Abstract
In this study 3-D poly(ethylene glycol) (PEG) hydrogel arrays were used to screen for the effects of fibroblast growth factor-2 (FGF2), combined with multiple hydrogel matrix parameters, on human mesenchymal stem cell (hMSC) viability and spreading. In particular, we examined the effects of FGF2 while co-varying hydrogel matrix degradability, cell adhesion ligand type, and cell adhesion ligand density. FGF2 significantly improved viability of hMSCs in a dose-dependent manner in both non-degrading and degrading PEG hydrogels in the absence of extracellular matrix (ECM)-derived cell adhesion ligands. The presence of a small molecule that inhibits autophosphorylation of the FGF2 receptor blocked the effects of FGF2 on hMSC viability in PEG hydrogels, both in the presence and absence of the RGDSP ligand. FGF2 effects on hMSC viability were less pronounced when FGF2 was presented in combination with the RGDSP cell adhesion ligand or the IKVAV cell adhesion ligand in non-degrading PEG hydrogels. Importantly, spread hMSC morphologies were observed and quantified in a select subset of hydrogel networks, which were degradable and included both FGF2 and RGDSP. These results indicate that the hydrogel arrays described here can be used to efficiently study the influence of soluble and insoluble hydrogel matrix parameters on stem cell behavior, and to identify synthetic, 3-D environments that promote specific hMSC behaviors.
Keywords: extracellular matrix, mesenchymal stem cells, fibroblast growth factor, poly(ethylene glycol), bFGF, tissue engineering
Introduction
A variety of recent studies in biomaterials science and tissue engineering have focused on three-dimensional (3-D) culture of stem cells in hydrogels. A goal of these studies is to determine the influence of extracellular matrix (ECM)-derived signals on stem cell behavior, and to ultimately generate materials in which stem cell phenotype is controlled. A subset of these 3-D culture studies has focused on creating well-defined signaling environments within stem cell-laden hydrogels. For example, poly(ethylene glycol) (PEG) hydrogels have been used as a platform to present small molecules1, peptides2,3, polysaccharides4, or proteins3 to stem cells while minimizing non-specific interactions with other serum-derived or cell-secreted biological molecules, as the PEG chains do not intrinsically interact with proteins5. For example, Anseth and coworkers have used PEG hydrogels as a model to demonstrate that the ECM-derived cell adhesion peptide Arg-Gly-Asp (RGD)6, the polysaccharide heparin4, and the corticosteroid dexamethasone1 can each influence the viability and differentiation of mesenchymal stem cells. Therefore, PEG hydrogels, and other similarly “bioinert” hydrogel networks (e.g. alginate7, agarose8), have served as adaptable platforms to study the effects of ECM-derived signals on stem cell behavior in 3-D culture. However, it remains a challenge to efficiently identify the optimal signaling environments that encourage stem cell viability, lineage-specific differentiation, and tissue formation.
A particular challenge facing 3-D stem cell culture studies is the large number of ECM-derived signals and signal combinations that can influence stem cell phenotype. Previous studies in 3-D culture indicate that cell adhesion ligands (e.g. RGD6, YIGSR9, IKVAV2), growth factors (e.g. TGF-β310, TGF-β111, BMP212), and material degradation13,14 each influence hMSC viability and differentiation. Furthermore, combinations of these distinct signals are likely to uniquely influence stem cell phenotype, as demonstrated recently in 2-D cell culture studies that have simultaneously explored the effects of osteogenic induction media and defined substrate compliance on hMSCs15. Taken together, previous studies indicate that a broad range of signals and signal combinations can affect stem cell phenotype. However, due to practical limitations, hydrogel-based stem cell culture formats have typically focused on examining the effects of only a single ECM-derived signal or a small subset of signals. Although these previous studies provide important insights into the signals that can influence stem cell behavior, they are not capable of exploring the broad ranges of signal amplitudes or complex signal combinations that may be most relevant to in vivo environments. Therefore, there is a need to develop experimental approaches that enable investigators to rapidly examine complex, biologically-relevant signaling environments.
To address current limitations in 3-D stem cell culture, we and others have recently developed 3-D cell culture systems with enhanced throughput capabilities. One approach has used photolithographic methods to generate spatially patterned hydrogel structures with distinct regions that contain specific cell types16, cell adhesion ligands17, or ECM chemistries17. For example, Pishko and coworkers have used photopolymerization within microchannels17 or spots18 to generate PEG microstructures, and demonstrated that multiple mammalian cell types remain viable in hydrogels for up to 7 days. Khademhosseini and coworkers have used soft lithography to generate agarose or PEG microstructures loaded with viable cells19. Jongpaiboonkit et al. recently used an automated liquid handling approach to generate PEG hydrogel arrays, which present an adaptable milieu of ECM-derived signals to multiple cell types, including hMSCs2,20. Taken together, these previous studies demonstrate that it is possible to create spatially patterned hydrogels for 3-D stem cell culture, and that these hydrogels are promising platforms for enhanced throughput stem cell culture studies.
In this study we used 3-D PEG hydrogel arrays to screen for the influence of the soluble protein fibroblast growth factor-2 (FGF2) on human mesenchymal stem cell (hMSC) phenotype (Fig. 1). FGF2 has been previously shown to promote survival and proliferation of undifferentiated MSCs21-24, and has therefore been commonly used as a culture supplement for MSC expansion. In addition, previous studies have indicated a key role for FGF2 in MSC migration25 and in maintenance of MSC differentiation potential26-28. Based on the important role of FGF2 in defining MSC phenotype in 2-D culture and in vivo, we hypothesized that FGF2 could have a significant, dose-dependent influence on hMSC viability and morphology in 3-D hydrogel matrices. Therefore, this study examined the effects of various concentrations of soluble FGF2 on hMSCs, while co-varying network degradability, cell adhesion ligand type, and cell adhesion ligand density (Fig. 1). We specifically examined the fibronectin-derived Arg-Gly-Asp-Ser-Pro (RGDSP) sequence and the laminin-derived Ile-Lys-Val-Ala-Val (IKVAV) sequence based on our previous observation that these ligands promote hMSC viability in 3-D PEG hydrogels in a dose-dependent manner2,14. The results presented here provide an initial demonstration that FGF2 influences hMSC viability and spreading in a 3-D hydrogel matrix.
