Abstract
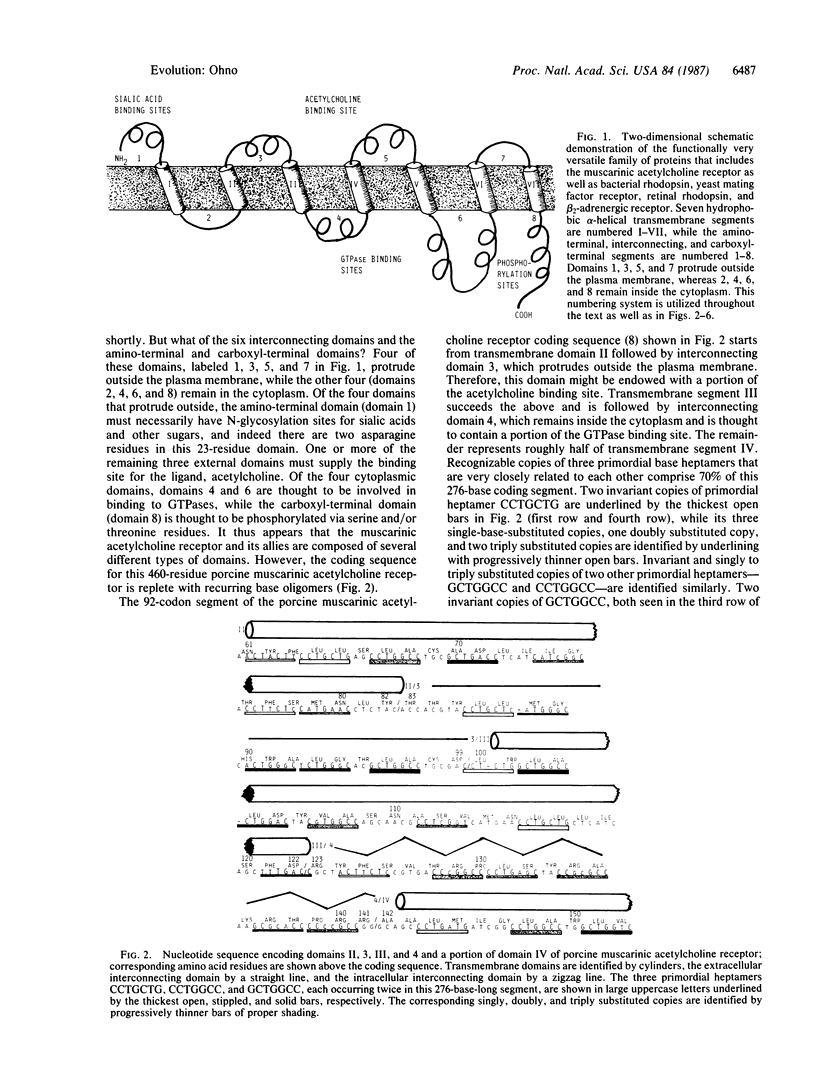
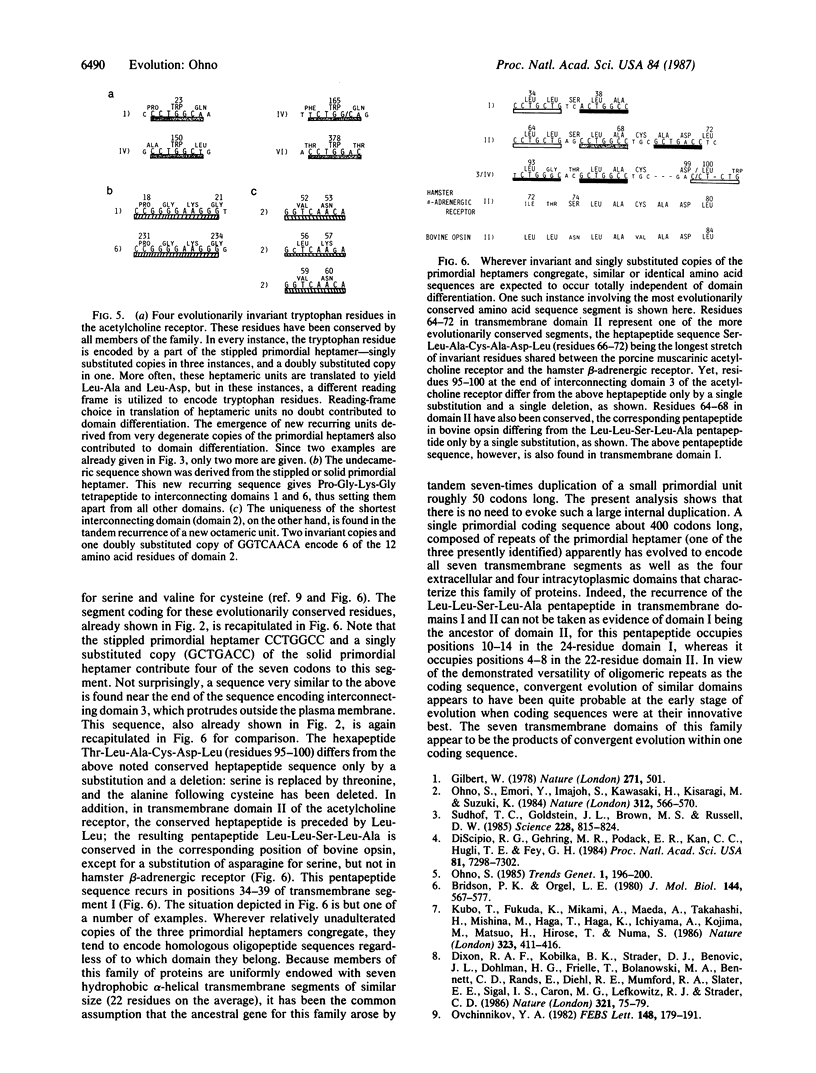
One of the more popular concepts to emerge in recent years is that new proteins evolved by domain exchanges between preexisting proteins. The presence of introns within eukaryotic genes is thought to enhance such exchanges. Yet domain exchanges must necessarily be the secondarily developed process in evolution, for they would have been effective only after multitudes of domains came into being. Many of the proteins with functionally divergent domains were established before the division of prokaryotes from eukaryotes; i.e., soon after the creation of life on this earth. I attribute the extreme innovativeness of early coding sequences to their construction; i.e., being repeats of oligomeric units. The rhodopsin family of proteins--with seven hydrophobic, alpha-helical transmembrane domains, four extracellular domains, and four intracytoplasmic domains--indeed arose before the division of prokaryotes from eukaryotes and later gave rise to muscarinic acetylcholine receptor and beta-adrenergic receptor among others. In this paper, I show that the entire coding sequence for porcine muscarinic acetylcholine receptor is still replete with copies of three heptameric units that are very closely related to each other. Original heptameric units are more stringently conserved in parts encoding the seven transmembrane domains, whereas new repeating units are comingled with the old in parts encoding extracellular and intracytoplasmic domains.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bridson P. K., Orgel L. E. Catalysis of accurate poly(C)-directed synthesis of 3'-5'-linked oligoguanylates by Zn2+. J Mol Biol. 1980 Dec 25;144(4):567–577. doi: 10.1016/0022-2836(80)90337-x. [DOI] [PubMed] [Google Scholar]
