Summary
In an attempt to understand ribosome-induced GTP hydrolysis on eEF2, we have determined a 12.6-Å cryo-EM reconstruction of the eEF2-bound 80S ribosome in the presence of aluminum fluoride (AlF4−) and GDP, with AlF4− mimicking the γ-phosphate during hydrolysis. This is the first visualization of a structure representing a transition state complex on the ribosome. Tight interactions are observed between the factor’s G-domain and the large ribosomal subunit, as well as between domain IV and an intersubunit bridge. In contrast, some of the domains of eEF2 implicated in small subunit binding display a large degree of flexibility. Furthermore, we find support for a transition state model conformation of the switch I region in this complex where the reoriented switch I interacts with a conserved rRNA region of the 40S subunit formed by loops of the 18S RNA helices 8 and 14. This complex is structurally distinct from the eEF2-bound 80S ribosome complexes previously published, and analysis of this map sheds light on the GTPase-coupled translocation mechanism.
Keywords: AlF4−, GDP, GTPase, Ribosome, Translocation
Introduction
Translocation is a major step in the elongation phase of translation during which, following peptide-bond formation, a coordinated movement by one codon of the mRNA·(tRNAs) complex occurs in the inter-subunit space of the ribosome. A- and P-site bound tRNAs move to the P and E sites, respectively, thus reinitiating the elongation cycle. The transition of the ribosome from the pre- to the post-translocational state (to be referred to as pre- and post-state) is catalyzed by a elongation factor (EF-G for prokaryotes, and eEF2 for eukaryotes), in a reaction stimulated by hydrolysis of GTP to GDP and inorganic phosphate 1.
Structural insight into the translocation mechanism has been derived on the basis of cryo-EM studies combined with X-ray crystal structures of the translocation promoting elongation factors EF-G 2,3 and eEF2 4. Cryo-EM studies on the bacterial ribosome complexes 5 and recent studies by single molecule FRET 6 established that binding of EF-G induces large-scale conformational changes in the ribosome, described as ratchet-like subunit rearrangement (RSR). Apparently, RSR has a pivotal role in the translocation mechanism for moving the mRNA with tRNAs.
Much less information has been available related to the eEF2-induced translocation in eukaryotic ribosomes until recently 1. Cryo-EM structures of eEF2-bound 80S ribosomes suggest overall functional similarity between eEF2 in the eukaryotic and EF-G in the prokaryotic system.8,9 The crystal structures of elongation factors 4 and structures of factor-bound ribosome complexes determined by cryo-EM 7; 8; 9 uncovered evidence of large conformational changes within the elongation factors, and suggested that the factor actively participates in the translocation mechanism. In parallel with the structural data, a wealth of kinetic data related to tRNA translocation are available in the prokaryotic system 10; 11; 12; 13; 14 which also indicated the occurrence of significant conformational changes in the ribosome as well as the elongation factors during the translocation process 15. Despite many biochemical, kinetic, and structural studies, the translocation mechanism has only been partially unraveled in molecular details 1.
With the aim of studying the GTPase coupled mechanism of translocation, and taking advantage of the fact that the unprogrammed 80S ribosome stimulates GTP hydrolysis in EF-G/eEF2 (Rodnina et al, 1997), we have formed a 80S:eEF2 complex stabilized by aluminium tetrafluoride (AlF4−) and GDP and determined its cryo-EM structure at 12.6 Å resolution. AlF4− mimics the gamma phosphate of GTP during hydrolysis in GTPases (see figure 1a,b), as documented in several crystal structures of small GTPases 16; 17. By this rationale GDP-AlF4− presumably traps ribosome-bound eEF2 in a transition state of GTP hydrolysis in the present complex.
Figure 1. Transition state eEF2 on the 80S ribosome.
(a,b) Schematic representations of the transition state of GTP during hydrolysis. The terminal phosphate is in a planar configuration (a), which is mimicked by AlF4− in our structure (b).
