Abstract
Toxoplasma gondii is among the most successful parasites, with nearly half of the human population chronically infected. Recently a link between the T. gondii Hsp90 chaperone machinery and parasite development was observed. Here, the T. gondii Hsp90 co-chaperones p23 and Hip were identified mining the Toxoplasma- database (www.toxodb.org). Their identity was confirmed by domain structure and blast analysis. Additionally, analysis of the secondary structure and studies on the chaperone function of the purified protein verified the p23 identity. Studies of co-immunoprecipitation (co-IP) identified two different types of complexes, one comprising at least Hip-Hsp70-Hsp90 and another containing at least p23-Hsp90. Indirect immunofluorescence assays showed that Hip is localized in the cytoplasm in tachyzoites and as well in bradyzoites. For p23 in contrast, a solely cytoplasmic localization was only observed in the tachyzoite stage whereas nuclear and cytosolic distribution and colocalization with Hsp90 was observed in bradyzoites. These results indicate that the T. gondii Hsp90-heterocomplex cycle is similar to the one proposed for higher eukaryotes, further highlighting the implication of the Hsp90/p23 in parasite development. Furthermore, co-IP experiments of tachyzoite/bradyzoite lysates with anti-p23 antiserum and identification of the complexed proteins together with the use of the curated interaction data available from different source (orthologs and Plasmodium databases) allowed us to construct an interaction network (interactome) covering the dynamics of the Hsp90 chaperone machinery.
Keywords: Toxoplasma gondii, development, Hsp90 heterocomplex, p23, protein prediction interaction, bioinformatic
1. Introduction
Toxoplasma gondii is a ubiquitous, obligate, intracellular protozoan parasite that infects a large number of warm-blooded animals. In humans, T. gondii follows an asexual replication cycle, characterized by two stages: rapidly growing ‘tachyzoites’ and latent ‘bradyzoite’ tissue cysts. Tachyzoites are responsible for acute disease and congenital neurological birth defects, while the slowly dividing bradyzoite form may remain latent within the tissues for many years, representing a potential threat to immune-compromised patients. Both developmental stages are essential for disease and parasite propagation.
Stress has been shown to induce bradyzoite formation and heat shock proteins (Hsp) are likely to play an important role during stage conversion [1]. The expression of the heat shock proteins Hsp60, 70 and 90 is increased during conversion from tachyzoites to bradyzoites [1,2]. In this context, Radke et al., [3] performed serial analysis of gene expression (SAGE) to define the T. gondii transcriptome of the intermediate-host life cycle that leads to the formation of the bradyzoite/tissue cyst. In their study, an increase in Hsp90 mRNA occurs within the first 24 h of bradyzoite development, suggesting that Hsp90 mRNA may be an early bradyzoite marker. In this context, we recently showed that subcellular localization of the T. gondii Hsp90 is also developmentally regulated [2]. Furthermore, geldanamycin, a benzoquinone ansamycin antibiotic capable of binding and disrupting the function of Hsp90, blocked the conversion from tachyzoite to bradyzoite and vice versa, suggesting an important role of this protein in the regulation of stage inter-conversion [2]. Due to lack of drugs capable of eliminating tissue cysts, up to now there is no effective treatment for chronic toxoplasmosis. Thus, the T. gondii Hsp90 emerges as an interesting target for drug development also because of its appearing pleiotropic role, including invasion and replication [2,4].
Hsp90 does not act as a regular chaperone in the folding of non-native proteins. Instead, it binds to substrate proteins (client proteins) that are in a near-native state, at an advanced stage of folding [5]. In addition to protein folding activity, Hsp90 has an alternative function associated with the assembly of multi-protein complexes and their turnover. In the cell, Hsp90 is chaperoning more than 100 ‘client proteins’, most of them involved in signal transduction, regulation of the cell cycle or regulation of transcription and thereby influencing development and evolution [6]. In higher eukaryotes, Hsp90 is regulated by further proteins, so called co-chaperones, which participate in dynamic multi-chaperone complexes [6,7]. Co-chaperones can regulate the ATP-hydrolysis of Hsp90, influence its affinity for client proteins [8], target it to its client protein [9,10] or to a particular subcellular compartment [9–11]. In studies based on glucocorticoid receptor (GR) maturation co-chaperones were determined to be involved in achieving efficient Hsp90-heterocomplex assembly: Hsp90, Hsp70, Hsp organizing protein (Hop), p23, an Hsp90-binding co-chaperone and Hsp40 [6]. Another co-chaperone, the Hsp70 interacting protein (Hip), has also been purified by co-immunoprecipitation (co-IP) [12]. Mechanistically, Hip was detected in early Hsp90-heterocomplex (formed by Hsp40-Hip-Hsp70-client protein-Hop-Hsp90). By contrast, p23 enters at late stage of the cycle, leading to complete inhibition of the ATPase activity and increasing the apparent affinity of Hsp90 for ATP [8,13–15].
Despite the observation that the Hsp70/Hsp90 cycle may be involved in apicomplexan parasites propagation, only Plasmodium Hip and p23 co-chaperones have been identified and preliminary characterized so far [16,17]. Here, we set out to elucidate the role of Hsp90-heterocomplex during T. gondii differentiation. We studied Hip and p23 interactions in T. gondii and assessed subcellular localization of Hip and p23 during tachyzoite-bradyzoite conversion. Additionally, basic structural and functional characteristics of p23 were determined to further confirm the identity of this Hsp90 co-chaperone. Finally, putative interactors of p23 and Hsp90 during tachyzoite and bradyzoite stages were identified by mass spectrometry analysis following co-IP.
2. Materials and Methods
2.1 In silico analysis
In order to identify proteins of T. gondii Hsp70/Hsp90 machinery we searched Toxodb (www.toxodb.org) for Hsp90 and its putative binding proteins on the basis of the respective domains: Hsp90, DNAJ, TPR, P23 and Aha. To identify Hip and p23 proteins, the Plasmodium berghei Hip, p23, Rattus norvegicus Hip and human p23 (AN: Q08168, XP_680236, P50503 and L24804, respectively) sequences were used to blast (www.toxodb.org blastp). Several amino acids and their respective DNA were retrieved from ToxoDB web page. Identity was further analyzed by Blast programs (www.ncbi.nlm.nih.gov.ar). β-sheet was deduced by using the http://protevo.eb.tuebingen.mpg.de/toolkit. Mitochondria and chloroplast transit peptides were analzyed by Predotar software at http://urgi.versailles.inra.fr/predotar/predotar.html)
2.2 cDNA cloning, sequencing and sequence analysis
Putative Hip and p23 open reading frames (ORF) were amplified from cDNA. Total RNA was extracted using TRIzol®Reagent (InvitrogenTM Life Technologies) according to the manufacturer’s instructions. Two micrograms of total RNA from tachyzoites was reverse transcribed to cDNA with 200 U of M-MLV reverse transcriptase (Invitrogen Life Technologies) and oligo(dT)12–18- For PCR the following primers were used: Hip: sense 5’-GGATCCATGGCGGCGCTCAGCCCTCA and antisense 5’-GTCGACTTACATAGGAGAGCCAGTTG; p23: sense 5’-CTGCAGATGGCGGACGCACAAGTTCA and antisense 5’-GTCGACTCAAGCCTGCGCAGCTTCTG. BamHI and KpnI sites were included in the sense and antisense primers respectively (underlined sequence). These fragments were cloned in pGEM T easy vector (Promega). The 5’Untranslated region (UTR) of both p23 and hip and 3’UTR of p23 mRNA were sequenced by reverse transcriptase-PCR using new primers: for p23 (sense (5’-CCGTCGCGACTTTTTTTGCGA) and an antisense (5’- AACACGACGCCCGTCAGCTCC). For hip 5’UTR sequence a new sense primer was designed (5’- GGACGTGTTTAATCCATTCT).
2.3 Expression and Purification of recombinant T. gondii Hsp70/Hsp90 co-chaperones
The open reading frames of T. gondii hip and p23 genes were sub-cloned into the prokaryotic expression vector pRSET A (Invitrogen Life Technologies) downstream and in frame with a sequence that encodes an N-terminal 6xHis-tag. BamHI and KpnI restriction sites were engineered into the 5´ and 3´primers respectively. Escherichia coli BL21 (DE3) were freshly transformed with the expression plasmids. Cultures were grown to OD600 = 0.6 before protein expression induction by the addition of isopropyl thio-β-D-galactoside (2 mM). After overnight incubation at 30°C, the cells were harvested and purified in soluble form using a commercial HisTrap 5ml column (GE Healthcare) according to the manufacturer’s instructions.
2.4 Circular dichroism spectroscopy
The content of structural elements was determined by Far-UV-CD spectroscopy, using a Jasco J-715 spectropolarimeter (Jasco, Groβumstadt, Germany). The measurements were carried out in quartz cuvettes with 0.02 cm pathlength at a protein concentration of 0.3 mg/ml. Far UV-spectra were recorded in 50 mM Tris/HCl, 2 mM EDTA, pH 8.0 at 20°C; 16 spectra were accumulated and all spectra were buffer-corrected. Structural elements were calculated using the CD spectra deconvolution software CDNN [18 R. and Jaenicke, R. (1992) Protein Eng. 5, 191–195)].
2.5 Chaperone activity
Chaperone activity of T. gondii p23 was assayed as described for human p23 [19]. To examine the influence of T. gondii p23 on the thermal aggregation of citrate synthase (CS), a solution of 30 µM CS (monomer) was diluted 1:30 in 40 mM HEPES/KOH pH 7.5, equilibrated at 44°C, in the presence and in the absence of p23. Aggregation kinetics was measured in a Cary 50 spectrophotometer (Varian, Darmstadt, Germany) following the change in light scattering at 360 nm.
To determine the influences of p23 on the thermal inactivation of CS, CS was incubated in the absence or presence of T. gondii p23 in 40 mM Hepes/KOH, 0.5 mM DTT, pH 7.5 at 45°C. CS activity was assayed taking aliquots at time points indicated in 50 mM Tris/HCl, 2 mM EDTA, pH 8.0 at 25°C as described previously [19,20]
2.6 Production of antibodies
C3H mice were immunized with 10 µg of Hip and p23 recombinant proteins in complete Freund’s adjuvant (Sigma-Aldrich™ de Argentina S.A) and boostered every two weeks with three successive injections with identical doses of the respective recombinant protein in incomplete Freund’s adjuvant (Sigma-Aldrich™ de Argentina S.A.).