Figure 1.
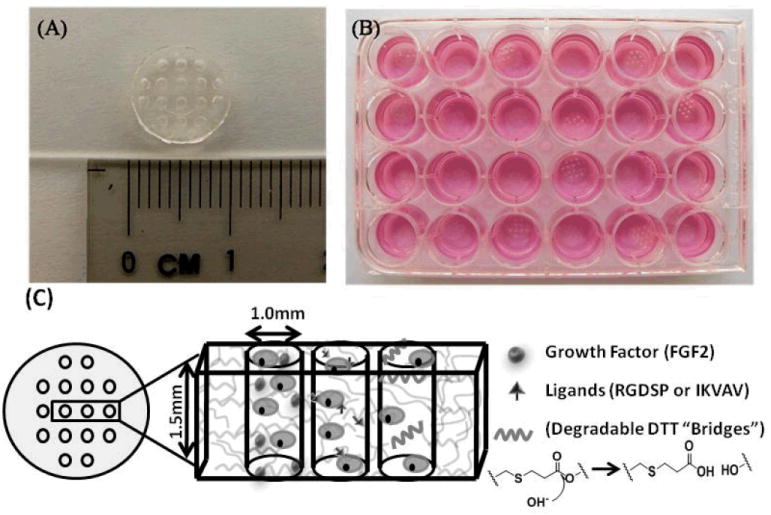
A) 8kDa PEG hydrogel array “background” showing an array of 16 cylindrical spots. B) Image demonstrating 24 hydrogel arrays within 24-well tissue culture plate. C) Schematic demonstrating incorporation of stem cells, growth factors, peptide ligands, and hydrolytically degradable DTT “bridges” into hydrogel array spots.
Materials and Methods
Preparation of synthetic PEG hydrogel arrays
8kDa Poly(ethylene glycol) (PEG) was purchased from Sigma-Aldrich. The synthesis of PEG-diacrylate (PEGDA) was performed as described elsewhere29. PEG hydrogel arrays were prepared as described previously2,20. First, hydrogel array “background” (Fig. 1) was prepared by mixing 10 wt% PEGDA and 0.05%w/v photoinitiator Irgacure 2959 (I2959, BASF, Ludwigshafen, Germany) in serum free medium, and was passed through a 0.22 μm filter (Fisher Scientific, Fair Lawn, NJ) for sterilization. Then the polymer solution was added to a teflon mold containing 16 cylindrical posts (1 mm diameter, 1.25 mm depth), and crosslinked via exposure to UV radiation (λ = 365 nm, intensity = 4.5 mW/cm2) for 5 min. The array spots were then automatically filled with the aforementioned polymer solution using an automated liquid handler, with hMSCs and various biological molecules at various concentrations (as detailed below).
In some experiments, PEG hydrogel arrays were designed to degrade hydrolytically over time using an approach described elsewhere2,14,20. Briefly, 8kDa PEGDA chains were reacted with 2.5 or 5mM of dithiothreitol (DTT, Fisher Scientific, Fair Lawn, NJ) in serum-free medium at 37°C for 60 minutes to form water-soluble, acrylate-terminated (PEG-DTT)n-PEG conjugates. hMSCs were then added to the polymer solution, and the solution was photocrosslinked by UV radiation as described above to create PEG hydrogels with degradable “DTT bridges” included. The ester bonds adjacent to thioether groups in these bridges degrade hydrolytically, and the concentration of the bridges therefore dictates the hydrolytic degradation rate of PEG network2,14,20. As a result, PEG hydrogel arrays containing DTT bridges are referred to as “degrading” networks in subsequent sections of this manuscript, while PEG hydrogel arrays without DTT bridges are referred to as “non-degrading”.
In some experiments, acrylate-terminated PEG chains or (PEG-DTT)n-PEG conjugates were reacted with peptides containing the fibronectin-derived cell adhesion ligand RGDSP, or the laminin-derived cell adhesion ligand IKVAV to generate cell-interactive hydrogels. In these experiments a 10X excess of acrylate-terminated PEG chains (Mw=8kDa) or (PEG-DTT)n-PEG conjugates were incubated with a CGGRGDSP and/or a CGGIKVAV peptide for 90 minutes in PBS (37°C, pH=7.4) to allow for Michael-type addition of the cysteine sulfhydryl group to the acrylate group, as described previously14,30. The efficiency of the reaction was quantified by measuring free thiol content in a 2.5mM CGGRGDSP solution before and after its reaction with a 10X molar excess of 8k PEGDA using the Measure-IT Thiol assay (Invitrogen, Carlsbad, CA). The resulting solutions contained PEG-diacrylate and acrylate-PEG-CGGRGDSP and/or acrylate-PEG-CGGIKVAV molecules, which were subsequently photo-crosslinked to form cell-interactive hydrogels.
A standard protocol for solid phase peptide synthesis using Fmoc-chemistry was followed for the synthesis of CGGRGDSP and CGGIKVAV. Peptides were synthesized on Rink Amide resin (.72meq functional amine group/g) (Novabiochem, San Diego, CA) at a scale of 0.2 mmol on a C036s automated peptide synthesizer (CSBio, Menlo Park, CA). First, amino acid couplings were performed by introducing a 2.5X molar excess of Fmoc-protected amino acids (Novabiochem, San Diego, CA) activated with N-Hydroxybenzotriazole • H2O (Advanced Chemtech, Louisville, KY) and N,N-diisopropylcarbodiimide (Anaspec, San Jose, CA) to the resin in sequencing grade dimethylformamide (DMF) (Fisher Scientific, Fair Lawn, NJ). Prior to each coupling, Fmoc-protecting groups were removed using a 20% solution of Piperidine by volume (Sigma-Aldrich, St. Louis, MO) in DMF. Upon completion of synthesis, peptides were cleaved from the resin using a solution of 95% Trifluoracetic Acid (Fisher Scientific, Fair Lawn, NJ), 2.5% Triisopropylsilane (Aldrich, St. Louis, MO), and 2.5% H20 and precipitated into 4°C ethyl ether (Fisher Scientific, Fair Lawn, NJ). The peptides were precipitated 3X in ethyl ether and left for two days to dry. The molecular weights of the peptides were verified on a Bruker REFLEX II MALDI-ToF mass spectrometer (Bruker, Billerica, MA) and via HPLC (Shimadzu Scientific Instruments Inc., Columbia, MD). In some experiments, hydrogel array spots were prepared in the presence of soluble FGF2 at various concentrations. In these experiments, the FGF2 was added as a soluble component to the hydrogel precursor solution.
