important in affecting habitat occupancy by ticks.77,84 Unfortunately, the results of these studies are inconsistent and often contradictory, and no consensus has arisen concerning the best climatic variables for monitoring disease risk.85,86
One of the key limitations to our ability to specify climatic indicators of tick-borne disease risk is the sheer number of possible candidate variables combined with data sets that span only several to a dozen years.85 A critical research need, therefore, is to maintain tick population monitoring programs for sufficient periods that the fit of different climatic models to the data can be compared. Ideally, climatic variables initially selected as candidates for monitoring could be identified with field experiments to test specific mechanisms by which those variables affect tick demography. These experiments would need to be spatially distributed along gradients of temperature and precipitation regimes, that is over hundreds to thousands of kilometers, in order to be maximally useful in predicting spatial spread and interannual, site-specific variation in LD risk.
Other efforts to predict temporal changes in risk or incidence of tick-borne diseases have included monitoring of specific host species. Ixodes scapularis and its close relatives are host-generalists that feed from a wide variety of different mammals, birds, and sometimes reptiles. Nevertheless, some host species are particularly permissive of tick feeding, promoting tick survival, whereas others groom off and kill many ticks.87 The most permissive hosts, such as white-footed mice (Peromyscus leucopus) also tend to be the most efficient reservoirs for the LD bacterium and other pathogens.87,88 Consequently, population densities of white-footed mice can be excellent leading indicators of the abundance of infected ticks and hence disease risk. In turn, mouse abundance in some habitats is highly predictable from acorn production by oaks (Quercus).85,89,90 Schauber et al.91 demonstrated that human incidence rates of LD (specifically, deviations from long-term trends) in the northeastern United States can be predicted almost two years in advance from data on local acorn production.
Monitoring mouse population abundance can be accomplished with moderate effort and is likely to provide useful data on interannual variation in risk of LD, babesiosis, and possibly anaplasmosis cases. A challenge to monitoring mouse abundance for predicting disease risk is the need for incorporating spatial variation in mouse population dynamics, which can be extreme. In contrast, monitoring of acorn production can be accomplished at many locations with minimal effort at each one.89 However, ticks and the pathogens they transmit are not restricted to oak-dominated forests, so the results of such monitoring efforts will not be spatially comprehensive.91 Monitoring of the tick populations themselves, or preferably abundances of ticks that are infected with zoonotic pathogens, would seem likely to provide epidemiologically highly relevant information, although few if any efforts to maintain long-term monitoring exist.
A limitation to such vector monitoring is that data would represent contemporaneous risk rather than acting as a leading indicator and would therefore not be conducive to preventative measures. Because public education about risk is crucial in the avoidance and prevention of tick-borne diseases, monitoring of leading indicators is more likely to serve the public health. Avoidance is particularly important given the lack of available vaccines for tick-borne zoonoses and given problems with diagnosis and treatment. Monitoring programs that capitalize on these strong ecological connections might provide early warning systems for preventing tick-borne zoonoses.
Discussion and conclusions
Here, we have reviewed an array of directly transmitted and vector-borne zoonoses, briefly highlighting their natural histories, demands for monitoring and means by which these demands could be addressed. While each zoonotic system is unique in its characteristics and specific monitoring demands, some commonalities can be drawn among these examples. For all zoonoses reviewed here, understanding of their dynamics and prediction of human incidence would be improved by implementation of additional identified monitoring (Table 1). For SNV, AI, RRV and LD extension of existing or implementation of new host monitoring programs would yield valuable insights. For WNV and LD implementation of monitoring of vector abundance and identification of environmental variables that consistently predict their abundance are needed. Additionally, understanding the pathogen dynamics and prediction of human incidence would be improved for AI, RRV and WNV by more coordination among public health and research groups with variable expertise and data.
Owing to the importance of monitoring in understanding and predicting spillover of zoonoses to humans, critical evaluations of environmental monitoring are valuable to continually maximize management outcomes. This critical review aims to encourage discussion among researchers and public health officials, ultimately promoting improvements in our understanding of zoonoses, their management, and the greater health of the public.
Environmental impact.