(c) Gel electrophoresis analysis. eEF2 was incubated with Tlan80S in the presence of either GDP, GDP and Sordarin, or GDP and AlF4−, and the amount of eEF2 bound following pelleting of ribosomes was quantified. The eEF2 band is indicated on the gel. The faster-migrating bands correspond to ribosomal proteins. (d) Cryo-EM reconstruction of the AlF4 complex. The inset shows a close-up view of the intersubunit space with eEF2 (red) binding regions. Weak densities at the 40S (yellow) interacting regions are marked with dashed circles, and strong interactions of the domains G’ and V with the stalk base region of the 60S (blue) are marked with asterisks. Landmarks for 40S: lf, left foot; rf, right foot; sh, shoulder; and for 60S: CP, central protuberance; sb, stalk base; st, extended stalk. Densities corresponding to the different domains of eEF2 are marked.
The first visualization of a transition state complex reveals that the GTP hydrolysis is accompanied by a coordinated interplay between factor and ribosome facilitating the completion of the translocation process and release of the factor. Underlying the current understanding of the translocation process is the hypothesis that a reverse ratcheting of the small subunit is necessary to complete the tRNA translocation 1; 5. The dynamic nature of some of the eEF2 domains seen in the present complex apparently assists in initiating the back-rotation of the ratcheted 40S subunit conformation by allowing the severing their contacts with this subunit.
Results
Cryo-EM Reconstruction of the 80S·eEF2·AlF4−GDP Complex
Using ribosomes and eEF2, both purified from the thermophilic fungus Thermomyces lanuginosus 18, we were able to form a complex by adding GDP and increasing concentrations of AlF4− (Figure 1c). The occupancy of eEF2 stalled by AlF4− and GDP was approximately 60–70% of the presumably maximum occupancy in the case of eEF2 trapped with the fungicide Sordarin.
Cryo-EM was used to generate a three-dimensional map of eEF2 bound to the 80S ribosome in the presence of AlF4− and GDP (Figure 1d). The resolution of the map was determined to be 12.6 Å (FSC = 0.5 criterion). The density corresponding to the ligand eEF2 is seen in the usual binding region of the elongation factors at the entrance of the intersubunit space (Figure 1d), 7; 8; 9; 19. We will be referring to this map in the following as the AlF4 complex. In order to confirm the role of AlF4− in stabilizing eEF2 on the ribosome, we visualized a complex made by incubating eEF2 and 80S ribosomes in the presence of only GTP (which quickly hydrolyzes to GDP). No density corresponding to the eEF2 is visible in this control map, suggesting that the occupancy of the ligand is very poor in that case. This observation corroborates the results of the binding assay shown in Figure 1c (showing only ~16% eEF2 binding to the ribosome in the presence of GDP), and confirms that AlF4− stabilizes eEF2 on the ribosome.
To analyze the functional state of the AlF4 complex we have compared this map with previously determined cryo-EM maps of the following complexes: (1) the 80S ribosome from T. lanuginosus, , referred to as ‘Tlan80S’ 18, (2) Tlan80S in complex with eEF2 stalled by the GTP analogue guanosine-(β-γ-imido)-triphosphate (GDPNP), referred to as ‘GDPNP complex’ 9, and (3) the Sordarin-stalled eEF2:80S ribosome from S. cerevisiae, referred to as ‘Sordarin complex’ 8. It is worth noting, in this context, that the binding of eEF2 to Tlan80S and Scer80S is very similar 9; 18.