Biological characterization of hydrogel arrays
(i) Cell culture
hMSCs (Cambrex Bio Science, Walkersville, MD, passage 6) were cultured in Minimum Essential Medium, Alpha (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Cambrex), 2.2 g/l NaHCO3 (ACROS Organics, Morris Plains, NJ), and 100 units/ml of penicillin/streptomycin. Cell cultures were maintained at 37°C/5% CO2 and media was replaced every 3-4 days. hMSCs were grown at low density using the method described previously by Sotiropoulou et al.24 to maintain hMSC multipotential.
(ii) Cell-seeding within PEG hydrogel arrays
Cells were photoencapsulated in a 10 wt% polymer solution (final concentration) in serum free media at a seeding density of 5×105 cells/ml. The cell/polymer solution (1μl) was pipetted in the wells of the hydrogel array using a Gilson automated liquid handler (Model: 223 Sample Changer) and ‘Trilution LH version 1.2’ control software (Gilson Inc., Middleton, WI) at a rate of approximately 5 spots per minute. Upon ultraviolet light exposure for 3 min, PEG-based hydrogels were cross-linked and cells were physically entrapped within the networks. The arrays were then placed in media and cultured at 37°C and 5% CO2, replacing media every 2-3 days.
To provide insight into FGF2 diffusion out of hydrogel arrays over time, a protein transport experiment was performed. The protein Lysozyme, chosen as a model protein due to its similar molecular weight (14.7kDa) to FGF2 (17.2kDa), was fluorescently-labeled with NHS-rhodamine using the manufacturer’s protocol (Pierce, Rockford, IL). Rhodamine labeled lysozyme was placed in 10,000 molecular weight cutoff dialysis tubing (Pierce, Rockford, IL) and dialyzed against ultrapure H2O (18 MΩ resistivity). 100μM Rhodamine-Lysozyme was loaded into hydrogel wells (n=4), which were then placed in phosphate buffered saline. Fluorescent micrographs were acquired using an inverted, compound fluorescence microscope (IX51, Olympus) at 40X magnification at predetermined times, and the buffer was replaced after each image acquisition. The fluorescence intensity was measured using ImageJ software (Freeware, NIH, Bethesda, MD).
To confirm that the effects observed in the presence of FGF2 were indeed due to FGF2 signaling, a subset of cell culture experiments were performed in the presence of a soluble inhibitor of FGF2 receptor autophosphorylation (PD 173074 molecule, Sigma-Aldrich, Milwaukee, WI). The PD 173074 molecule inhibits FGF2 signaling by binding to the receptor tyrosine kinase domain of FGF receptors (FGFRs), including FGFR131 and FGFR332, and preventing their autophosphorylation33. PD 173074 has been shown to specifically inhibit the effects of FGF2 on neural cell proliferation and differentiation34 and adipose-derived mesenchymal stem cell proliferation and differentiation potential35. In our FGF2 inhibition experiments, 100nM of the soluble PD173074 molecule was included in the culture medium used to make the background arrays as well as the medium bathing the arrays. Fresh medium containing 100nM PD173074 was added to the arrays 1 and 3 days post encapsulation. We chose 100nM as the concentration of the soluble inhibitor because this concentration of inhibitor was above the IC50 (5nM) of the PD 173074 molecule, but below the concentration at which this inhibitor is toxic to cells based on our toxicity study (see Supplementary Fig. 2 and associated text for details).
(iii) Cell viability within hydrogel arrays
After photoencapsulation, the arrays were removed from culture at various time points (1, 3, 5, and 7 days) and were stained using the LIVE/DEAD assay (Invitrogen Corp., Carlsbad, CA) according to the manufacturer’s instructions. This assay identifies esterase activity in live cells via green fluorescence emission from calcein AM and nuclear permeability in dead cells via red fluorescence emission from ethidium homodimer-1. Cell viability was visualized using the micrographs from the inverted microscope and the total percentage of viable cells was determined by ImageJ software analysis of live and dead cells in photomicrographs for at least 4 images per condition. In some conditions, cell morphology in hydrogel array spots was also characterized qualitatively using the same fluorescence microscope. The results are expressed as mean ± standard deviations for 4 samples per condition. Differences between data sets were assessed by one-way ANOVA analysis. In some cases, Tukey’s two-way analysis was performed using the R software package. Regardless of the test, a p-value less than 0.05 was considered a statistically significant difference.
(iv) hMSC Spreading within Hydrogel Arrays
hMSC spreading was quantified using techniques used in previous studies to measure 3-D cell spreading: measurements of hMSC area4 and aspect ratio36. First, hMSC area was measured in four experimental conditions: 1) 0mM DTT+ 0mM RGD+ 0ng/ml FGF2, 2) 5mM DTT+ 0mM RGD+ 0ng/ml FGF2, 3) 5mM DTT+ 2.5mM RGD+ 0ng/ml FGF2, 4) 5mM DTT+ 2.5mM RGD+ 100ng/ml FGF2. More than 15 LIVE/DEAD-stained hMSCs in 40X fluorescent micrographs were analyzed per condition using ImageJ software and presented in histogram form. Next, hMSC aspect ratio was quantified in conditions in which spreading was observed. Here the aspect ratio is defined as the ratio between the longest end-to-end distance of each cell and the distance of the perpendicular axis of the same cell. More than 5 LIVE/DEAD-stained hMSCs in 200X fluorescent micrographs were analyzed per condition using ImageJ software and are presented in histogram form.
Results
Modeling FGF2 Diffusion out of Hydrogel Arrays
Lysozyme was used as a model protein to gain insight into the diffusion of FGF2 out of individual spots in the hydrogel arrays. Lysozyme diffused out of the hydrogel arrays over time, as expected (Supplementary Fig. 1). After 6 hours 44.1±5.1% of the original lysozyme remained in the array spots, and after 7 days 14.9±1.2% of the original lyzozyme remained (Supplementary Fig. 1). Due to the similarities between the molecular weights of lysozyme (14.7kDa) and FGF2 (17.2kDa), it is likely that FGF2 diffusion out of hydrogel array spots would follow a similar trend.