We will emphasize how specific examples of existing or newly created environmental monitoring could be integrated with disease surveillance to enhance understanding of the ecology and epidemiology of infectious diseases and solve public health problems. Specific examples in this review will include directly transmitted (Sin Nombre virus, Avian Influenza) and vector-borne (Ross River virus, West Nile virus, Lyme Disease, Anaplasmosis and Babesiosis) zoonoses. A common theme among these examples is the need for improved integration between researchers and governmental organizations, and better knowledge and access to data spanning multiple disciplines. The ideas brought forth in this review from an array of zoonotic pathogens across the globe, will make this review a significant contribution to scholarship, with clear and controversial value.
Acknowledgments
The authors would like to thank two anonyms reviewers for their valuable comments, which helped improve this paper. Financial support was provided by NIH grant P20 RR16455-06-07,08 from the INBRE-BRIN program of the National Center for Research Resources, NSF grants DEB 0444585, EF-0813035 and EF-0914866, and the U.S. Centers for Disease Control and Prevention, Atlanta, GA, through cooperative agreement.
Footnotes
Published as part of a special issue dedicated to Emerging Investigators.
References
- 1.Weaver SC. Arch Virol Suppl. 2005:33. doi: 10.1007/3-211-29981-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Nature. 2008;451:990. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills JN. Arch Virol Suppl. 2005;19:45. doi: 10.1007/3-211-29981-5_5. [DOI] [PubMed] [Google Scholar]
- 4.Mas-Coma S, Valero MA, Bargues MD. Revue Scientifique Et Technique-Office International Des Epizooties. 2008;27:443. [PubMed] [Google Scholar]
- 5.Gratz NG. Annu Rev Entomol. 1999;44:51. doi: 10.1146/annurev.ento.44.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Daszak P, Cunningham AA, Hyatt AD. Acta Trop. 2001;78:103. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- 7.Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Ecol Lett. 2006;9:467. doi: 10.1111/j.1461-0248.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- 8.Patz JA, Daszak P, Tabor GM, Aguirre AA, Pearl M, Epstein J, Wolfe ND, Kilpatrick AM, Foufopoulos J, Molyneux D, Bradley DJ Working Grp Land Use Change. Environ Health Perspect. 2004;112:1092. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelthaler DM, Mosley DG, Cheek JE, Levy CE, Komatsu KK, Ettestad P, Davis T, Tanda DT, Miller L, Frampton JW, Porter R, Bryan RT. Emerging Infect Dis. 1999;5:87. doi: 10.3201/eid0501.990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjelle B, Glass GE. J Infect Dis. 2000;181:1569. doi: 10.1086/315467. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay M, Oliveira N, Jasinska E, Johansen C, Harrington S, Wright AE, Smith D. Emerging Infect Dis. 1996;2:117. doi: 10.3201/eid0202.960206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson AP, Hudson PJ. In: Ecology of Infectious Diseases in Natural Populations. Grenfell BT, Dobson A, editors. Cambridge University Press; Cambridge: 1995. p. 52. [Google Scholar]
- 13.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Science. 1993;262:914. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 14.CDC. Morbidity and Mortality Weekly Report. 1993;42:421. [PubMed] [Google Scholar]
- 15.Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, Gage KL, Rollin PE, Sarisky J, Enscore RE, Frey JK, Peters CJ, Nichol ST. J Infect Dis. 1994;169:1271. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 16.Parmenter RR, Brunt JW, Moore DI, Ernest MS. Sevilleta LTER Publication no. 41. Albuquerque: Sevilleta Long-Term Ecological Research Program, Department of Biology, University of New Mexico; 1993. Report to the Federal Centers for Disease Control and Prevention, Atlanta, Georgia; p. 1. [Google Scholar]