Effect of GTP Hydrolysis on Switch I Conformation
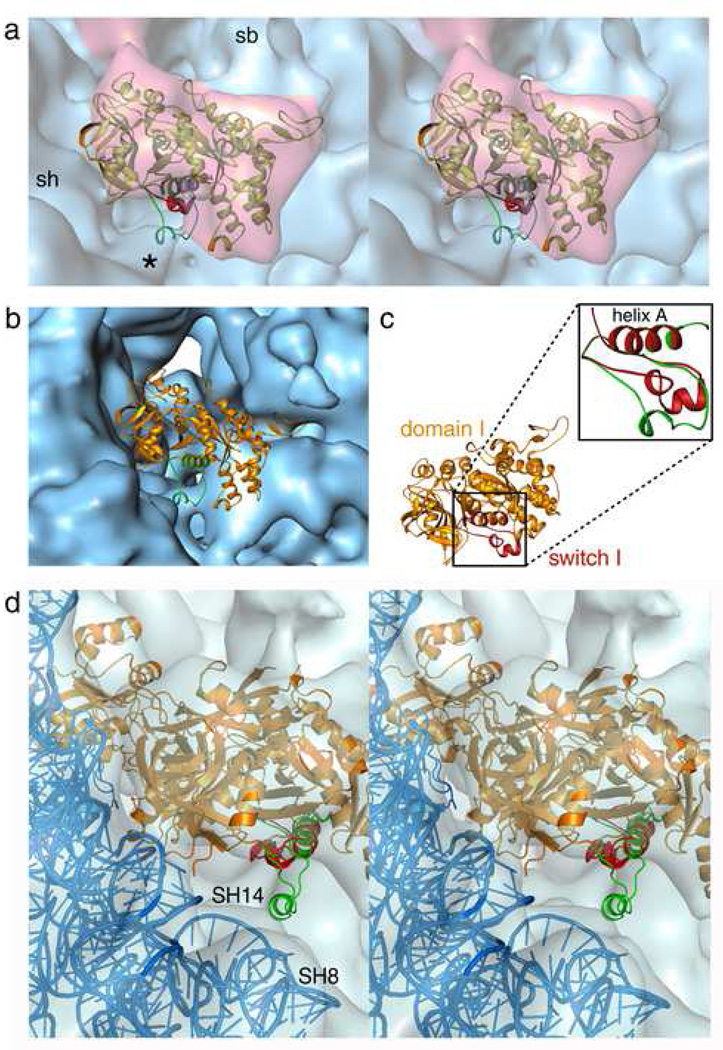
Upon GTP hydrolysis, while the core of the G domain remains unaltered, several conserved regions, particularly the switch I and switch II, and P-loop regions 20, are known to change conformation in G-proteins including elongation factors 21; 22; 23, indicating the existence of a common mechanism of GTP hydrolysis on the ribosome 9; 24; 25. Docking of the GTP form of switch 19 into the map of the AlF4 complex shows no density supporting this particular conformation of switch I (Figure 2), indicating that the initially well-ordered switch I loop has changed conformation in response to activation of GTP hydrolysis, as represented by the AlF4 transition state complex.
Figure 2. Conformational changes in the Switch I region.
(a) Stereo view of the domain I (G, G’) model (golden) fitted into the density attributed to eEF2 (semi-transparent red) in the AlF4 complex. The switch I structure in the GTP-form (red), and transition state model (green), clearly shows that the GTP-form is not fitted into the density, whereas extra density (marked with asterisks (*) corresponding to the short helical structure of the transition state model is visible at the shoulder region of the 40S subunit. (b) The same model of domain I docked onto the entrance of the intersubunit space of the control (empty) Tlan80S map to show that the extra density marked with asterisk in panel (a) in AlF4 complex is not associated with the control map. (c) Alignment of the switch I model in GTP-form (red) with the helix A of eEF2 domain I (golden). Inset: close-up view of the aligned models of switch I in GTP-form (red), and in transition state (green). (d) Stereo view of the eEF2 (orange), and the 40S model (blue) coordinates (pdb code: 1S1H) fitted into the sordarin post-complex map (semi-transparent blue 8). The switch I structure in GTP-form (red), and transition state model (green), both aligned to the domain I (helix A), clearly show that neither the GTP-form nor the transition state model fits into the density, and no extra density corresponding to the short helical structure of the transition state model is visible at the shoulder region of the 40S subunit.
We have modeled the switch I conformation for the GTPase transition state (Figure 2c,3a), based on available crystal structures of the heterotrimeric G-protein Gi-alpha, which has a switch I region of similar sequence as the translation GTPases (Figure 3a). The model shows part of the amino-acid sequence as a short helix, supported by extended coil structures of the preceding and following sequences. Direct alignment of this model structure with the eEF2 structure, docked into the AlF4 complex (see Methods), places the short helix in contact with the shoulder region on the 40S subunit (Figure 2a). Interestingly, we observe an extra density consistent with the short helical structure at the shoulder of the 40S subunit, at the junction of helices 8 and 14 of the 18S rRNA (Figure 3b), which is clearly missing in the empty Tlan80S map (Figure 2b) as well as in GDPNP complex9. This observation indicates that the model structure of the switch I loop represents the conformation of the switch I region in the AlF4 complex quite well, and we therefore postulate that this model structure likely represents a GTPase transition state relevant to both EF-Tu/eEF1A and EF-G/eEF2.