FGF2 effects on hMSC viability
The presence of soluble FGF2 significantly enhanced hMSC viability in non-degrading PEG hydrogels (Fig. 2). In the absence of FGF2, cell viability dramatically decreased from 82.1±1.9% at day 1 to 45.5±4.8% at day 7. The addition of 0.1, 1, 10, 100, and 200 ng/ml FGF2 significantly enhanced viability at day 5 and day 7 of culture. The influence of FGF2 appeared to be most pronounced when either 10 or 100 ng/ml FGF2 was included, and this dose-dependence is particularly evident at day 3. Importantly, the effects of 100 ng/ml FGF2 on hMSC viability were blocked in the presence of the PD 173074 (Sigma-Aldrich, Milwaukee, WI) soluble inhibitor of FGF2 receptor autophosphorylation (Fig. 3). In the presence of this inhibitor, hMSC viability decreased to less than 40% after 5 days in culture, both in the presence and absence of 100 ng/ml FGF2. A toxicity experiment confirmed that this inhibitor is not toxic to hMSCs at the concentration explored here (Supplementary Fig. 2). The presence of soluble FGF2 also significantly enhanced hMSC viability in degrading PEG hydrogels (Fig. 4). This enhancement was more substantial in degrading PEG hydrogels (Fig. 4) when compared to non-degrading PEG hydrogels (Fig. 2). In addition, the effect was clearly dose dependent in degrading hydrogels, as the influence of FGF2 was significant at days 5 and 7 only when FGF2 was included at higher concentrations (10 or 100 ng/ml). Taken together, these results indicate that the presence of FGF2 enhances hMSC viability in a dose-dependent manner in both degrading and non-degrading PEG hydrogels.
Figure 2.
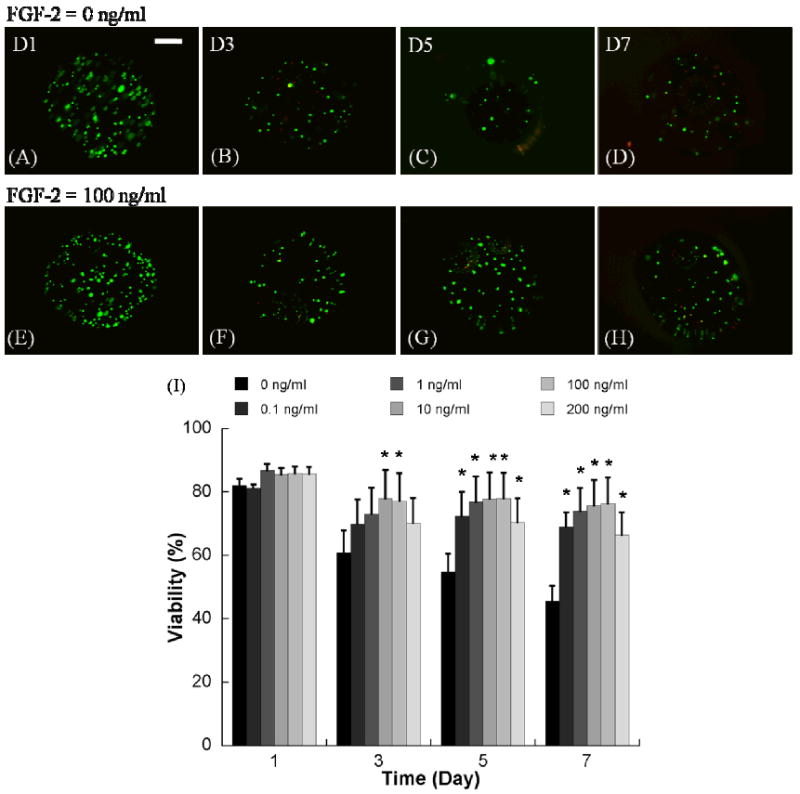
Representative images of hMSCs incorporation into non-degrading PEG hydrogel arrays with various FGF-2 concentrations A-D) 0 ng/ml, and E-H) 100 ng/ml. (Scale bar = 100 μm). I) Viability of hMSCs cultured in non-degrading PEG hydrogel array spots with various FGF-2 concentrations. *Denotes significant difference from 0 ng/ml condition at the same time point, ANOVA p<0.05.
Figure 3.
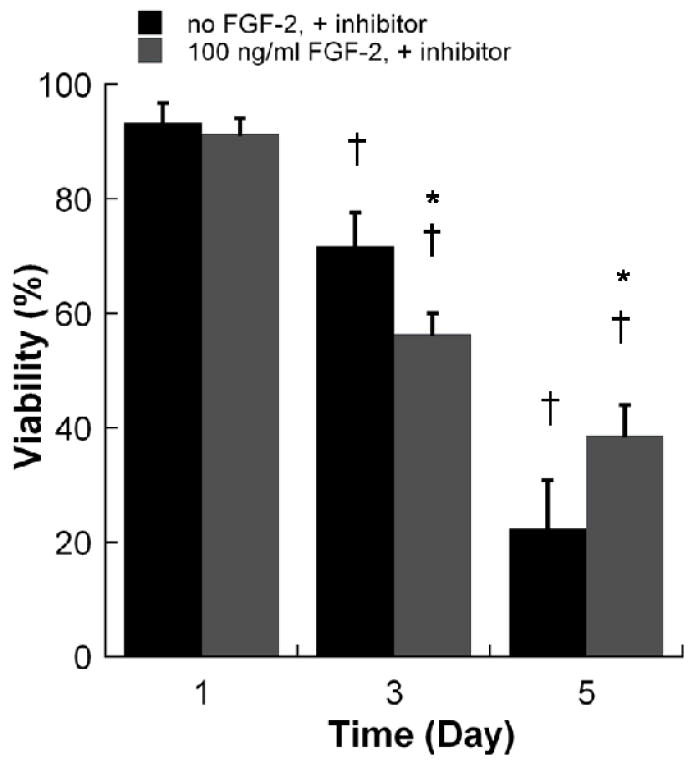
Viability of hMSCs in non-degrading PEG hydrogel arrays containing 100 ng/ml FGF-2 inhibitor with variable concentrations of FGF-2. *Denotes a significant difference compared to 0 ng/ml condition at the same time point. †Denotes a significant difference from the D1 value for the same experiment condition, ANOVA p<0.05.