- 17.Mills JN, Ksiazek TG, Peters CJ, Childs JE. Emerging Infect Dis. 1999;5:135. doi: 10.3201/eid0501.990116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglass RJ, Calisher CH, Bradley KC. Vector-Borne Zoonotic Dis. 2005;5:189. doi: 10.1089/vbz.2005.5.189. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong LR, Zaki SR, Goldoft MJ, Todd RL, Khan AS, Khabbaz RF, Ksiazek TG, Peters CJ. J Infect Dis. 1995;172:1166. doi: 10.1093/infdis/172.4.1166. [DOI] [PubMed] [Google Scholar]
- 20.Zeitz PS, Butler JC, Cheek JE, Samuel MC, Childs JE, Shands LA, Turner RE, Voorhees RE, Sarisky J, Rollin PE, Ksiazek TG, Chapman L, Reef SE, Komatsu KK, Dalton C, Krebs JW, Maupin GO, Gage K, Sewell CM, Breiman RF, Peters CJ. J Infect Dis. 1995;171:864. doi: 10.1093/infdis/171.4.864. [DOI] [PubMed] [Google Scholar]
- 21.Calisher CH, Wagoner KD, Amman BR, Root JJ, Douglass RJ, Kuenzi AJ, Abbott KD, Parmenter C, Yates TL, Ksiazek TG, Beaty BJ, Mills JN. J Wildl Dis. 2007;43:1. doi: 10.7589/0090-3558-43.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Mills JN, Yates TL, Ksiazek TG, Peters CJ, Childs JE. Emerging Infect Dis. 1999;5:95. doi: 10.3201/eid0501.990111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglass RJ, VanHorn R, Coffin KW, Zanto SN. J Wildl Dis. 1996;32:527. doi: 10.7589/0090-3558-32.3.527. [DOI] [PubMed] [Google Scholar]
- 24.Luis AD, Douglass RJ, Mills JN, Bjørnstad ON. J Anim Ecol. 2010;79:462. doi: 10.1111/j.1365-2656.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 25.Carver S, Mills JN, Kuenzi A, Flieststra T, Douglass R. Vector-Borne Zoonot Dis. 2010;10:517. doi: 10.1089/vbz.2009.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglass RJ, Wilson T, Semmens WJ, Zanto SN, Bond CW, Van Horn RC, Mills JN. Am J Trop Med Hyg. 2001;65:33. doi: 10.4269/ajtmh.2001.65.33. [DOI] [PubMed] [Google Scholar]
- 27.Madhav NK, Wagoner KD, Douglass RJ, Mills JN. Vector-Borne Zoonotic Dis. 2007;7:353. doi: 10.1089/vbz.2006.0605. [DOI] [PubMed] [Google Scholar]
- 28.Clay CA, Lehmer EM, Jeor SS, Dearing MD. PLoS One. 2009;4:e6467. doi: 10.1371/journal.pone.0006467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuenzi AJ, Douglass RJ, Bond CW. Emerging Infect Dis. 2000;6:386. doi: 10.3201/eid0604.000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otteson EW, Riolo J, Rowe JE, Nichol ST, Ksiazek TG, Rollin PE, St Jeor SC. Am J Trop Med Hyg. 1996;54:127. doi: 10.4269/ajtmh.1996.54.127. [DOI] [PubMed] [Google Scholar]
- 31.Kuenzi AJ, Douglass RJ, White D, Bond CW, Mills JN. Am J Trop Med Hyg. 2001;64:137. doi: 10.4269/ajtmh.2001.64.137. [DOI] [PubMed] [Google Scholar]
- 32.Kuenzi AJ, Douglass RJ, Bond CW, Calisher CH, Mills JN. J Wildl Dis. 2005;41:473. doi: 10.7589/0090-3558-41.3.473. [DOI] [PubMed] [Google Scholar]
- 33.Douglass RJ, Semmens WJ, Matlock-Cooley SJ, Kuenzi AJ. J Wildl Dis. 2006;42:813. doi: 10.7589/0090-3558-42.4.813. [DOI] [PubMed] [Google Scholar]
- 34.Childs JE, Krebs JW, Ksiazek TG, Maupin GO, Gage KL, Rollin PE, Zeitz PS, Sarisky J, Enscore RE, Butler JC, Cheek JE, Glass GE, Peters CJ. Am J Trop Med Hyg. 1995;52:393. doi: 10.4269/ajtmh.1995.52.393. [DOI] [PubMed] [Google Scholar]
- 35.Shechmeister IL. In: Disinfection, Sterilization, and Preservation. Block SS, editor. Lea and Febiger; Philadelphia: 1991. p. 553. [Google Scholar]