Figure 3. Transition-state model of switch I, and its interactions in the AlF4 complex.
(a) Model building of the switch 1 region in the transition state. The structure of Giα1 (PDB 1AS2), based on which the model was built, is shown in a wheat cartoon representation. The switch 1 region, the GDP nucleotide and γ phosphate (liberated, but trapped) are shown in red. A large insertion domain is present in switch 1 of Giα1. The G-domain of eEF2 (from PDB 1N0U) is superimposed on Giα1 and shown in slate-blue. Modelling of the transition state conformation of eEF2 is based on this conformation. The small helix of switch 1, which is generally observed in translation GTPases, docks into an additional density observed in our 80S:eEF2:GDP:AlF4− complex.
(b) Interaction of the transition state switch I conformation (green) in the AlF4 complex with the junction of small subunit rRNA helices 8 (blue) and 14 (red) shown in stereo representation. The short helix of the switch I structure (marked with asterisk) is placed close to the conserved tetraloops of the rRNA helices.
Since Sordarin stalls the eEF2-bound ribosome in a post-GTP hydrolysis state, we closely examined the density map of the Sordarin complex8 with the transition state model of the switch I region (Figure 2d). No density corresponding to the aforementioned short helix is seen at the shoulder region of the small subunit.
In contrast to the switch I, densities corresponding to the switch II and P-loop, the other two nucleotide binding regions, are inseparable from the rest of the density in the present resolution of the map, making any conformational changes in these regions indistinguishable at the current resolution.
Structural dynamics of eEF2
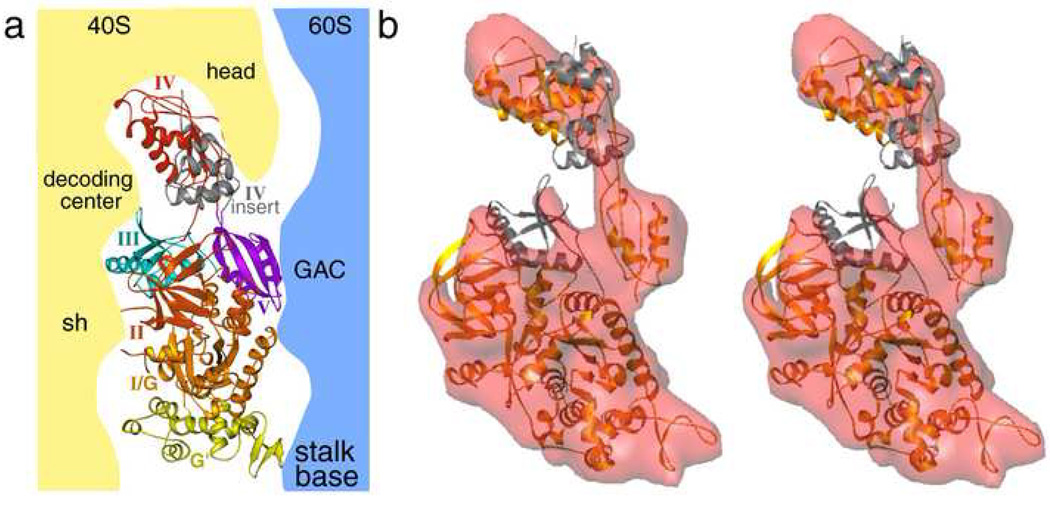
The eEF2 crystal structure 4 revealed that the overall five-domain architecture of EF-G is conserved in its eukaryotic counterpart. In addition, eEF2 possesses insertions in domains I (G and G’ domains) and IV. In previously-determined GDPNP and Sordarin complexes 8; 9 the interaction between eEF2 and the ribosome were seen to involve both ribosomal subunits and all five domains of the factor (Figure 4a).
Figure 4. Docking of the eEF2 atomic model into the cryo-EM density.