2.7 Parasite in vitro differentiation and manipulation
Parasite differentiation and co-localization analysis were further described elsewhere [2]. Briefly, tachyzoites of avirulent PK strain were grown in vitro in human foreskin fibroblasts (HFF) and induced to differentiate into bradyzoites for 4 days in low CO2 concentration (0.03%) and pH 8.1. Bradyzoite induction was assessed and followed by murine mAb αp34 T82C2 (specific marker of bradyzoite P34 surface antigen) a gift from J. F. Dubremetz, Université de Montpellier II, France or by rabbit polyclonal anti-BAG1 (specific marker of cytoplasmic BAG1 small heat shock protein) obtained in our laboratory [21]. Antibodies specific to the tachyzoite surface protein SAG1 (murine mAbαp30 T4IE5) a gift from J. F. Dubremetz, (Université de Montpellier II, France), to the cytoplasmic/nuclear Hsp90 (rabbit polyclonal anti-Hsp90, C-terminal region) [2,22]. Following incubation with primary antibodies, cells were washed three times with PBS, and then incubated with the corresponding secondary antibodies (1:4000). Alexa fluor goat anti-rabbit 350 or Alexa fluor goat anti-mouse 488 (Invitrogen Argentina Ltda.). Cover slips were washed three times and mounted in Fluoromont G (Southern Biotechnology Associates) and viewed using a Zeiss Axiovert 35 inverted microscope equipped with a 100 W Hg-vapor lamp and epifluorescence filter sets or a Nikon Model Eclipse E600 (magnification 100X, numerical aperture 1,40 at 24°C). Propidium iodide (PI, Sigma-Aldrich™ Argentina S.A) used to stain the nucleus. Fluorescences were recorded separately, given a colour (red, green or blue) and the images were analyzed by Image-Pro Plus version 5.1.0.20 and merged using Adobe Photoshop.
2.8 Immunoblot and co-immunoprecipitation (co-IP) analysis
In order to carry out immunoblot studies in both tachyzoites and induced bradyzoites, medium was removed and the cells were washed once with PBS, the monolayer was scraped and passed five times through a 27-gauge needle, bradyzoites were also passed once through a 30-gauge needle to release parasites from the host cells. The parasites were then centrifuged at 1,800 rpm for 10 min at room temperature and resuspended in sterile PBS and counted in a Neubauer hemocytometer chamber. Both stages of parasites were purified from the host cell material by passage through a 3 mm-pore size filter (Amersham Hybond, GE Healthcare Argentina S.A.).
In order to perform Western blot, proteins from parasites and human foreskin fibroblasts (HFF) were extracted in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, electrophoresed and transferred on nitrocellulose. Non-specific binding sites were blocked with 5% non-fat-dried milk in PBS containing 0.05% Tween-20 (PBS-T) and the membranes were then incubated (1 h, room temperature) with polyclonal antibodies diluted 1:500. The membranes were washed with PBS-T prior to incubation with alkaline phosphatase–conjugated anti-rabbit or anti-mouse secondary antibodies, diluted 1:5,000 (Santa Cruz Biotechnology). Immunoreactive protein bands were visualized by NBT-BCIP (Sigma-Aldrich™ Argentina S.A).
To perform co-IP studies, tachyzoites and in vitro induced bradyzoites of T. gondii PK (Me49) were incubated 2 h at 4°C in lyses buffer (0.1% [v/v] NP40, 10 mM Tris-HCl pH 7.4, 50 mM NaCl, 4 mM EDTA, sodium molybdate 20mM, Sigma inhibitors cocktail [1 mM PMSF, 104 mM 4-{2-aminoethyl}benzenesulfonyl fluoride, 80 mM aprotinin, 2.1 mM leupeptin, 3.6 mM bestatin, 1.5 mM pepstatin A, and 1.4 mM E-64]) and pelleted 20 min at 12,000 g at 4°C. In parallel, the antibody (1:500 to 1,000) was incubated with protein A/G-Sepharose (Sigma-Aldrich™ de Argentina S.A) in lysis buffer for 2 h at 4 °C. Supernatants of parasite lysates were then incubated in lysis buffer plus antibody (1:500 to 1,000) and protein A/G-Sepharose for 2 h at 4°C in gentle rocking. After the resin was washed four times with lysis buffer and 2 times with wash buffer (0.5% [v/v] NP40, 50 mM Tris-HCl pH=8,3, NaCl 1M), the antigen was eluted by boiling in sample buffer and separated by SDS-PAGE.
2.9 Mass spectrometry analysis
Tachyzoite and bradyzoite proteins were pulled down with the anti-p23 and -Hsp90 antibodies, separated on a 10% polyacrylamide gel and stained with Coomassie Blue R-250 (Fig. S1). Negative controls with pre-immune sera did not result in any visible protein bands. Bands of interest were excised (Fig. S1), and de-stained with three progressive washes of 25 mM NH4HCO3 in 50% acetonitrile , and then followed by reduction with 200 µl of 10 mM dithiothreitol (DTT) in 100 mM NH4HCO3 for 1 h at 56 °C and alkylation with 55 mM iodoacetamide in 200 µl of 100 mM NH4HCO3 in the dark for 30 min. The hydrated gel slice was covered with 30 µl of trypsin solution (10 ng/µl) and incubated overnight at 37°C. A small amount of 10% acetic acid was then added to the tube to stop the digestion. The sample was then centrifuged at 3,000 r.p.m., and the supernatant was saved for analysis by electrospray ionization (ESI) liquid chromatography-mass spectrometry (LC-MS). A fused-silica microcapillary LC column (12 cm x 75 µm id) packed with C18 reversed phase resin (Magic C18AQ, 5-µm particle size, 20-nm pore size, Michrom Bioresourses, Auburn, CA) was used with nanospray ESI. The nanospray-ESI was fitted onto a linear quadrupole ion trap (LTQ) mass spectrometer (Thermo Electron, San Jose, CA) that was operated in a collisional-induced dissociation mode to obtain both MS and MS/MS spectra. Samples of tryptic peptides were loaded onto the microcapillary column and separated by applying a gradient from 5–80% acetonitrile in 0.5% acetic acid at a flow rate of 250 nl/min for 55 min. Mass spectrometry data were acquired in data-dependent acquisition mode, in which a full MS scan was followed by 10 MS/MS spectra of the most abundant ions.
After an LC-MS run was completed and spectra obtained, the spectra were searched against the ToxoDB 4.1 protein database (www.toxodb.org) using SEQUEST (Bioworks software package, version 3.3, Thermo Electron,). The search parameters permitted ± 1.0 Da peptide MS tolerance, and ± 1.0 Da MS/MS tolerance. Oxidation of methionine and carboxymethylation of cysteines were allowed as variable modifications. Up to two missed tryptic cleavages of peptides were considered. The cutoff for SEQUEST assignment was a cross-correlation score (Xcorr) greater than 1.9, 2.5, and 3.8 for peptide charge states of 1, 2, and 3, respectively; and a delta-correlation score (ΔCn) >0.1.
2.10 Interactome map
After collecting data from proteomics experiment, a network of Hsp90 and p23 interactors was constructed using network visualization and the modeling tool of Cytoscape (http://www.cytoscape.org) [23]. A putative name was assigned to each interactor according to ToxoDB 4.1 and Uniprot databases (http://beta.uniprot.org/). Subcellular localization information of proteins was obtained from ToxoDB 4.1, or predicted according to gene ontology data of orthologs of the putative Toxoplasma proteins (obtained from Uniprot and ToxoDB). With this information and using Cerebral (a Cytoscape pluging) [24], a network basing on intracellular localization was arranged. All interactions were compared mining Hsp90 and p23 interactomes present in public protein-protein interaction (PPI) databases like POINT (http://point.bioinformatics.tw) and the specific database of protein interaction network for Plasmodium falciparum (http://apidb.org/apidb/showQuestion.do?questionFullName=GeneQuestions.GenesByFunctionalInteraction), derived from yeast two-hybrid studies [25,26]. Based on the Cytoscape generated diagrams, Hsp90 and p23 interactomes were schematized in a tachyzoite and bradyzoite draw.
3. Results
3.1 Identification of novel Hsp90 genes and Hsp90-cochaperones of T. gondii
In order to determine the number of Toxoplasma Hsp90 proteins, a database search in the Toxoplasma genome project (www.toxodb.org) was performed. Four putative proteins with Hsp90 signature were found: the previously described Hsp90 (Accesion AY344115) detected as extracellular, cytosolic and nuclear [2,4], the Hsp90-protein described by Saksouk et al. [27] which co-immunoprecipitate (co-IP) with parasite histones (Accesion AY53805), and the not previously described genes 83.m01184 and 583.m05357. In silico analysis AY53805 revealed a signal peptide (possible cleavage site between amino acids 24 and 25), an endoplasmic reticulum retention motif (HDEL) in the C-terminus (66.7 % of endoplasmic reticulum prediction value). Therefore, it could be assumed that this protein is a T. gondii GRP94. The other two potential new proteins with Hsp90 signature (“gene IDs” 83.m01184 and 583.m05357) included mitochondrial and mitochondrial/chloroplast transit peptides in the N-terminal region of their sequences and lacks the C-terminal region of Hsp90, which contains the highly conserved MEEVD motif that terminates Hsp90 and is the binding site for the Hop co-chaperone during the Hsp90-heterocomplex cycle [28]. The gene ID 83.m01184 also showed homology (identity of 42%) to Hsp75/TNF alpha receptor (TRAP1), whereas the identity with Hsp90 (AY344115) is lesser than 28%.
T. gondii cytoplasmic Hsp70 and immunophilin FBCP were already described, and putatives Hsp40, Hip, Hop, Aha1, p23 and PP5 Hsp90-co-chaperones were also identified after searching the ToxoDB database (Table 1).
Table 1.