Figure 4.
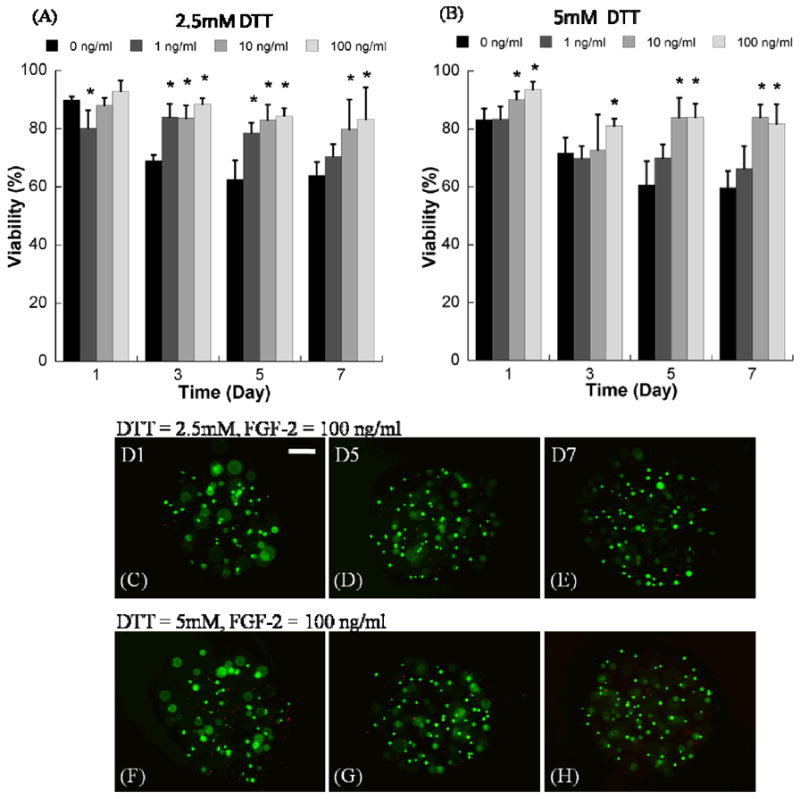
Quantitative analysis of hMSC viability within degrading PEG hydrogel arrays with variable degradability and variable soluble FGF-2 concentration. A) hMSC viability in degrading PEG hydrogel arrays prepared with 2.5mM DTT with variable concentrations of FGF-2, B) hMSC viability in degrading PEG hydrogel arrays prepared with 5mM DTT with variable concentrations of FGF-2. *Denotes significant difference from 0 ng/ml concentration at the same time point, ANOVA p<0.05. (C-H) Representative live/dead images of hMSC cultured in degrading PEG hydrogel arrays containing 100 ng/ml FGF-2, C-E) 2.5mM DTT, and F-H) 5mM DTT. (Scale bar = 100 μm).
Conjugation of cell adhesion peptides to PEG-diacrylate
To ensure the cell adhesion peptides were efficiently incorporated into the hydrogel networks, we quantified the free thiol content of 2.5mM CGGRGDSP before and after its reaction with a 10X molar excess of 8k PEGDA – the same excess used in our hydrogel array processing. 0.01±0.005% of the original thiol groups were present after the reaction, indicating that the coupling of the peptide to the polymer was approximately 99% efficient. This result suggested that nearly all of the peptide ligand was incorporated into the hydrogel network.
Combined effects of FGF2 and cell adhesion ligands on viability in PEG hydrogels
The presence of soluble FGF2 significantly enhanced hMSC viability in non-degrading PEG hydrogels containing the RGDSP cell adhesion ligand (Fig. 5). hMSC viability was less pronounced than they were in the absence of CGGRGDSP, Cell viability remained relatively high after 7 days in PEG hydrogels with 0.1mM (63.1±5.0%) or 2.5mM (69.5±3.4%) RGDSP ligand, even in the absence of FGF2. In the presence of 10 or 100 ng/ml FGF2, cell viability was significantly enhanced at days 5 and 7 in PEG hydrogels containing either 0.1mM or 2.5mM RGDSP. Optimal cell viability at day 7 was observed in the presence of 2.5mM RGDSP and 100 ng/ml FGF2 (82.7±5.2%), and this viability was not significantly different when compared to the initial viability at day 1 (80.4±4.4%). Therefore, RGDSP and FGF2 significantly enhanced hMSC viability within non-degrading PEG hydrogels when presented either alone or in combination.
Figure 5.
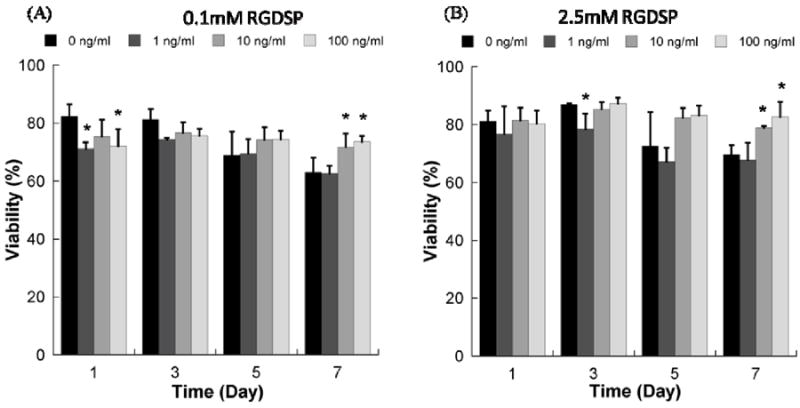
Viability of hMSCs cultured in non-degrading PEG hydrogel array spots containing A) 0.1mM RGDSP ligand with variable concentrations of FGF-2, and B) 2.5mM RGDSP ligand with variable concentrations of FGF-2. *Denotes a significant difference compared to 0 ng/ml condition at the same time point, ANOVA p<0.05.