(a) Interactions of different domains of eEF2 with ribosomal subunits (40S, yellow; 60S, blue) as seen in GDPNP complex. (b) Stereo view of the fitting of the eEF2 crystal structure (golden) in the density (semi-transparent red) attributed to the factor isolated from the cryo-EM map of the AlF4 complex. Densities corresponding to the domains III and IV-insert regions (colored gray) are not well defined.
To interpret the conformation of the eEF2 in molecular terms, we have generated a quasi-atomic model of the ribosome-bound eEF2 in the AlF4 complex, by docking the eEF2 crystal structure into the segmented density that corresponds to the factor (Figure 4b). In the AlF4 complex the strong densities corresponding to domains I (G and G’) and V are clearly visible, showing contacts with the 60S subunit (also see Figure 1b). In contrast, the densities corresponding to the insertion regions in domain IV, which interacts with the head region (helix 33 of 18S rRNA), and domain III, which contacts the decoding region of the 40S subunit (rpS23 (S12p)), are either weak or fragmented (Figure 1b, 4b), resulting in a very approximate docking of these domains (See Figure 4b). Furthermore, while a strong density is visible for the bulk of domain II (packed with domain I), the densities for the regions that interact with the helices 5 and 15 of 18S rRNA appear weaker. The sites of interaction between eEF2 domains and ribosomal subunits in the various complexes are summarized in Supplementary Table 1. The observation of missing densities for specific domains prompts us to conclude that in the AlF4 complex the domains implicated in 40S subunit binding possess a high degree of flexibility, causing the densities to appear less defined.
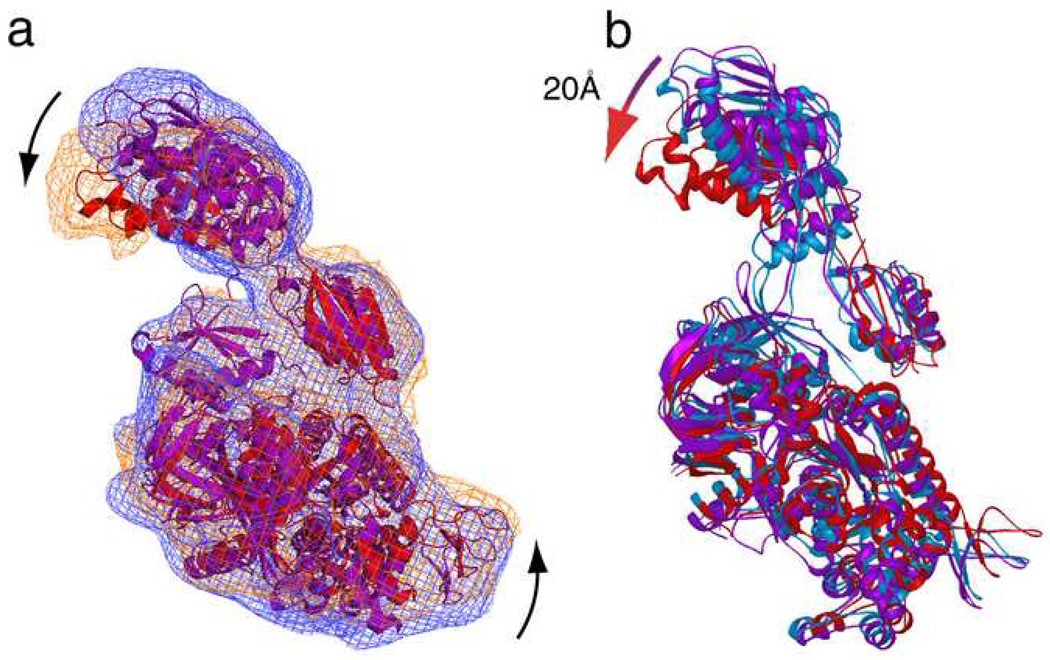
The density corresponding to the core of domain IV is fused with the 40S subunit density at the decoding site, suggesting the existence of close contacts (Figure 1b). Juxtaposition of the isolated densities as well as the fitted coordinates of eEF2 in the GDPNP 9 and AlF4 complexes reveals that, while the cores of domains I and II are very well aligned, significant positional changes are confined primarily to the G’ domain and domain IV (Figure 5a). Our structure shows that during GTP hydrolysis the G' domain moves toward the GAC, whereas the tip of domain IV moves in the opposite direction. The direction of displacements in domains G’ and IV from the GDPNP to the AlF4 complex is very similar as in the displacements observed by Taylor and coworkers 9 from the GDPNP complex to the Sordarin complex, but the range of the movement, particularly in the tip of domain IV, is much larger now (Figure 5b). Sordarin apparently traps eEF2 between the GDPNP state and the AlF4 state, indicating that the Sordarin complex likely may not reflect an actual post state of GTP hydrolysis.