- 36.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A. Nature. 2009;459:1122. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 37.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Microbiol Rev. 1992;56:152. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus A, Fouchier RAM. Science. 2006;312:384. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 39.Kawaoka Y, Chambers TM, Sladen WL, Webster RG. Virology. 1988;163:247. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 40.Vandegrift KJ, Sokolow SH, Daszak P, Kilpatrick AM. Ann N Y Acad Sci. 2010 doi: 10.1111/j.1749-6632.2010.05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Proc Natl Acad Sci U S A. 2006;103:19368. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan Y, Peiris JSM, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Proc Natl Acad Sci U S A. 2002;99:8950. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munster VJ, Baas C, Lexmond P, Bestebroer TM, Guldemeester J, Beyer WEP, de Wit E, Schutten M, Rimmelzwaan GF, Osterhaus A, Fouchier RAM. J Clin Microbiol. 2009;47:666. doi: 10.1128/JCM.01625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell RC. Annu Rev Entomol. 2002;47:1. doi: 10.1146/annurev.ento.47.091201.145100. [DOI] [PubMed] [Google Scholar]
- 45.Harley D, Sleigh A, Ritchie S. Clin Microbiol Rev. 2001;14:909. doi: 10.1128/CMR.14.4.909-932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyd AM, Hall RA, Gemmell RT, Kay BH. Am J Trop Med Hyg. 2001;65:777. doi: 10.4269/ajtmh.2001.65.777. [DOI] [PubMed] [Google Scholar]
- 47.Boyd AM, Kay BH. Arbovirus Res Aust. 2001;8:14. [Google Scholar]
- 48.Kay BH, Boyd AM, Ryan P, Hall RA. Am J Trop Med Hyg. 2007;76:417. [PubMed] [Google Scholar]
- 49.Tong S, Hu W, McMichael AJ. Trop Med Int Health. 2004;9:298. doi: 10.1046/j.1365-3156.2003.01175.x. [DOI] [PubMed] [Google Scholar]
- 50.Hu WB, Nicholls N, Lindsay M, Dale P, McMichael AJ, Mackenzie JS, Tong SL. Am J Trop Med Hyg. 2004;71:129. [PubMed] [Google Scholar]
- 51.Kelly-Hope LA, Purdie DM, Kay BH. J Med Entomol. 2004;41:1116. doi: 10.1603/0022-2585-41.6.1116. [DOI] [PubMed] [Google Scholar]
- 52.Woodruff R, Guest C, Garner G, Becker N, Lindsay MF. Epidemiology. 2003;14:S94. doi: 10.1097/01.ede.0000229467.92742.7b. [DOI] [PubMed] [Google Scholar]
- 53.Woodruff RE, Guest CS, Gainer MG, Becker N, Lindsay M. Epidemiology. 2006;17:569. doi: 10.1097/01.ede.0000229467.92742.7b. [DOI] [PubMed] [Google Scholar]
- 54.Gard GP, Marshall ID, Woodroofe GM. Am J Trop Med Hyg. 1973;22:551. doi: 10.4269/ajtmh.1973.22.551. [DOI] [PubMed] [Google Scholar]
- 55.Lindsay MDA, Breeze AL, Harrington SA, Johansen CA, Broom AK, Gordon CJ, Maley FM, Power SL, Jardine A, Smith DW. Arbovirus Res Aust. 2005;9:194. [Google Scholar]
- 56.McManus TJ, Marshall ID. Arbovirus Res Aust. 1986;4:127. [Google Scholar]
- 57.Mudge PR, Lim RS, Moore B, Radford AF. Med J Aust. 1980;2:626. doi: 10.5694/j.1326-5377.1980.tb77070.x. [DOI] [PubMed] [Google Scholar]
- 58.Russell RC, Cloonan MJ, Wells PJ, Vale TG. J Med Entomol. 1991;28:796. doi: 10.1093/jmedent/28.6.796. [DOI] [PubMed] [Google Scholar]
- 59.Aaskov JG, Ross PV, Davies CE, Innis MD, Guard RW, Stallman ND, Tucker M. Med J Aust. 1981;2:17. [PubMed] [Google Scholar]
- 60.Russell RC. J Vector Ecol. 1998;23:1. [PubMed] [Google Scholar]
- 61.Carver S, Bestall A, Jardine A, Ostfeld RS. Vector-Borne Zoonotic Dis. 2009;9:51. doi: 10.1089/vbz.2008.0040. [DOI] [PubMed] [Google Scholar]
- 62.Johansen CA, Broom AK, Lindsay MDA, Maley FM, Power SL, Gordon CJ, Smith DW. Arbovirus Res Aust. 2005;9:159. [Google Scholar]
- 63.Arnold GW, Grassia A, Steven DE, Weeldenburg JR. Wildl Res. 1991;18:561. [Google Scholar]
- 64.Caughley G, Shepherd N, Short J. Kangaroos: their ecology and management in the sheep rangelands of Australia. Cambridge University Press; Cambridge: 1987. [Google Scholar]
- 65.Menkhorst P, Knight F. A field guide to the mammals of Australia. Oxford University Press; Melbourne, Australia: 2001. [Google Scholar]