Identification of T. gondii Hsp90 and Hsp90-heterocomplex related proteins
| Gene ID Release 5 |
Gene ID Release 4 |
Domains*/signal sequences | Chromosome | Protein identification+ (GenBank) |
|---|---|---|---|---|
| TGME49_088380 TGGT1_035020 TGVEG_082150 |
80.m00001 | Histidine kinase-like ATPase, Hsp90, HtpG, C-terminal EEVD |
IX | Hsp90 (AN: AY344115) |
| TGME49_044560 TGGT1_045380 TGVEG_104350 |
49.m00060 | Histidine kinase-like ATPase, Hsp90, HtpG, C-terminal KDEL |
VI | GRP94 putative (AN: AAY53805) |
| TGME49_092920 TGGT1_073160 TGVEG_014980 |
83.m01184 | Histidine kinase-like ATPase, Hsp90, HtpG N-terminal mitochondria/chloroplast transit peptide |
Ia | Hsp75 or TRAP1 putative |
| TGME49_110430 TGGT1_086650 TGVEG_094510 |
583.m05357 | Histidine kinase-like ATPase, Hsp90, HtpG, N-terminal mitochondria/chloroplast transit peptide |
XI | Hsp90Chloroplast putative |
| TGME49_073760 TGGT1_112840 TGVEG_021100 |
59.m00003 | Hsp70, DnaK, actin-like ATPase, C-terminal EEVD |
VIII | Hsp70 (AN: AAC72002) |
| TGME49_ 111240 TGGT1_087560 TGVEG_095340 |
583.m05418 | 1 DNAJ, G-Rich region, DnaJ_CXXCXGXG repeats/Zinc finger CR-type profile, 1 DNAJ_C, HPD sequence |
XI | TgYdJ1 putative |
| TGME49_032660 TGGT1_114860 TGVEG_022980 |
44.m00052 | 1 TPR, 1 STI1, C-terminal DP repeat |
VIII | Hip putative |
| TGME49_052220 TGGT1_001140 TGVEG_055940 |
52.m00012 | 3 TPR, C-terminal DP repeat | III | Hop putative |
| TGME49_121520 TGGT1_064560 TGVEG_059010 |
645.m00320 | 1 P23/CS | Ib | p23 putative |
| TGME49_ 112200 TGGT1_088450 TGVEG_096300 |
583.m05469 | 1 TPR, 1 Protein phosphatase 2A homologue/ Calcineurin- like phosphoesterase |
XI | Immunophilin PP5-like putative |
| TGME49_083850 TGGT1_035880 TGVEG_032190 |
76.m00007 | 1 FKBP_C, 1 TPR, 1 cyclophilin |
V | Immunophilin FBCP (AN: AAX51680) |
| TGME49_010800 TGGT1_124070 TGVEG_008760 |
27.m00826 | Aha1_N, Activator of Hsp90 ATPase |
IV | Aha 1 putative |
Release 4
NCBI Conserved Domain Search
based on domains, signal sequences and/or blastp identity
TGME49, T. gondii strain Me49, TGGT1, T. gondii strain GT1 and TGVEG, T. gondii strain VEG
3.2 Cloning and sequence characterization of T. gondii Hip as early marker for the Hsp90-heterocomplex cycle
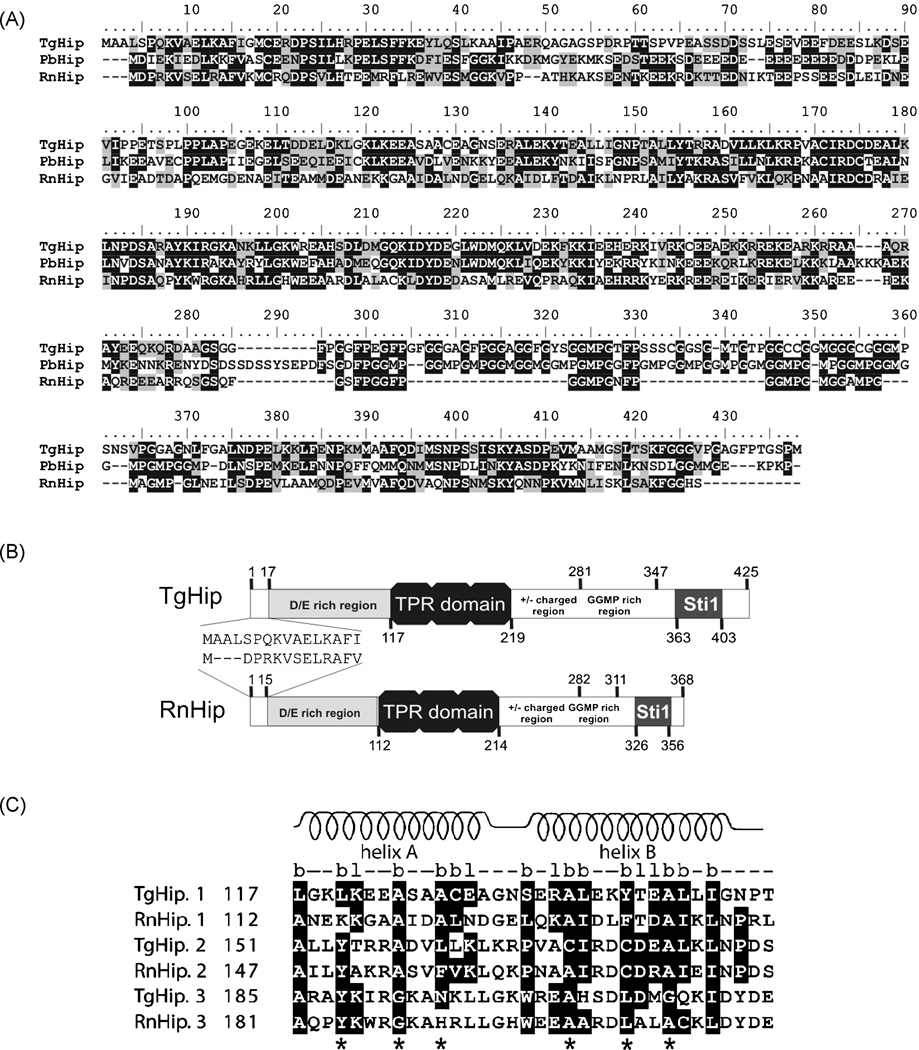
In higher eukaryotes, Hip was shown to be associated to Hsp70 early in the Heterocomplex-Hsp90 [9], becoming a stage marker for the Hsp90-heterocomplex cycle. The cDNA for gene 44.m00052 present an ORF of 1,278-bp which encodes for a protein of 425 amino acids with a theoretical mass of 45.1-kDa. Hip proteins comprise a series of consecutive modules [12]. Comparison to the R. norvegicus, P. berghei and T. gondii Hip proteins showed high overall similarity (Fig. 1A). T. gondii Hip showed a highly conserved structure (Fig. 1B), revealing the first 15 residues necessary for homo-oligomerization, a negatively charged residues region preceding the TPR domain and the Sti1 domain containing two “DPEV”-motifs (Fig. 1B). In the “GGMP” rich region, two “GGMP”, two “GGFP” and several “GGXX” repeats are present (Fig. 1A).
Figure 1. Characterization of T. gondii Hip co-chaperone.
A. Sequence alignment generated by clustal W (Bioedit program). Letters in black indicate identical residues; letters in grey indicate conserved residues. Gaps are indicated by dashes and were introduced to improve the alignment. TgHip, T. gondii Hip (Accession number [AN]: DQ680015), PbHip, P. berghei Hip (AN: Q08168), RnHip, R. norvegicus Hip (AN: P50503). TgHip and RnHip sequences showed 36% of identity and 55% of positives, whereas with PbHip identity was of 45% and positives were 58% (Blast2 program at www.ncbi.nlm.nih.gov/Blast). B. Schematic representation of T. gondii and R norvegicus Hip proteins with the putative protein motifs. Numbers denote the amino acid positions of motifs of the protein. C. Sequence alignment of the T. gondii and R. novergicus´s tetratricopeptide units from the TPR domains of Hip proteins. Consensus primary structure of the TPR domains: “b”: hydrophobic aminoacids, “l”: hydrophilic aminoacids. Above, the schematic representation of α helices determined by prediction of secondary structure is indicated. Conserved amino acids are marked in black. Stars indicate the most important positions in this structure (residues 4, 8, 11, 20, 24 and 27). The amino acid in position 32 is normally a hydrophobic residue (e.g. proline).
The TPR motif is a 34-amino acid sequence occurring three times in the protein in slight variation. It forms α-helices (A and B) that fold back on each other resulting in a compactly folded domain [29]. There is a consensus pattern of hydrophobic and hydrophilic residues (b and l in Fig. 1C), positions 8, 20 and 27 being small hydrophobic residues. Position 32 is frequently a proline [30]. Analysis of the three TgHip TPR motifs showed that they contain nearly all of the conserved residues and predicted two α-helices (Fig. 1C).