Figure 5. Comparison of eEF2 conformations in AlF4, GDPNP and sordarin complexes.
(a) Isolated densities and corresponding fitted models of eEF2 for GDPNP (Taylor et al 2007) (density, blue mesh; model, purple) and AlF4 (density, orange mesh; fitted model, red) complexes. Positional displacements of G’ and tip of domain IV are marked with arrows. (b) The eEF2 models for the GDPNP (purple), Sordarin (blue), and AlF4 (red) complexes are superimposed.
The movement of domain IV triggered by GTP hydrolysis places the tip of eEF2 domain IV in the AlF4 complex onto inter-subunit bridge B2a 26 (Figure 6), which, in the eukaryotic ribosome, is made by minor-groove interactions between SH44 of 18S rRNA and LH69 of 28S rRNA 27 (rRNA helices are denoted as 'SH' for the 18S rRNA from the small subunit, or 'LH' for the 28S rRNA from the large subunit).
Figure 6. Interaction of domain IV with the intersubunit bridge b2a.
The interaction of eEF2 domain IV (colored red) with the bridge b2a (SH44, golden; LH69, blue) in the AlF4 complex is shown in a stereo representation. The tip of the domain IV contacts the ribosome where SH44 and the apical loop of LH69 meet to form the bridge b2a.
Discussion
Our results fit very well into the model for the translocation mechanism proposed based on current knowledge of the process 1; 9. According to this model, once the tRNAs are settled in the P and E sites, the small subunit must release the mRNA·tRNA complex and ratchet back on its own for completion of the translocation process. The parts of the domains that are not well-defined in the AlF4 complex are those components that lock the head and body of the 40S subunit and thereby stabilize their relative positions, implying that the stabilizing contacts are weakened as a result of GTP hydrolysis so as to allow the back-ratcheting. However, another interpretation is that the partial disorder is due to the absence of A- and P-site tRNA in our complex, and that under physiologically more relevant conditions, with a tRNA•mRNA actually being translocated, eEF2 may attain a specific conformation in the transition state. Nevertheless our data clearly demonstrate that eEF2 undergoes a conformational change directly coupled to GTP hydrolysis, since the GDPNP and Sordarin complexes both adopt well-defined conformations.
The complexes, stalled either by Sordarin or AlF4−, in the presence of GDP, represent intermediate states following GTP hydrolysis. However, our analysis shows that the two reagents employed capture two structurally distinct states. The fungicide Sordarin binds at the interface of domain III, IV, and V 4, and thereby restricts the movement of these domains. As revealed by the comparison of eEF2 conformations in all three states, Sordarin does not allow the full-scale movement of the domains G’ and IV accompanied by the GTP hydrolysis, and thereby traps eEF2 on the ribosome in an intermediate GTP hydrolysis state. It may be pointed out in this context that, in addition to its interaction with the 40S subunit, eEF2 domain III is found to contact the SRL of the large subunit in the Sordarin complex 8, but this interaction is seen neither in GDPNP, nor in AlF4 complexes. Furthermore, an intersubunit bridge B6 (involving helix 14 of the 18S rRNA of the 40S subuit and rpL23 (L14p) of the 60S subunit), placed close to this interaction site, is reported missing in the Sordarin complex 8, whereas this bridge is clearly visible in the GDPNP as well as in AlF4 complex. It has been postulated 8 that, in the Sordarin complex, the domain III-SRL interaction -- which prevents domain III from moving away from the SRL after GTP hydrolysis and thereby prevents the dissociation of eEF2 from the ribosome -- might play a crucial role in the inhibitory mechanism of Sordarin. Our observation further supports the idea that eEF2 domain III is reoriented and engaged in an unusual position in the Sordarin complex, thus inhibiting the release of the factor.