- 66.Strahan R. The Australian Museum complete book of Australian mammals. Angus & Robertson; London: 1988. [Google Scholar]
- 67.Hubalek Z, Halouzka J. Emerging Infect Dis. 1999;5:643. doi: 10.3201/eid0505.990505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kilpatrick AM, LaDeau SL, Marra PP. Auk. 2007;124:1121. [Google Scholar]
- 69.Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Proc R Soc London, Ser B. 2006;273:2327. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kilpatrick AM, Kramer LD, Campbell S, Alleyne EO, Dobson AP, Daszak P. Emerging Infect Dis. 2005;11:425. doi: 10.3201/eid1103.040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamer GL, Kitron UD, Brawn JD, Loss SR, Ruiz MO, Goldberg TL, Walker ED. J Med Entomol. 2008;45:125. doi: 10.1603/0022-2585(2008)45[125:cpdcab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 72.Kilpatrick AM, Meola MA, Moudy RM, Kramer LD. PLoS Pathog. 2008;4:e1000092. doi: 10.1371/journal.ppat.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reisen WK, Fang Y, Martinez VM. J Med Entomol. 2006;43:309. doi: 10.1603/0022-2585(2006)043[0309:EOTOTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 74.Koenraadt CJM, Harrington LC. J Med Entomol. 2008;45:28. doi: 10.1603/0022-2585(2008)45[28:feoroc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 75.Petersen LR, Hayes EB. N Engl J Med. 2004;351:2257. doi: 10.1056/NEJMp048261. [DOI] [PubMed] [Google Scholar]
- 76.Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P. PLoS Biol. 2006;4:e82. doi: 10.1371/journal.pbio.0040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brownstein JS, Holford TR, Fish D. Emerging Infect Dis. 2004;10:1129. doi: 10.3201/eid1006.030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soverow JE, Wellenius GA, Fisman DN, Mittleman MA. Environ Health Perspect. 2009;117:1049. doi: 10.1289/ehp.0800487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeGaetano AT. Int J Biometeorol. 2005;49:345. doi: 10.1007/s00484-004-0242-2. [DOI] [PubMed] [Google Scholar]
- 80.Killilea M, Swei A, Lane R, Briggs C, Ostfeld R. EcoHealth. 2008;5:167. doi: 10.1007/s10393-008-0171-3. [DOI] [PubMed] [Google Scholar]
- 81.Jones CJ, Kitron UD. J Med Entomol. 2000;37:408. doi: 10.1603/0022-2585(2000)037[0408:POISAI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 82.Subak S. Am J Epidemiol. 2003;157:531. doi: 10.1093/aje/kwg014. [DOI] [PubMed] [Google Scholar]
- 83.Ogden NH, Bigras-Poulin M, O’Callaghan CJ, Barker IK, Lindsay LR, Maarouf A, Smoyer-Tomic KE, Waltner-Toews D, Charron D. Int J Parasitol. 2005;35:375. doi: 10.1016/j.ijpara.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 84.Estrada-Pena A. Environ Health Perspect. 2002;110:635. doi: 10.1289/ehp.110-1240908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ostfeld RS, Canham CD, Oggenfuss K, Winchcombe RJ, Keesing F. PLoS Biol. 2006;4:e145. doi: 10.1371/journal.pbio.0040145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ostfeld RS. Lyme disease: the ecology of a complex system. Oxford University Press; 2011. in press. [Google Scholar]
- 87.Keesing F, Brunner J, Duerr S, Killilea M, LoGiudice K, Schmidt K, Vuong H, Ostfeld RS. Proc R Soc London, Ser B. 2009;276:3911. doi: 10.1098/rspb.2009.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. Proc Natl Acad Sci U S A. 2003;100:567. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ostfeld RS, Jones CG, Wolff JO. BioScience. 1996;46:323. [Google Scholar]
- 90.Elkinton JS, Healy WM, Buonaccorsi JP, Boettner GH, Hazzard AM, Smith HR, Liebhold AM. Ecology. 1996;77:2332. [Google Scholar]
- 91.Schauber EM, Ostfeld RS, Evans AS. Ecol Appl. 2005;15:575. [Google Scholar]