3.3 Cloning and structural characterization of T. gondii p23
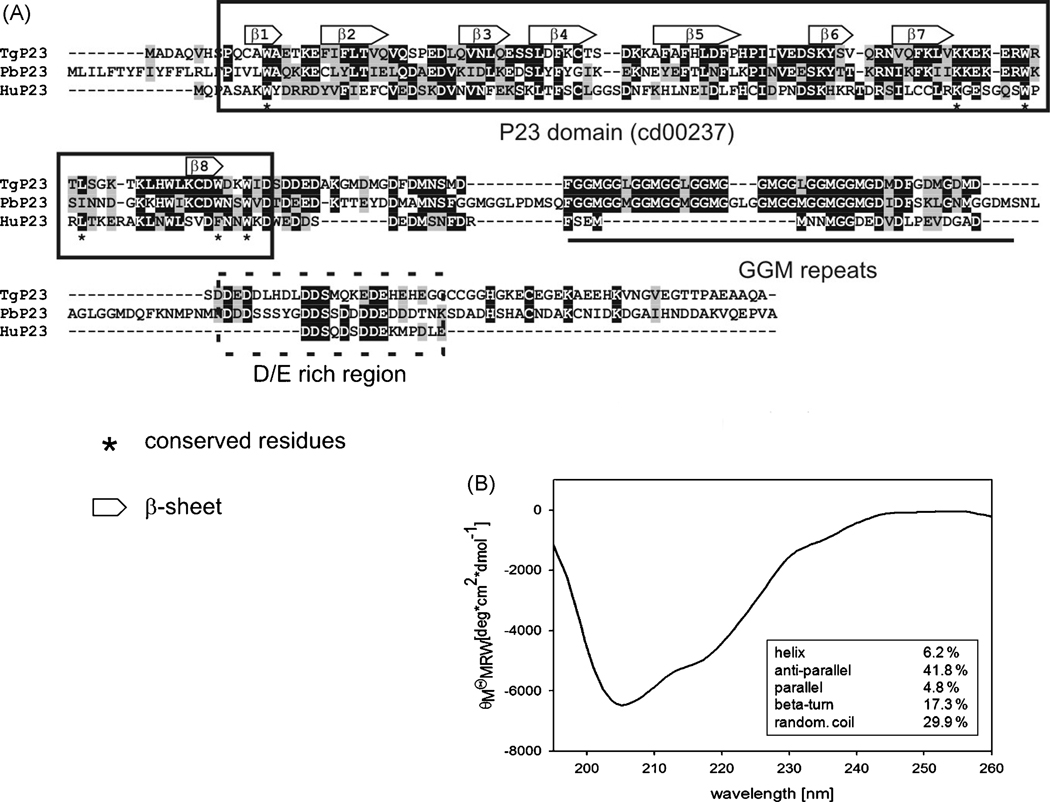
Searching Toxoplasma sequence databases with Plasmodium and human p23 (AN: XP_680236 and L24804 respectively) resulted in the identification of three putative p23-containing proteins: 645.m00320 (TGME49_121520: annotated as p23 co-chaperone), 541.m01195 (TGME49_105820: annotated as Calcyclin binding protein) and 44.m02666 (TGME49_031590: annotated as SGS domain-containig protein). The two latter have one additional SGS domain, a feature of SGT1 and calcyclin binding proteins. The ID 645.m00320 sequence presented the highest identity values when compared with P. berghei and human p23 (38% and 32% respectively), meanwhile 541.m01195 and 44.m02666 presented identity values ranging between 23% to non-significant identity). Based on these data it represented the most likely candidate to be the T. gondii p23 related to Hsp90-heterocomplex [17]. Therefore, the respective cDNA was cloned and sequenced. It showed an ORF of 681-bp encoding a 226-aminoacid protein (Fig. 2A) with a theoretical mass of 24.8-kDa. The amino acid sequence and its similarity with P. berghei p23 and Human p23 proteins are shown in Figure 2 (A). As expected, p23 is composed by an N-terminal anti-parallel β-sandwich domain and a C-terminal unstructured acidic region [31]. Yeast p23-like homologues presented also a “GGM/A” situated within the C-terminal region [32,33]. T. gondii p23 presented a characteristic p23 domain which showed the 8 putative β-sheets and presented a C-terminal region rich in acidic residues (Fig. 2A). It also contained a “GGM” rich region. By evolutionary tracing (ET), Zhu and Tygat [34] identified conserved residues along the p23 sequences (8W, 79K, 86W, 89L, 103F/W and 106W in human p23), which are also present in T. gondii p23 (asterisks in Fig. 2A).
Figure 2. Characterization of T. gondii p23.
A. Sequence alignment was generated by clustal W (Bioedit program). Letters in black indicate identical residues; letters in grey indicate conserved residues. Gaps are indicated by dashes and were introduced to improve the alignment. Tgp23, T. gondii p23 (Accesion number [AN]: DQ680014), Pbp23, P. berghei p23 (AN: XP_680236), Hup23, human p23 (AN: L24804). TgHip and Pbp23 sequences showed 40% of identity and 57% of positives, whereas with Hup23 it was of 28% of identity and 45% of positives (Blast2 program at www.ncbi.nlm.nih.gov/Blast). Domains, β-sheet regions and conserved residues are indicated in figure. β-sheet was deduced by using the http://protevo.eb.tuebingen.mpg.de/toolkit. BT. gondii p23 shows the characteristic, (β-sheet rich secondary structure. The far-UV-CD spectrum of the native T. gondii p23 was recorded at a protein concentration of 0.3 mg/ml in 50 mM Tris/HCl, pH 8.0 at 20°C. Inset: Calculated percentage of secondary structure elements.
With the aim to analyze the protein properties of T. gondii p23 we cloned the gene, expressed the respective protein in E. coli, and purified it to homogeneity. In order to investigate the secondary structure of T. gondii p23 experimentally, we analyzed the protein by far UV CD spectroscopy. The spectrum revealed a minimum between 204 and 208 nm indicative of a protein containing mainly β-sheets and a significant content of random coil (Fig. 2B). Using the deconvolution software CDNN, a content of ~ 46 % β-sheet, 17 % β-turn, 30 % random coil and only ~ 6 % α-helix was calculated. These data are in agreement with the gene-based predictions of a β-sandwich structure and structural data on human and yeast p23 [31,35,36].
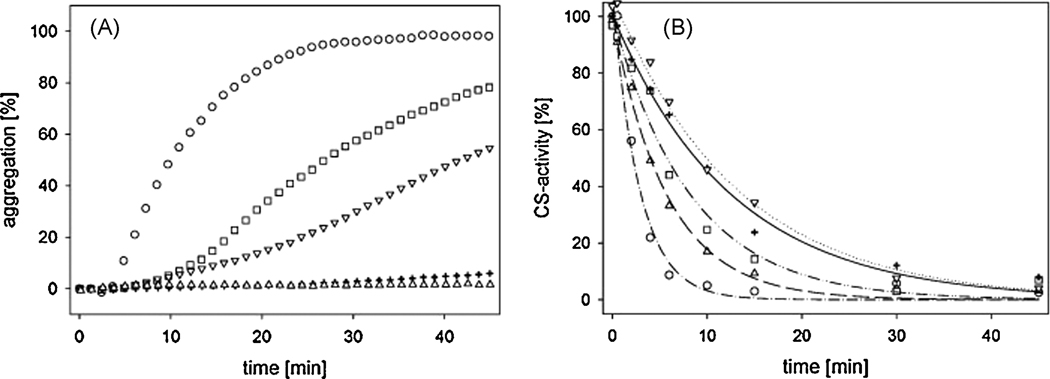
Despite of its modulating interaction with Hsp90, a functional key characteristic of p23 is its ability to act as a molecular chaperone itself. To test whether T. gondii p23 exhibits chaperone activity, as previously shown for its human or yeast homologues, [31,37] we performed thermal unfolding assays using citrate synthase (CS) as a model substrate. CS gets inactivated and aggregates irreversibly at temperatures above 40°C [19]. As shown in figure 3A, the spontaneous aggregation of CS, as monitored by light scattering at 44°C, reached a maximum value after approximately 40 min. Increasing concentrations of T. gondii p23 led to a concentration-dependent inhibition of CS aggregation. Interestingly, molar ratios of 2:1 p23:CS (monomers) already resulted in half maximum and ratios of 4:1 p23:CS in total suppression of the aggregation of CS. In comparison to human or yeast p23, which show half maximal aggregation suppression at ratios of ~5:1 and 10:1 respectively, this indicates a significantly higher (~2–5 fold) chaperone activity for the T. gondii protein [31,37].
Figure 3. Determination of T. gondii p23 chaperone activity.
A: Influence of p23 on the thermal aggregation of CS. CS (final concentration: 1.0 µM) was diluted into a thermostated solution (43°C) containing 1.0 µM (□), 2.0 µM (◊), 4.0 µM (∇) or 8.0 µM (Δ) p23. Circles (◦) represent the spontaneous aggregation of CS at 43°C. The kinetics of aggregation was determined by measuring the light scattering of the samples at 400 nm. B: Influence of p23on the thermal inactivation of CS. Inactivation of CS (0.15 µM) at 43°C in the absence (◦) and in the presence of increasing concentrations of p23; 0.075 µM (+), 0.15 µM (□), 0.3 µM (◊) or 0.6 µM (∇).
A second interesting functional feature of previously described p23 proteins is their ability to slow down the inactivation of model substrates. In contrast to aggregation, inactivation is an early step on the unfolding pathway of substrates like CS. To investigate if T. gondii p23 is also able to bind early unfolding intermediates of CS, we analyzed the influence of the T. gondii p23 on CS inactivation. Similar to human and yeast p23, the T. gondii protein was able to slow down the inactivation reaction significantly in a concentration-dependent manner (Fig. 3B). Interestingly, already sub-stoichiometric amounts of p23 influenced CS inactivation. The maximum effect could be seen at molar ratios of 4:1 p23:CS (monomers). This observation implicates a transient binding behavior of p23 to CS.
Taken together, compared to human and yeast p23, T. gondii p23 shows highly similar structural properties but overall seems to be an even stronger chaperone in vitro.
3.4 Hip and p23 are integrated in different Hsp90 complexes
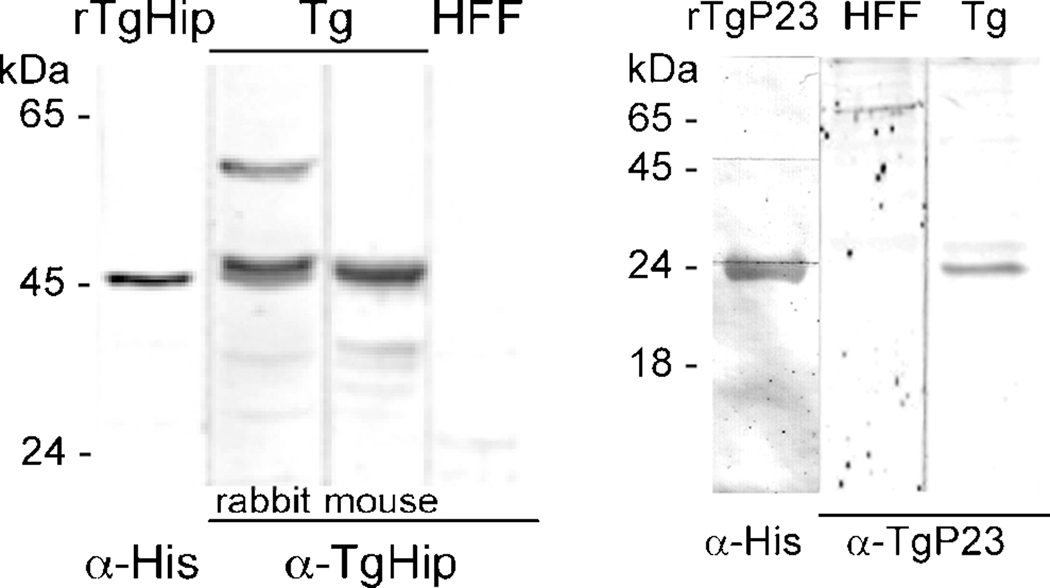
To investigate if T. gondii p23 interacts with Hsp90 and is integrated in similar interaction schemes than human or yeast p23 we first generated anti-p23 and as well anti-Hip antibodies to be able to address their protein-protein interactions by Western blot, co-immunoprecipitation (Co-IP) and immune-fluorescence analysis (IFA). Western-blot analysis showed a single band of 46-kDa which corresponds to the predicted protein size in tachyzoite lysate and coinciding with the one recognized by anti-His antibody in extracts of purified recombinant Hip (Fig. 4). Similarly, anti-T. gondii p23 antibody reacted with a single band of 25-kDa on tachyzoite lysate, also matching with the one recognized by anti-His antibody in extracts of purified recombinant Tgp23 (Fig. 4).