In contrast, the presence of soluble FGF2 resulted in a slight, but not significant, increase in hMSC viability in non-degrading PEG hydrogels containing the IKVAV cell adhesion ligand (Fig. 6). The presence of soluble FGF2 also did not significantly enhance hMSC viability in degrading PEG hydrogels containing the RGDSP cell adhesion ligand (Fig. 7). hMSC viability in degrading PEG hydrogels in the presence of RGDSP was either unaffected (Fig. 7A-C) or negatively affected (Fig. 7D) by the presence of FGF2.
Figure 6.
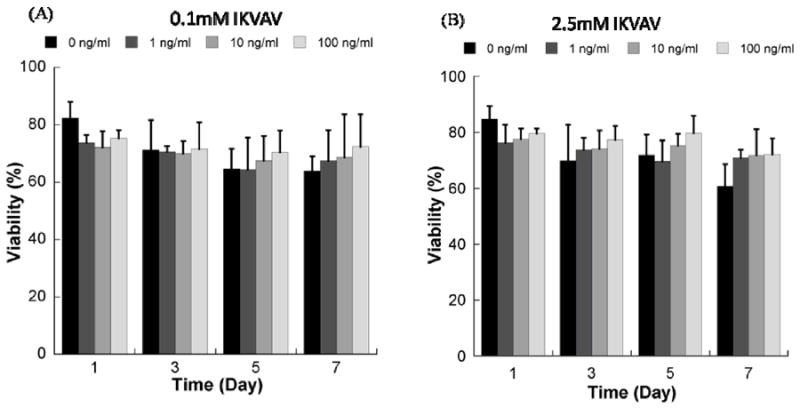
Viability of hMSCs cultured in wells of non-degrading PEG hydrogel arrays containing A) 0.1mM IKVAV ligand with variable concentrations of FGF-2, and B) 2.5mM IKVAV ligand with variable concentrations of FGF-2. *Denotes a significant difference compared to 0 ng/ml condition at the same time point, ANOVA p<0.05.
Figure 7.
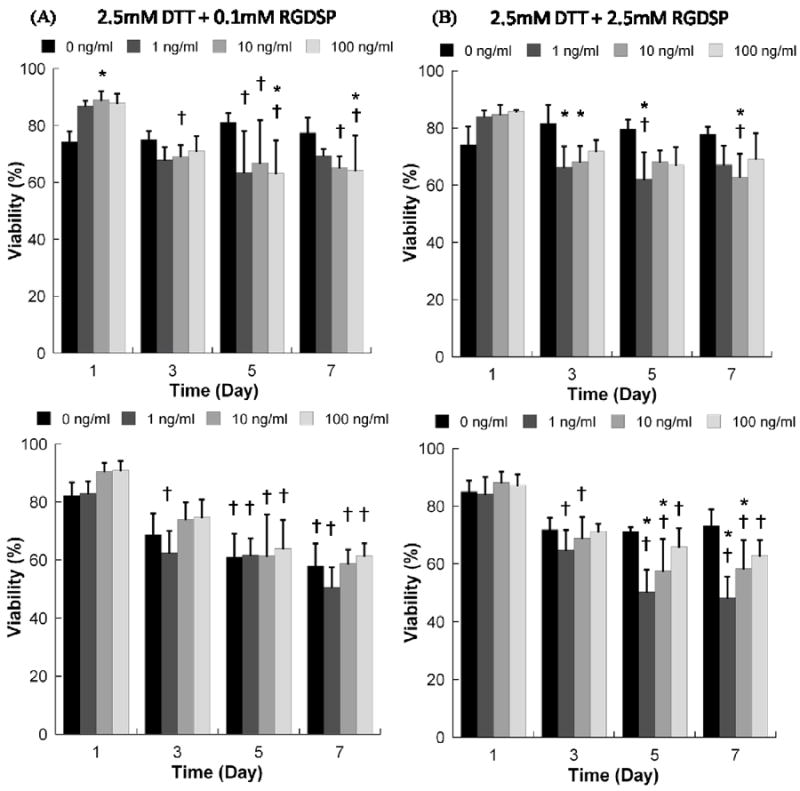
Viability of hMSCs cultured within hydrogel arrays with varying degradability, RGDSP concentration, and FGF2 concentration. A) 2.5mM DTT + 0.1mM RGDSP; B) 2.5mM DTT + 2.5mM RGDSP; C) 5mM DTT + 0.1mM RGDSP; D) 5mM DTT + 2.5mM RGDSP. * Denotes a significant difference compared to 0 ng/ml condition at the same time point. †Denotes significant difference from D1 value for the same condition, p<0.05.
hMSC spreading in PEG hydrogels
hMSC behavior in all PEG hydrogels without adhesion ligands and in all non-degrading PEG hydrogels displayed a relatively small, rounded morphology; both in the presence and absence of FGF2 (see Figs. 2, 4, 8). This rounded morphology is consistent with a variety of previous studies of cell encapsulation in PEG networks (e.g. ref. 14). However, we observed uncommon hMSC morphologies in a select subset of experimental conditions, in which cells had a relatively large cross-sectional area and displayed extensions consistent with cell “spreading” (See Figs. 8D+ 9). The spreading of hMSCs was more pronounced in PEG hydrogels that were degrading and contained both 2.5mM RGDSP and FGF2, as demonstrated by increased hMSC area and aspect ratio after 7 days in culture (Figs. 8-9). hMSC spreading was observed at all time points studied in experimental conditions containing 5mM DTT, 2.5mM RGD, and 100ng/ml FGF2 (Fig. 8D).
Figure 8.