As suggested by Taylor and coworkers 1; 9, the movement of the tip of domain IV most likely results in the disruption of the critical connection formed during the decoding process between the mRNA-tRNA subcomplex with the flipped-out bases of helix44 located in the decoding center of the small subunit 28. Uncoupling of this link in turn allows the head of the small subunit to rotate in the direction of translocation, a movement that presumably drags the hybrid tRNAs to the final, translocated positions 9. Moreover, the interaction of the tip with the bridge b2a may have an instrumental role in assisting back-ratcheting of the small subunit, keeping the b2a interactions (between SH44 and LH69) unperturbed.
Finally, the interaction of the reoriented switch I structure in the transition state with the 40S subunit is intriguing. The packing interaction between GAAA and UACG tetraloops of helices 8 and 14 near the subunit interface, where the switch I structure interacts, is highly conserved 29. Moreover a recent mutational study 30 detected a deleterious mutation at the conserved tetraloop of helix 8 (A161G) having inhibitory effect on translation. It is therefore tempting to speculate that this conserved rRNA region plays a crucial role in the activity of GTP-dependent factors on the ribosome.
In summary, our analysis of the AlF4 complex provides glimpses of the trajectory of conformational changes in the eEF2 structure initiated by the hydrolysis of GTP, by showing a global change in the arrangement of domains and a local change in the switch I structure as the latter adopts a transition-state conformation. Thus the present study offers structural evidence that the AlF4 complex represents a transition state of eEF2 trapped at the moment of GTP hydrolysis. The stepwise engagement of eEF2 with the ribosome may be seen as a confirmation of the ‘locking-unlocking’ scenario previously proposed 31,32, according to which association and dissociation of the elongation factor to ribosome are accompanied by intermediate steps involving partial breaking and re-making of contacts between the ligand and target molecule.
Materials and Methods
80S ribosomes and eEF2 were purified from the fungus T. lanuginosus 18. Binding of eEF2 to 80S procedure is as follows: 5 µg of 80S ribosomes were incubated with 2 µg of eEF2 in 100 µl of buffer A (20 mM Hepes-NH3 pH=7.2, 100 mM KCl, 20 mM MgCl2). The final nucleotide concentration was 100 µM and that of Sordarin was 100 µM. The AlF4 concentration was varied from 0–400 µM. The reactions were incubated on ice for 20 minutes and then placed on top of a 50 µl 30% sucrose cushion in buffer A containing the same composition with regard to nucleotide, Sordarin and AlF4− as the reaction.
The grids for cryo-electron microscopy were prepared following standard procedures: micrographs were recorded on a Philips FEI (Eindhoven, The Netherlands) Technai F20 field emission gun electron microscope at 200 kV and a magnification of 50,000 (±2%) following standard low-dose procedures. The micrographs were digitized with a step size of 14 µm on a Zeiss/Imaging scanner (Z/I Imaging Corporation, Huntsville, AL), the final pixel size corresponded to 2.82 Å on the object scale.
The image processing using SPIDER 33 included a 3D projection alignment procedure with correction of the contrast transfer function and enhancement of the high-resolution Fourier amplitudes based on X-ray solution scattering data. The density corresponding to the ligand (eEF2) was low compared to the ribosome, indicating lower than 100% occupancy. We therefore used a supervised classification method (method described in ref 34) to identify a subpopulation with high occupancy of the ligand. For the final reconstruction a total number of 28,242 ribosome particles were selected to obtain the refined 3D cryo-EM map of the Tlan80S-AlF4-GDP complex (AlF4 complex). The resolution was estimated as 12.6Å using a cutoff of 0.5 in the Fourier shell correlation (supplementary Fig 1). We reconstructed a ~19Å control map following the exact same procedure of AlF4 complex except for the addition of AlF4−, and this map does not show any density corresponding to the eEF2.