Figure 4. Detection of native Hip and p23 proteins.
Western-blot with murine anti-T. gondii Hip (α-TgHip), murine anti-T. gondii p23 (α-Tgp23) and murine anti-Histidines (α-His) antibodies. Tg: T. gondii PK strain lysate. Protein extract from uninfected HFF cells were also assayed. Preimmune serum samples did not show reactivity (data not shown). Migration of size marker is indicated (in kilo Daltons [kDa]).
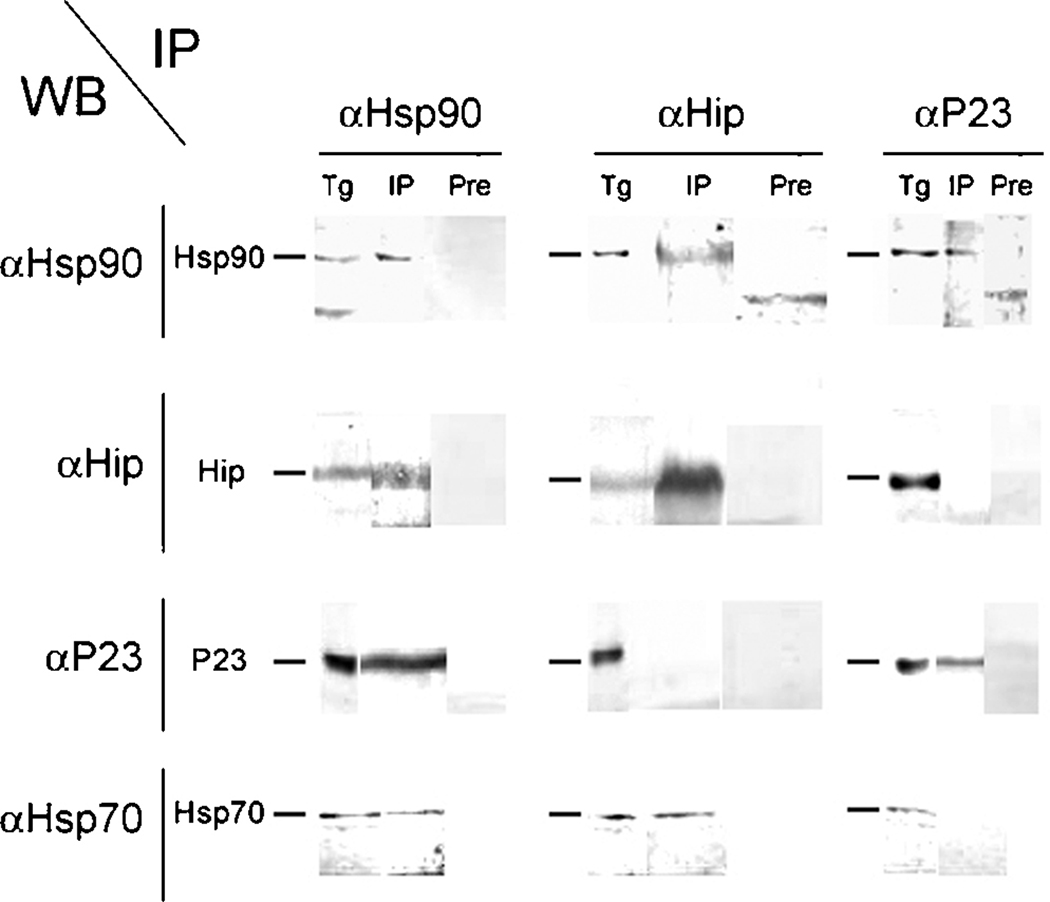
In order to demonstrate that the Hsp90-heterocomplex maturation pathway occurring in T. gondii is similar to that observed in higher eukaryotes, co-IP assays were performed using anti-Hsp90, anti-Hip and anti-p23 antibodies and analyzed by Western blot with all of them. A commercial antibody against Hsp70 was included to detect this protein in the analyzed complexes. Pull down of Hsp90, Hip and p23 proteins (Fig. 5) revealed that Hip is associated to Hsp70 and Hsp90 but not with p23, whereas p23 interacted with Hsp90 but not with Hip or Hsp70 (Fig. 5).
Figure 5. Co-IP analysis of Hsp90, Hip and p23 proteins.
Toxoplasma gondii PK strain lysates (Tg) were used for immunoprecipitation (IP) using either preimmune (Pre) or specific αHsp90, αHip and αp23 antisera (vertical columns). Pulled down materials were analyzed by western blot with specific sera antibodies and a commercial anti-Hsp70 antibody (horizontal columns).
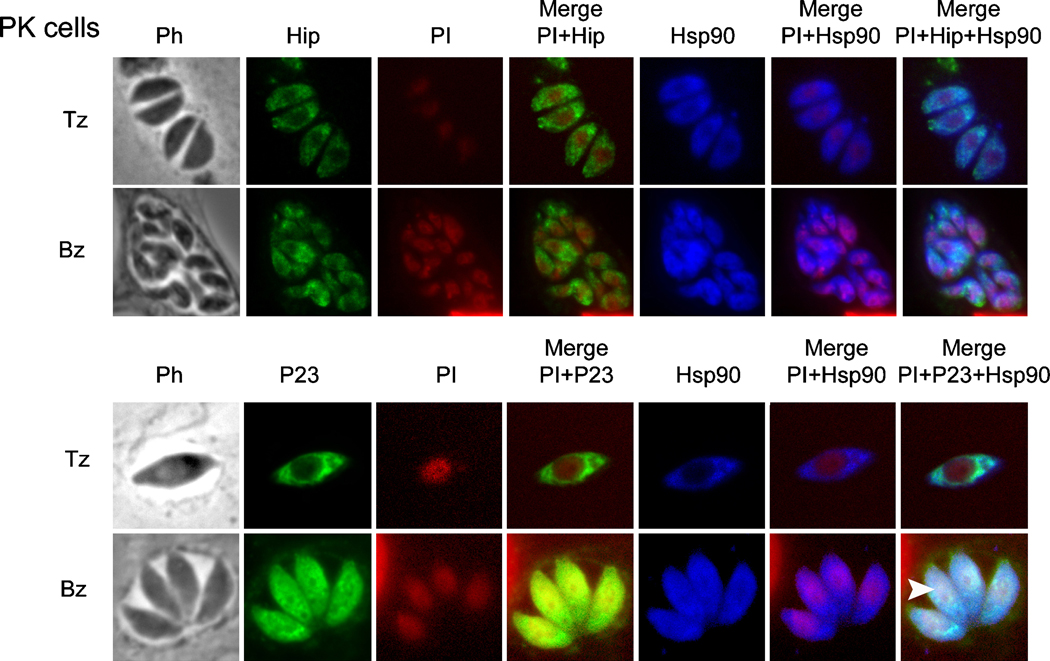
3.5 p23, but not Hip, translocates to the nucleus during bradyzoite development
Recently we have reported that cytoplasmic Hsp90 translocates to the nucleus when T. gondii differentiates to bradyzoite [2]. In order to investigate if some of the Hsp90 co-chaperones behave similarly, IFAs on intracellular tachyzoite and bradyzoites were carried out (Fig. 6). Intracellular, non-virulent T. gondii PK cells were grown in tachyzoites and bradyzoites conditions (37°C, 5% CO2 and 37°C, 0.03% CO2, pH 8.1, respectively). Hip showed a cytoplasmic localization in tachyzoite and bradyzoite stages (Fig. 6), without any co-localization with propidium iodide stained nuclei. Co-localization of Hip with cytoplasmic Hsp90, revealed by the overlayed fluorescence signal, confirmed the potential interaction between both proteins (Fig. 6).
Figure 6. Indirect immune-fluorescence assay (IFA) and subcellular localization of Hip and p23 in intracellular PK tachyzoites (Tz) and bradyzoites (Bz).
PK Tz were grown in vitro in human foreskin fibroblasts (HFF) and induced to Bz.[2]. To induce Bz formation in PK strain, parasites were grown 4 days at pH 8.1 and low CO2). IFA was performed on fixed intracellular parasites with antibodies against T. gondii Hip (murine serum), -p23 (murine serum) and -Hsp90 (rabbit serum) antibodies [2]. Propidium iodide (PI) staining reveals the location of the nucleus (red color). Mouse anti-P30 (α-SAG1) or anti-P34 serum (α-p34, mAb T82C2)[59] were used as marker of Tz and Bz respectively (data not shown). Rabbit anti-BAG1 polyclonal serum was used as cytoplasmic bradyzoite marker. The efficiency of in vitro conversion to Bz was 82–88%. White arrows heads are signaling the localization of the nucleus in both panels.
For p23, a cytoplasmic localization was observed in tachyzoite samples whereas in bradyzoites both, a cytosolic and nuclear distribution was observed. Furthermore, co-localizing with the cytoplasmic/nuclear Hsp90 was indicated by the overlayed fluorescence (Fig. 6).