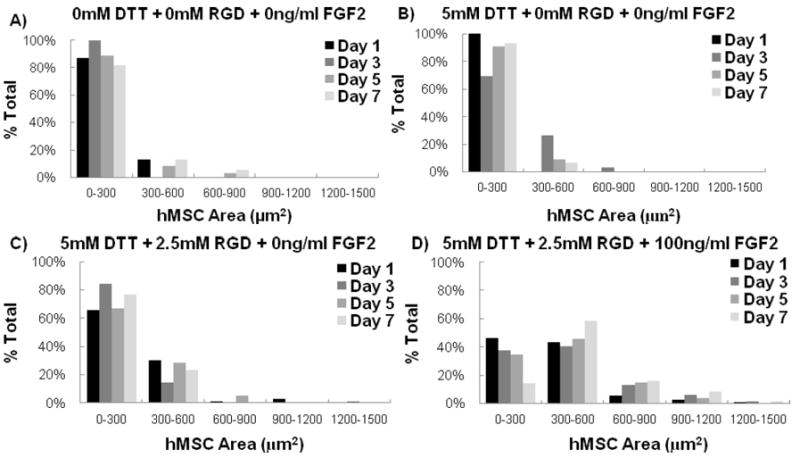
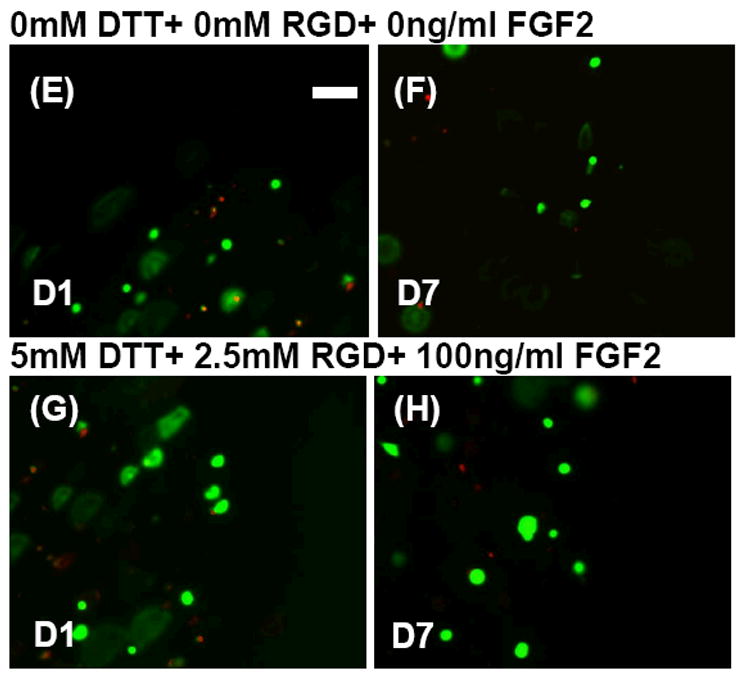
The relative frequency of hMSCs within specified ranges of projected cell area. Data are presented for hMSCs grown in array spots with A) 0mM DTT+0mM RGD+ 0ng/ml FGF2 B)5mM DTT+ 0mM RGD+ 0ng/ml FGF2 C) 5mM DTT+2.5mM RGD+ 0ng/ml FGF2 and D) 5mM DTT+ 2.5mM RGD+ 100ng/ml FGF2. Representative live/dead images of hMSC cell areas in array spots containing 0mM DTT+ 0mM RGD+ 0ng/ml FGF2 E) 1 day and F) 7 days post encapsulation. Also included are representative live/dead images of hMSC cell areas in array spots containing 5mM DTT+ 2.5mM RGD+ 100ng/ml FGF2 G) 1 day and H) 7 days post encapsulation.
Figure 9.
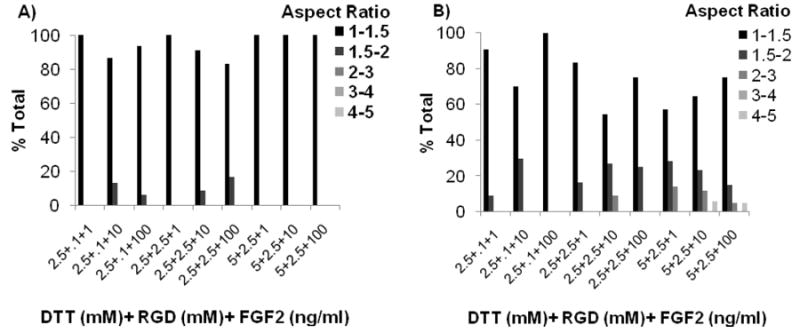
The relative frequency of hMSCs within specified ranges of cellular aspect ratio. Data were collected after A) 1 day and B) 7 days in 3-D cell culture environments that promoted hMSC spreading.
Discussion
FGF2 has been identified as a critical factor for survival and self-renewal of various stem cell types, including neural stem cells37 and embryonic stem cells38 in standard cell culture conditions. The influence of FGF2 on MSC proliferation and differentiation potential has also been well-characterized in 2-D cell culture. Specifically, FGF2 has recently been shown to promote expansion of undifferentiated hMSC populations21,22,24 and suppress hMSC senescence23. Therefore, we hypothesized that FGF2 could be an important factor in applications that require viable, undifferentiated hMSCs, including tissue engineering applications. Here we demonstrate that hydrogel arrays formed via an automated approach detailed previously2,20 can be used to screen the effects of FGF2 and various hydrogel matrix parameters on hMSC viability and spreading.
Soluble FGF2 significantly enhanced hMSC viability in both non-degrading PEG hydrogels (Fig. 2) and in degrading PEG hydrogels (Fig. 4). A PD 173074 inhibitor of FGF receptor auto-phosphorylation blocked the effects of FGF2 on hMSC viability (Fig. 3), indicating that enhanced hMSC viability can likely be directly attributed to FGF2 signaling. The effects of FGF2 observed here are consistent with recent 2-D hMSC culture studies23, which have shown that FGF2 promotes hMSC survival and suppresses senescence. Other previous studies in 2-D culture have also demonstrated that FGF2 controls hMSC migration25 and promotes maintenance of chondrogenic27, osteogenic26, and adipogenic28 differentiation potential. Therefore, FGF2 may be a particularly important factor in applications that require high hMSC viability and multilineage differentiation potential. Further studies will be needed to determine whether the previously observed effects of FGF2 on hMSC differentiation potential can be extended to 3-dimensional culture systems such as the ones explored in the current study.