We used the X-ray structure of eEF2 from S. cerevisiae 4, which has high sequence homology with T. lanuginosus eEF2, to manually dock into the cryo-EM density map of AlF4 complex using the software ‘O’ 35. Each of the domains were fitted individually, as a rigid body, except the domain I (G and G’) and II, which were fitted as a single unit. The quasi-atomic model of the 80S ribosome (pdb code: 1S1H, 1S1I) for the Sordarin complex 8 was also used to determine the molecular interactions between eEF2 and ribosome.
The switch I region is absent in the available crystal coordinates of eEF2. We have taken the coordinates of this region for the GTP state from a crystal structure of the EF-Tu ternary complex in complex with GDPNP (pdb code: 1TTT). The switch I conformation for the GTPase transition state was modeled based on available crystal structures of the G-protein Gi-alpha (pdb code 1AS2) making allowance for deletions and additions as appropriate. The coordinates of the switch I region (for both the GTP state, and the transition state) were aligned to the helix A of the eEF2 coordinates which is located next to the switch I sequence.
The graphic visualizations were done with the programs IRIS Explorer (The Numerical Algorithms Group Ltd. Downers Grove, Illinois, USA), PyMol (DeLano Scientific, San Carlos, CA), and Ribbons 36.
Supplementary Material
Acknowledgments
We thank Diana Lalor for helping in cryo-EM data processing, Michael Watters for preparing the illustrations, and Derek Taylor for valuable discussions. This work was supported by the Human Frontier Science Program (to P.N.) and by the Howard Hughes Medial Institute and grants NIH R37 GM 29169 and R01 GM 55440 (to J.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The 12.6 Å and 15.3 Å cryo-EM maps of the AlF4 complex (80S.eEF2.AlF4−.GDP) have been deposited in the 3D-EM database, EMBL-European Bioinformatics Institute, Cambridge, UK (http://www.ebi.ac.uk/msd-srv/emsearch/index.html) with accession codes EMD-5015 and EMD-5016, respectively. The eEF2 density was segmented out from the 12.6Å cryo-EM map and deposited in the same Data Bank with accession code EMD-5017.
References
- 1.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci U S A. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 5.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 6.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieden HJ, Gromadski K, Rodnin D, Rodnina MV. Mechanism of elongation factor (EF)-Ts-catalyzed nucleotide exchange in EF-Tu. Contribution of contacts at the guanine base. J Biol Chem. 2002;277:6032–6036. doi: 10.1074/jbc.M110888200. [DOI] [PubMed] [Google Scholar]
- 11.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 12.Rodnina MV, Savelsbergh A, Wintermeyer W. Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol Rev. 1999;23:317–333. doi: 10.1111/j.1574-6976.1999.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 13.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 14.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci U S A. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- 16.Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 17.Wittinghofer A. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr Biol. 1997;7:R682–R685. doi: 10.1016/s0960-9822(06)00355-1. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson J, Sengupta J, Gursky R, Nissen P, Frank J. Comparison of fungal 80 S ribosomes by cryo-EM reveals diversity in structure and conformation of rRNA expansion segments. J Mol Biol. 2007;369:429–438. doi: 10.1016/j.jmb.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 20.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 21.Abel K, Yoder MD, Hilgenfeld R, Jurnak F. An alpha to beta conformational switch in EF-Tu. Structure. 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- 22.Liljas A, A AE, al-Karadaghi S, Garber M, Zheltonosova J, Brazhnikov E. Crystallographic studies of elongation factor G. Biochem Cell Biol. 1995;73:1209–1216. doi: 10.1139/o95-130. [DOI] [PubMed] [Google Scholar]
- 23.Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- 24.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 26.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 27.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 28.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 29.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 30.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc Natl Acad Sci U S A. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spirin AS. Ribosome as a molecular machine. FEBS Lett. 2002;514:2–10. doi: 10.1016/s0014-5793(02)02309-8. [DOI] [PubMed] [Google Scholar]
- 32.Bretscher MS. Translocation in protein synthesis: A hybrid structure model. Nature. 1968;218:3. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 33.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 34.Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Carson M. Ribbons 2.0. J. Appl. Crystallog. 1991;24:958–961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.