3.6 Interactome analysis of T. gondii p23
The protein composition of the p23 interactome during bradyzoite development was determined from a differentiated PK strain by immunoprecipitation and mass spectrometry analysis. In order to perform a comparative analysis, tachyzoite lysates of PK strain were also studied. Immunoprecipitation with p23 antibody was more efficient than with Hsp90 antibody (data not shown). Preimmune serum sample gave a clean immunoprecipitation-pattern (Fig. S1). In addition, the use of an anti-T. gondii Hsp20 antibody as independent control, did not pull down any protein (data not shown and [38]). Overall, 43 parasite proteins that co-IP with anti-p23 antibody either from tachyzoite or bradyzoite lysates were identified (Table 2, S1 and S2). Almost all of them are involved in ATP production/glycolysis (1–7) and translation (8–13) or are chaperones and other proteins with protein binding properties (24–33). Additionally, some further proteins from rhoptry, microneme, dense granules and histones were pulled down (Table 2, Table S3).
Table 2.
Proteomic analysis of proteins pulled down with anti-P23 antibody in tachyzoite and bradyzoite stages
| Function | Protein name | stage | Gene ID Release 5 |
Gene ID Release 4 |
Peptide freq (Tz/Bz) |
Interactome analysis (db)* |
|
|---|---|---|---|---|---|---|---|
| Glycolisis/ ATP generation |
|||||||
| 1 | Triose phosphate isomerase I | Bz | TGME49_025930 | 42.m00050 | 3 | ||
| 2 | Glyceraldehyde-3-phosphate dehydrogenase I | Bz | TGME49_089690 | 80.m00003 | 10 | ||
| 3 | Phosphoglycerate kinase I | Bz | TGME49_118230 | 641.m00193 | 7 | ||
| 4 | Enolase 2 (ENO2) | Tz,Bz | TGME49_068850 | 59.m03410 | 4/8 | yes | |
| 5 | Pyruvate kinase I | Bz | TGME49_056760 | 55.m00007 | 5 | yes | |
| 6 | Lactate dehydrogenase I (LDH1) | Tz,Bz | TGME49_032350 | 44.m00006 | 9/13 | yes | |
| 7 | Fructose-1,6-bisphosphate aldolase | Tz,Bz | TGME49_036040 | 46.m00002 | 6/9 | yes | |
| Translation machinery/ Protein metabolism |
|||||||
| 8 | Elongation factor 1alpha | Tz,Bz | TGME49_086420 | 76.m00016 | 14/14 | ||
| 9 | Elongation factor 2 | Tz,Bz | TGME49_005470 | 20.m03912 | 5/4 | yes | |
| 10 | Asparaginyl-tRNA synthase | Bz | TGME49_070510 | 59.m03518 | 2 | yes | |
| 11 | Tryptophanyl tRNA Synthase | Bz | TGME49_088360 | 80.m00063 | 4 | ||
| 12 | nascent polypeptide-associated complex | Tz | TGME49_005560 | 20.m03918 | 3 | ||
| 13 | Cytosol aminopeptidase | Bz | TGME49_090670 | 80.m00088 | 4 | ||
| Kinases/ Phosphorylases/ Phosphatases |
|||||||
| 14 | Rhoptry protein 4 (protein kinase Rop4)) | Bz | TGME49_095110 | 83.m02145 | 9 | ||
| 15 | Rhoptry protein 5 (protein kinase Rop5) | Bz | TGME49_108080 | 551.m00238 | 13 | ||
| 16 | Uridine Phosphorylase | Bz | TGME49_110640 | 583.m00630 | 3 | ||
| 17 | Nucleoside triphosphatase I | Tz,Bz | TGME49_077240 | 65.m00001 | 5/7 | ||
| 18 | Ser-Thr phosphatase 2C | Tz,Bz | TGME49_031850 | 44.m00037 | 1/7 | yes | |
| Cytoskeleton/motility/ Traffic |
|||||||
| 19 | Actin | Tz,Bz | TGME49_ 009030 | 25.m00007 | 3/15 | yes | |
| 20 | Myosin A | Tz | TGME49_ 035470 | 46.m00001 | 21 | yes | |
| 21 | Clathrin heavy chain | Tz | TGME49_090950 | 80.m02298 | 2 | ||
| Redox homeostasis | |||||||
| 22 | Thioredoxin | Bz | TGME49_009950 | 25.m00203 | 4 | ||
| 23 | Protein disulfide isomerase | Tz,Bz | TGME49_011680 | 27.m00003 | 17/21 | ||
| Chaperones/ protein binding |
|||||||
| 24 | Hsp101/ClpA | Tz | TGME49_057990 | 55.m04729 | 5 | ||
| 25 | Grp94 | Tz,Bz | TGME49_044560 | 18/4 | |||
| 26 | Hsp90 | Bz | TGME49_044560 | 80.m00001 | 7 | yes | |
| 27 | Hsp70 | Tz,Bz | TGME49_073760 | 59.m00003 | 11/8 | yes | |
| 28 | Organellar Hsp70 | Bz | TGME49_051780 | 50.m00085 | 2 | yes | |
| 29 | Hsp70/BiP | Tz,Bz | TGME49_111720 | 583.m00009 | 2/24 | yes | |
| 30 | Hsp60 | Bz | TGME49_047550 | 50.m00006 | 5 | ||
| 31 | Hsp20 | Tz,Bz | TGME49_032940 | 44.m02755 | 224/69 | ||
| 32 | 14.3.3 | Bz | TGME49_063090 | 55.m00015 | 5 | yes | |
| 33 | Hypotetical protein, WD repeat containing protein | Tz | TGME49_056860 | 55.m04655 | 1 | ||
| Others | |||||||
| 34 | Surface antigen SAG1 | Bz | TGME49_033460 | 44.m00009 | 9 | ||
| 35 | B-antigen | Tz,Bz | TGME49_062470 | 55.m04993 | 2 | ||
| 36 | Dense granule 7 (Gra7) | Bz | TGME49_003310 | 20.m00005 | 11 | ||
| 37 | Dense granule 5 (Gra5) | Tz | TGME49_086450 | 76.m00004 | 2 | ||
| 38 | Rhoptry protein 9 (Rop9) | Bz | TGME49_043730 | 49.m00048 | 7 | ||
| 39 | Microneme protein 2 (Mic2) | Tz,Bz | TGME49_001780 | 20.m00002 | 2/3 | ||
| 40 | Histone 4 (H4) | Bz | TGME49_044560 | 49.m03134 | 4 | ||
| 41 | Histone 2A variant Z (H2AZ) | Tz | TGME49_100200 | 145.m00002 | 4 | yes | |
| 42 | Hypotetical protein | Bz | TGME49_034380 | 46.m01601 | 4 | ||
| 43 | Hypotetical protein | Bz | TGME49_063850 | 55.m08199 | 2 | ||
1–43, list order number of identified proteins in toxodb
Plasmodium Y2H and orthologs data bases
Tz: tachyzoite
Bz:bradyzoite
Peptide freq.: peptide frequency
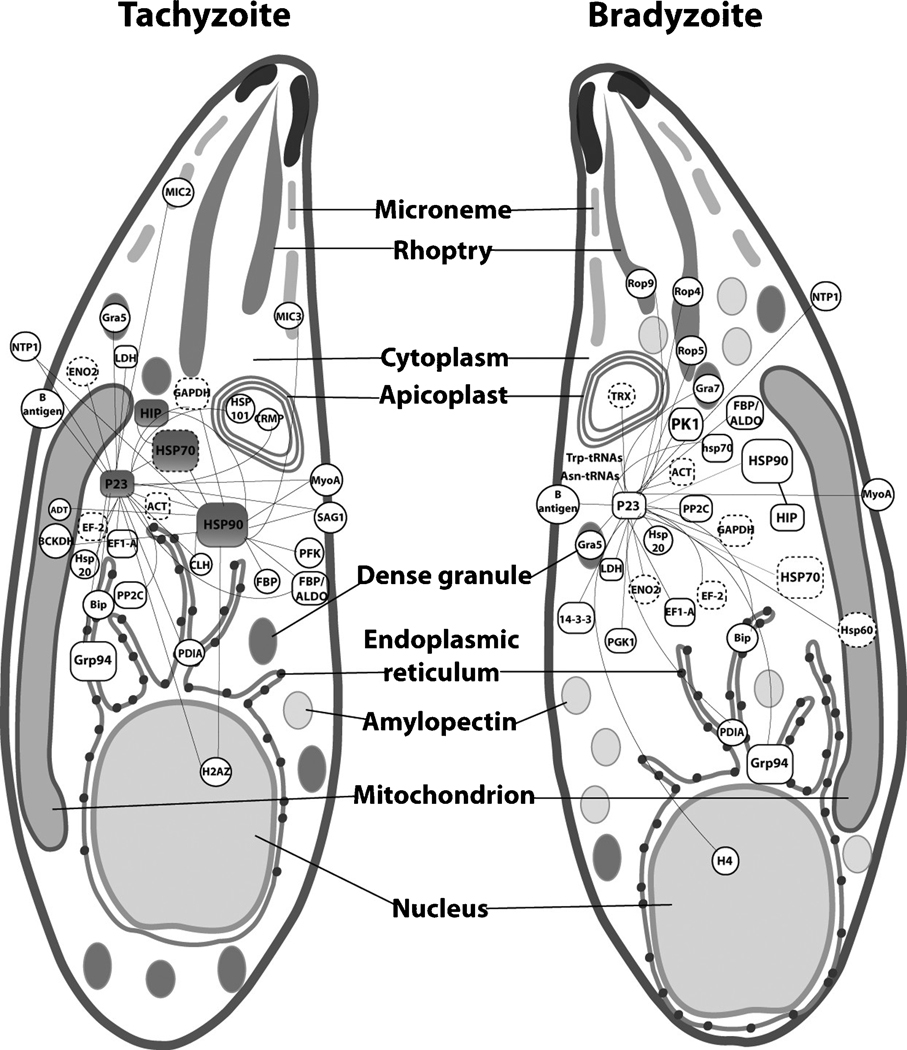
A protein-protein interaction (PPI) model of the T. gondii Hsp90/p23 proteins was created by combining our data sets obtained from T. gondii p23 IP-mass spectrometry analysis (Table S3), T. gondii Hsp90 IP-mass spectrometry (Table S3) and co-IP assays/microscopy co-localization with their subcellular compartmentalization and temporal synchronicity [tachyzoite or bradyzoite stages] (Fig. 7 and Cytoscape file S1). This PPI map was compared with Hsp90/p23 interactomes generated from: 1) the web-based resource POINT (http://poinet.bioinformatics.tw/Welcome.do) which integrates data from several protein prediction interactors (PPI) repositories (IntAct, Mint, DIP, BIND, BioGRID, etc.,) into interolog maps, which are based on the assumption that a pair of interacting proteins (interologs) in one species predicts an interaction of orthologous proteins in another species, and 2) from the PPI database for P. falciparum, derived from yeast two-hybrid studies [25,26]. Interactome attributes and interactors data are shown in Table S4. Among all the p23 interactors, 15 were also detected in PPI databases (Table 2). Taking together 23 interactors in tachyzoites and bradyzoites and Hsp90 interactors in tachyzoites (Fig. 5 and Table S3) we built an Hsp90/p23 interactome network (Fig. 7). In this network 77% are proteins conserved in other organisms, 65% present as far as mammals and another 12% conserved in addition in P. falciparum (Table S3). From this pool of evolutionary conserved interactors, 43% and 35% of the interactions found in this study were also found in the interolog PPI map and P. falciparum PPI databases, respectively (Fig. 7 and Table S3). The interactome maps also indicated a wide range of subcellular localizations and functions of putative Hsp90/p23 targets in both parasite stages (summarized in Fig. 7, Table S3 and Cytoscape file S1).