- DiScipio R. G., Gehring M. R., Podack E. R., Kan C. C., Hugli T. E., Fey G. H. Nucleotide sequence of cDNA and derived amino acid sequence of human complement component C9. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7298–7302. doi: 10.1073/pnas.81.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Kobilka B. K., Strader D. J., Benovic J. L., Dohlman H. G., Frielle T., Bolanowski M. A., Bennett C. D., Rands E., Diehl R. E. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986 May 1;321(6065):75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Kubo T., Fukuda K., Mikami A., Maeda A., Takahashi H., Mishina M., Haga T., Haga K., Ichiyama A., Kangawa K. Cloning, sequencing and expression of complementary DNA encoding the muscarinic acetylcholine receptor. Nature. 1986 Oct 2;323(6087):411–416. doi: 10.1038/323411a0. [DOI] [PubMed] [Google Scholar]
- Ohno S., Emori Y., Imajoh S., Kawasaki H., Kisaragi M., Suzuki K. Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature. 1984 Dec 6;312(5994):566–570. doi: 10.1038/312566a0. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA Rhodopsin and bacteriorhodopsin: structure-function relationships. FEBS Lett. 1982 Nov 8;148(2):179–191. doi: 10.1016/0014-5793(82)80805-3. [DOI] [PubMed] [Google Scholar]
- Südhof T. C., Goldstein J. L., Brown M. S., Russell D. W. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985 May 17;228(4701):815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]