Figure 7. Differential interactome of Hsp90/p23 complex in tachyzoites and bradyzoites.
Interactors are visualized in a scheme of T. gondii tachyzoite and bradyzoite. Combined interactome of Hsp90 and p23 proteins was built using the Cytoscape platform based on the data obtained from proteomic analysis and verified by: co-IP assays from the present study (grey interactors), mining Hsp90 and p23 interactomes present in the public PPI database POINT (http://point.bioinformatics.tw) and the specific database for P. falciparum (http://apidb.org/apidb/showQuestion.do?questionFullName=GeneQuestions) (rectangle and dashed contour, respectively). Cellular localization is inferred from Gene Ontology data. ACT, actin; ADT,ADP/ADT carrier; BCKDH, 2-oxoisovalerate dehydorgenase; CLH, clatrin heavy chain; CRMP, Cysteine repeat modular protein; EF1-A, elongation factor 1 alpha; EF-2, elongation factor 2; ENO2, enolase 2; FBP/aldo, Fructose 1,6 biphosphatase aldolase; GAPDH, glyceraldehyde-3-phosphate dehydorgenase; LDH, lactate dehydrogense 1, MyoA, myosin A; NTP1, nucleoside triphosphatase I; PDIA, protein disulfide isomerase; PKM, pyruvate kinase I; Trp and Asn-tRNAs, tryptophanyl and asparaginyl tRNA synthase.
4. Discussion
Recently we described the importance of T. gondii Hsp90 in the two asexual stages of interconversion (tachyzoite-bradyzoite) of the parasite [2]. Based on in silico analysis, the previously described Hsp90 [2] presented all the requisites to be a candidate to assemble into the Hsp90/70-heterocomplex in T. gondii, a prediction supported here by experiments that show that Hsp90 interacts with the respective co-chaperones Hip and p23. T. gondii Hip and p23 showed the typical hallmarks of proteins from these families. T. gondii p23 sequence and determined secondary structure was comparable to the sequence and structure of human or yeast p23 [32,33]. indicating conservation of the basic IgG like, globular, β sandwich fold throughout the protein family [35,36]. Moreover the T. gondii p23 chaperone activity showed to be stronger than its human and yeast counterparts, which could be important for the parasite during bradyzoite development usually triggered by stressful environments [39]. Both T. gondii Hip and p23 interacts with Hsp90, but not directly with each other. In higher eukaryotes Hip is a marker of the early Hsp90-heterocomplex, whereas p23 binds to Hsp90 dimmers at a later stage [8,13,14]. In comparison, a similar Hsp90-heterocomplex-cycle pathway is occurring in T. gondii.
Additionally, we showed that p23 translocates to parasite nucleus in the bradyzoite stage as it was observed for parasite Hsp90 [2]. Recently Picard et al [40] demonstrated by FRAP analysis that Hsp90 and p23 are always co-localizing, which is consistent with the notion that the bulk of p23 is engaged in a complex pattern of Hsp90-mediated interactions. Probably in T. gondii p23 and Hsp90 could be working as a complex rather than in separated ways. Several studies showed that in mammals and yeast, Hsp90 and its co-chaperone p23 are selectively recruited to gene expression regulatory complexes and that they play a role in telomere maintenance, suggesting extensive functions for these molecular chaperones in gene regulation and formation and maintenance of a particular state of the chromatin [41–44]. Direct interaction between Hsp90 and core histones or chromatin remodeling complexes has been suggested [45–47]. p23 pulled down with H2AZ and H4 in tachyzoite and bradyzoite stages respectively. Recently, Toxoplasma H2AZ showed to be associated to active chromatin whereas the variant H2AX and H4K20me1 (H4 mono-methylated in lysine 1) are enriched at silent promoters and its expression is increased during bradyzoite differentiation [48]. There might be a connection between Hsp90-heterocomplex and chromatin regulation in Toxoplasma as well.
Co-IP experiments identified several proteins that could be assisted by p23 during bradyzoite development. Some of these proteins could be contaminants (e.g. SAG1, a tachyzoite stage specific protein that appear in bradyzoite pulled down sample); therefore their interaction with p23 or p23/Hsp90 complex should be confirmed. The stage specific interaction also should be confirmed by additional experiments. On the other hand, some p23 interactors were not detected because of analyzing only coomassie stainable bands. It is interesting to note that although there is no antecedent of p23-IP analysis in other organisms, 15 out of the 43 proteins that pulled down in the IP performed with anti-p23 of T. gondii were also identified as p23-interacting proteins in other cell systems or interactome networks [49,50]. Since at least 8 proteins are specific of T. gondii (micronemes, rhoptry, dense granules and hypothetical proteins) the coincidence degree between our co-IP pool of conserved proteins and PPI databases (near 43%) suggest that our pull down experiment was highly specific. The complete set of Hsp90/p23 interactors that we found suggests that the Hsp90/p23 machine keeps its evolutionary conserved functions with slight specializations for the parasite biology [49,51].
Most of the putative p23-interacting proteins are involved in glycolysis, ATP production, protein synthesis, protein assistance (chaperones and foldases), some kinases and phosphatases (Table 2 and Table S2). Noteworthy, during bradyzoite development the parasite mitochondria is down-regulated [52,53], and accumulate amylopectin granules [54], indicating that in bradyzoites the predominant pathway to obtain energy is glycolysis. Moreover, it was demonstrated that pyruvate kinase and lactate dehydrogenase, presented a markedly higher activity in bradyzoites than in tachyzoites, and over-expression of T. gondii lactate dehydrogenase enzymes LDH1 and LDH2 enhances parasite differentiation under alkaline conditions [55], suggesting that lactate production is particularly important in this developmental form [54]. Recently a deoxyribose phosphate aldolase-like gene expressed in bradyzoite stage was identified [56]. Despite almost all of the enzymes involved in glycolisis are expressed in both parasite stages, some of them were found to be overexpressed in bradyzoite stage in Neospora caninum, a closely related parasite of T. gondii [57]. In this context, p23 could be assisting parasite glycolytic enzymes during bradyzoite conversion to ensure enough ATP generation in the new parasite scenario. Similarly, p23 could be assisting the translational machinery to generate new proteins during early bradyzoite transformation.
The interactome map generated could be an important tool for future studies in T. gondii as well as other cell systems. We could identify novel potential interactors, some of them specific proteins of T. gondii (Mic3, Rop4, Rop5, Rop9, Gra5, Gra7). Recently Gra5, but not other dense granule proteins, was shown to interact with Hsp90 at least in tachyzoite stage [58]. N. caninum Rop9 and Gra9 showed to be over expressed in bradyzoite stage [57]. Nevertheless, it should be noted that all of them could be in complexes throughout the secretory pathway, suggesting that Hsp90/p23 complex could be assisting the stabilization of these complexes.
The predicted pleiotropic role of parasite Hsp90 and in addition p23, is evidenced by the intracellular localization of its targets from the nucleus and apicoplast to the secretory organelles dense granules, ropthries and micronemes and by the wide range of annotated and predicted function of their interactors. Several cell cycle kinases, phosphatases and transcription factors are client proteins of the Hsp90/p23 complex [6]. Here, we identified some proteins of this type but their role during parasite differentiation remains currently uncertain. The inability to obtain classical cell cycle kinases or transcription factor-like proteins could be due to the fact that they are low abundance proteins.
Taken together, we showed that the Hsp90-hetercomplex cycle is present in T. gondii. Additionally we showed that T. gondii co-chaperone p23 has some differences in sequence structure and activity in comparison with human p23 that could be important for parasite biology. Finally, the p23 interactome network, presented here, will allow making predictions and hypothesis about the dynamics of the Hsp90/p23 chaperone machinery.
Supplementary Material
Acknowledgments
S. O. Angel (Researcher) and M. J. Figueras (Fellow) are members of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). We thank Georgina Davies Sala for assistance in parasite drawings and Maria Corvi for critical reading.
This work was supported by the Agencia Nacional para la Promoción de la Ciencia y la Tecnología (PICT 05-34415), the CONICET (PIP6336), and the Universidad Nacional General San Martin (UNSAM grant SA08/006) to S.A. as well as from DAAD/SECTY/PROALAR (DA0605) to J.B and S.A., and the Deutsche Forschungsgemeinschaft (SFB594) to J.B and M.H.
The proteomic analyses in this work were supported by NIH grants from the National Center for Research Resources IDeA Network of Biomedical Research Excellence program P20-RR016462 and P20-RR021905.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ferreira da Silva Mda F, Barbosa HS, Gross U, Luder CG. Stress-related and spontaneous stage differentiation of Toxoplasma gondii. Mol Biosyst. 2008;4:824–834. doi: 10.1039/b800520f. [DOI] [PubMed] [Google Scholar]
- 2.Echeverria PC, Matrajt M, Harb OS, et al. Toxoplasma gondii Hsp90 is a potential drug target whose expression and subcellular localization are developmentally regulated. J Mol Biol. 2005;350:723–734. doi: 10.1016/j.jmb.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. The transcriptome of Toxoplasma gondii. BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn HJ, Kim S, Nam HW. Molecular cloning of the 82-kDa heat shock protein (HSP90) of Toxoplasma gondii associated with the entry into and growth in host cells. Biochem Biophys Res Commun. 2003;311:654–659. doi: 10.1016/j.bbrc.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 5.Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of hsp90 with early unfolding intermediates of citrate synthase - implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- 6.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 7.Murphy PJ, Morishima Y, Chen H, et al. Visualization and mechanism of assembly of a glucocorticoid receptor•Hsp70 complex that is primed for subsequent Hsp90-dependent opening of the steroid binding cleft. J Biol Chem. 2003;278:34764–34773. doi: 10.1074/jbc.M304469200. [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin SH, Sobott F, Yao ZP, et al. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 9.Prapapanich V, Chen S, Smith DF. Mutation of Hip's carboxy-terminal region inhibits a transitional stage of progesterone receptor assembly. Mol Cell Biol. 1998;18:944–952. doi: 10.1128/mcb.18.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pratt WB, Galigniana MD, Harrell JM, DeFranco DB. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Prapapanich V, Chen S, Nair SC, Rimerman RA, Smith DF. Molecular cloning of human p48, a transient component of progesterone receptor complexes and an hsp70-binding protein. Mol Endocrinol. 1996;10:420–431. doi: 10.1210/mend.10.4.8721986. [DOI] [PubMed] [Google Scholar]
- 13.Freeman BC, Felts SJ, Toft DO, Yamamoto KR. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 2000;14:422–434. [PMC free article] [PubMed] [Google Scholar]
- 14.Richter K, Walter S, Buchner J. The co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J Mol Biol. 2004;342:1403–1413. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 15.Morishima Y, Kanelakis KC, Murphy PJ, et al. The Hsp90 cochaperone p23 is the limiting component of the multiprotein Hsp90/Hsp70-based chaperone system in vivo where it acts to stabilize the client protein: hsp90 complex. J Biol Chem. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- 16.Wiser MF, Jennings GJ, Uparanukraw P, van Belkum A, van Doorn LJ, Kumar N. Further characterization of a 58 kDa Plasmodium berghei phosphoprotein as a cochaperone. Mol Biochem Parasitol. 1996;83:25–33. doi: 10.1016/s0166-6851(96)02743-0. [DOI] [PubMed] [Google Scholar]
- 17.Wiser MF. A Plasmodium homologue of cochaperone p23 and its differential expression during the replicative cycle of the malaria parasite. Parasitol Res. 2003;90:166–170. doi: 10.1007/s00436-003-0835-4. [DOI] [PubMed] [Google Scholar]
- 18.Bohm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 19.Buchner J, Weikl T, Bugl H, Pirkl F, Bose S. Purification of Hsp90 partner proteins Hop/p60, p23, and FKBP52. Methods Enzymol. 1998;290:418–429. doi: 10.1016/s0076-6879(98)90035-0. [DOI] [PubMed] [Google Scholar]
- 20.Srere PA. Citrate-condensing enzyme-oxalacetate binary complex. Studies on its physical and chemical properties. J Biol Chem. 1966;241:2157–2165. [PubMed] [Google Scholar]
- 21.de Miguel N, Echeverria PC, Angel SO. Differential subcellular localization of members of the Toxoplasma gondii small heat shock protein family. Eukaryot Cell. 2005;4:1990–1997. doi: 10.1128/EC.4.12.1990-1997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas PA, Martin V, Nigro M, et al. Expression of a cDNA encoding a Toxoplasma gondii protein belonging to the heat-shock 90 family and analysis of its antigenicity. FEMS Microbiol Lett. 2000;190:209–213. doi: 10.1111/j.1574-6968.2000.tb09288.x. [DOI] [PubMed] [Google Scholar]
- 23.Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007;23:1040–1042. doi: 10.1093/bioinformatics/btm057. [DOI] [PubMed] [Google Scholar]
- 25.LaCount DJ, Vignali M, Chettier R, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 26.Date SV, Stoeckert CJ., Jr Computational modeling of the Plasmodium falciparum interactome reveals protein function on a genome-wide scale. Genome Res. 2006;16:542–549. doi: 10.1101/gr.4573206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saksouk N, Bhatti MM, Kieffer S, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 29.Groves MR, Barford D. Topological characteristics of helical repeat proteins. Curr Opin Struct Biol. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 30.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weikl T, Abelmann K, Buchner J. An unstructured C-terminal region of the Hsp90 co-chaperone p23 is important for its chaperone function. J Mol Biol. 1999;293:685–691. doi: 10.1006/jmbi.1999.3172. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oxelmark E, Knoblauch R, Arnal S, Su LF, Schapira M, Garabedian MJ. Genetic dissection of p23, an Hsp90 cochaperone, reveals a distinct surface involved in estrogen receptor signaling. J Biol Chem. 2003;278:36547–36555. doi: 10.1074/jbc.M305960200. [DOI] [PubMed] [Google Scholar]
- 34.Zhu S, Tytgat J. Evolutionary epitopes of Hsp90 and p23: implications for their interaction. FASEB J. 2004;18:940–947. doi: 10.1096/fj.04-1570hyp. [DOI] [PubMed] [Google Scholar]
- 35.Ali MM, Roe SM, Vaughan CK, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver AJ, Sullivan WP, Felts SJ, Owen BA, Toft DO. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J Biol Chem. 2000;275:23045–23052. doi: 10.1074/jbc.M003410200. [DOI] [PubMed] [Google Scholar]
- 37.Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 38.de Miguel N, Lebrun M, Heaslip A, et al. Toxoplasma gondii Hsp20 is a stripe-arranged chaperone-like protein associated with the outer leaflet of the inner membrane complex. Biol Cell. 2008;100:479–489. doi: 10.1042/BC20080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vonlaufen N, Kanzok SM, Wek RC, Sullivan WJ., Jr Stress response pathways in protozoan parasites. Cell Microbiol. 2008;10:2387–2399. doi: 10.1111/j.1462-5822.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 40.Picard D, Suslova E, Briand P-A. 2-color photobleaching experiments reveal distinct intracellular dynamics of two components of the Hsp90 complex. Exp Cell Res. 2006;312:3949–3958. doi: 10.1016/j.yexcr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 41.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 42.Cvoro A, Tzagarakis-Foster C, Tatomer D, Paruthiyil S, Fox MS, Leitman DC. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol Cell. 2006;21:555–564. doi: 10.1016/j.molcel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 43.Oxelmark E, Roth JM, Brooks PC, Braunstein SE, Schneider RJ, Garabedian MJ. The cochaperone p23 differentially regulates estrogen receptor target genes and promotes tumor cell adhesion and invasion. Mol Cell Biol. 2006;26:5205–5213. doi: 10.1128/MCB.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toogun OA, Dezwaan DC, Freeman BC. The Hsp90 molecular chaperone modulates multiple telomerase activities. Mol Cell Biol. 2008;28:457–467. doi: 10.1128/MCB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnaider T, Oikarinen J, Ishiwatari-Hayasaka H, Yahara I, Csermely P. Interactions of Hsp90 with histones and related peptides. Life Sci. 1999;65:2417–2426. doi: 10.1016/s0024-3205(99)00508-1. [DOI] [PubMed] [Google Scholar]
- 46.Feng Q, Yi P, Wong J, O'Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao R, Davey M, Hsu YC, et al. Navigating the chaperone network: integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 48.Dalmasso MC, Onyango DO, Naguleswaran A, Sullivan WJ, Jr, Angel SO. Toxoplasma H2A Variants Reveal Novel Insights into Nucleosome Composition and Functions for this Histone Family. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Pavithra SR, Kumar R, Tatu U. Systems analysis of chaperone networks in the malarial parasite Plasmodium falciparum. PLoS Comput Biol. 2007;3:1701–1715. doi: 10.1371/journal.pcbi.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsaytler PA, Krijgsveld J, Goerdayal SS, Rudiger S, Egmond MR. Novel Hsp90 partners discovered using complementary proteomic approaches. Cell Stress Chaperones. 2009 doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohne W, Heesemann J, Gross U. Reduced replication of Toxoplasma gondii is necessary for induction of bradyzoite-specific antigens: a possible role for nitric oxide in triggering stage conversion. Infect Immun. 1994;62:1761–1767. doi: 10.1128/iai.62.5.1761-1767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomavo S, Boothroyd JC. Interconnection between organellar functions, development and drug resistance in the protozoan parasite, Toxoplasma gondii. Int J Parasitol. 1995;25:1293–1299. doi: 10.1016/0020-7519(95)00066-b. [DOI] [PubMed] [Google Scholar]
- 54.Denton H, Brown SM, Roberts CW, et al. Comparison of the phosphofructokinase and pyruvate kinase activities of Cryptosporidium parvum, Eimeria tenella and Toxoplasma gondii. Mol Biochem Parasitol. 1996;76:23–29. doi: 10.1016/0166-6851(95)02527-8. [DOI] [PubMed] [Google Scholar]
- 55.Liwak U, Ananvoranich S. Toxoplasma gondii: over-expression of lactate dehydrogenase enhances differentiation under alkaline conditions. Exp Parasitol. 2009;122:155–161. doi: 10.1016/j.exppara.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Ueno A, Dautu G, Munyaka B, Carmen G, Kobayashi Y, Igarashi M. Toxoplasma gondii: Identification and characterization of bradyzoite-specific deoxyribose phosphate aldolase-like gene (TgDPA) Exp Parasitol. 2009;121:55–63. doi: 10.1016/j.exppara.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 57.Marugan-Hernandez V, Alvarez-Garcia G, Risco-Castillo V, Regidor-Cerrillo J, Ortega-Mora LM. Identification of Neospora caninum proteins regulated during the differentiation process from tachyzoite to bradyzoite stage by DIGE. Proteomics. doi: 10.1002/pmic.200900664. [DOI] [PubMed] [Google Scholar]
- 58.Aguirre AA, Keefe TJ, Reif JS, et al. Infectious disease monitoring of the endangered Hawaiian monk seal. J Wildl Dis. 2007;43:229–241. doi: 10.7589/0090-3558-43.2.229. [DOI] [PubMed] [Google Scholar]
- 59.Tomavo S, Fortier B, Soete M, Ansel C, Camus D, Dubremetz JF. Characterization of bradyzoite-specific antigens of Toxoplasma gondii. Infect Immun. 1991;59:3750–3753. doi: 10.1128/iai.59.10.3750-3753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.