In this and many other studies that have characterized the influence of exogenous growth factors on cells, it is true that serum-derived components could influence cell behavior and influence results. One possibility is that serum-derived FGF2 could have influenced hMSC viability. Although batch-to-batch variations in serum component concentrations have been widely reported, the FGF2 serum concentrations from other studies can help us to approximate the potential influence of serum-derived FGF2 on hMSCs in our study. Hentges et al. measured 0.014ng/ml FGF2 in DMEM-F12 media supplemented with 10% Fetal Calf Serum39, and Benoit et al. measured 0.003ng/ml FGF2 in hMSC proliferation media4. In our experiments 0, 0.1, 1, 10, 100, and 200ng/ml FGF2 were exogenously added to hMSCs in hydrogel array wells (Figs. 2-9). Therefore, the serum derived FGF2 likely constituted a small fraction of the total FGF2 in the hMSCs’ microenvironment, and may have only a restricted role in promoting hMSC viability and spreading. A second possibility is that other soluble factors in serum may have influenced cell survival in combination with RGDSP-mediated integrin signaling. Many different soluble factors influence hMSC viability including TGF-β310, TGF-β111, and BMP212. Additional 3-D cell culture studies in enhanced throughput experimental formats will be necessary to characterize the roles of these proteins in regulating hMSC survival in 3D matrices.
FGF2 diffusion out of the hydrogel arrays may have also influenced hMSC viability and spreading. To mimic the diffusion of exogenous FGF2 out of the hydrogels, we loaded hydrogel array spots with a rhodamine-labeled lysozyme, and the array spots’ local protein concentrations were measured at predetermined times (Supplementary Fig. 1). Lysozyme was chosen as a model for FGF2 because 14.7kDa lysozyme, which has a hydrodynamic radius of 19.5Å40, has a similar molecular weight to 17.2kDa FGF2, which has a hydrodynamic radius of 28Å41. After 6 hours there was 44.1±5.1% of the original protein in the wells. This level of protein diffusion out of the PEG based hydrogel arrays is consistent with other studies in which growth factors were loaded into hydrogels4,42. After 7 days of protein diffusion out of the hydrogel arrays there was still 14.9±1.2% of the exogenous protein remaining (Supplementary Fig. 1). This result suggests that in the case of array spots originally loaded with 100ng/ml of FGF2 there would be approximately 15ng/ml FGF2 still present in the array spots at the end of our cell culture studies. This FGF2 concentration is greater than the 1ng/ml FGF2 concentration that Bianchi et al. have used to promote hMSC survival and maintenance of telomere length in cell culture21, so it is unlikely that diffusion alone will decrease the growth factor concentration enough to inhibit growth factor effects. However, it is difficult to predict the active FGF2 concentration in hydrogel arrays over time due to FGF2’s short half-life in serum containing media43. Taken together, there are multiple factors which could influence the active concentration of FGF2 over time in these 3-D hydrogel arrays.
The effects of FGF2 on hMSC viability were less substantial in PEG hydrogels containing the RGDSP (Fig. 5) or IKVAV (Fig. 6) cell adhesion ligands when compared to hydrogels without ligands included. Although the effects of FGF2 on hMSC viability were statistically significant in a limited subset of hydrogels containing RGDSP, the effects were only evident at high FGF2 concentrations and at late time points (Fig. 5). In contrast, FGF2 did not significantly influence hMSC viability in any hydrogels containing IKVAV (Fig. 6). In a previous study we demonstrated that each of these ligands enhances hMSC viability in a dose-dependent manner in non-degrading PEG hydrogels2. Therefore, our results collectively suggest that the effects of FGF2 are less significant when hMSCs are able to interact with an adhesion ligand. This effect can potentially be explained by the aforementioned propensity of FGF2 to serve as a survival factor. hMSCs may be more responsive to FGF2 when they are in an environment that is not conducive to cell survival, such as a non-degrading PEG hydrogel devoid of cell adhesion ligands. Also, it is possible that the effects of FGF2 are simply more difficult to detect in the presence of adhesion ligands, since cell viability does not decrease as substantially over 7 days in ligand-containing hydrogels when compared to ligand-free hydrogels.
One unexpected finding in this study was that FGF2 negatively affected cell viability in degradable gels with RGDSP (Fig 7D). This observation could be attributed to the fact that integrin signaling and FGF2 signaling are interrelated. For example, integrins interact with FGF2 and fibroblast growth factor receptors (FGFRs) to influence their canonical signaling pathway. Functional outcomes of integrin-FGFR interactions differ by cell type. Rusnati et al. demonstrated that the α5ß1 integrin, the integrin that binds to RGDSP, interacts with immobilized FGF2, and FGF2-induced angiogenesis can be blocked with α5 integrin-specific inhibitors44. In addition, FGF2 has been shown to alter hMSC integrin expression. In one study, Varas et al. quantified the expression of α10ß1 and α11ß1 integrins on hMSCs grown in chondrogenic induction media with and without 10ng/ml FGF2. The expression of α10ß1 integrin increased when hMSCs were cultured in chondrogenic induction media, but its expression level was reduced by addition of FGF2 to the chondrogenic induction media. FGF2 also increased the expression of α11ß1 integrins45. In this previous study the expression level of the α5ß1 integrin that binds to RGDSP was not quantified, but this research is an important demonstration that FGF2 signaling can effect hMSC integrin expression.
It is noteworthy that hMSC viability was quantified in every cell culture condition with the LIVE/DEAD assay. Therefore, our results do not explicitly indicate how hMSC proliferation may have contributed to differences in hMSC viability. Previous studies have demonstrated that there is no cell proliferation in PEG-based hydrogels that do not degrade9. Alternatively, cells can proliferate in PEG gels that undergo extensive degradation9,36. Our previous studies indicate that the PEG-DTT hydrogels explored in this study do not undergo extensive degradation over 7 days14. Taken together, these results suggest that hMSC proliferation likely did not have a significant impact on the differences in viability observed here.
hMSC spreading was dependent on FGF2 concentration. Spreading was not observed in culture conditions with less than 10ng/ml FGF2. In contrast, 10ng/ml and 100ng/ml FGF2 promoted hMSC spreading equally (Figs. 8-9). This observation is potentially important, as recent studies have correlated hMSC spreading with differentiation potential in 3-D hydrogels. In particular, Levenston and coworkers recently demonstrated that chondrogenic differentiation was inhibited and osteogenic differentiation was enhanced when hMSCs were allowed to attach and spread within RGD-modified alginate46 or agarose47 hydrogels. In contrast, hMSCs differentiated more efficiently down the chondrogenic lineage in conditions that did not promote cell spreading46. An interesting aspect of our findings is that FGF2, which has been shown to preserve hMSC multipotency, promoted hMSC spreading. Although hMSC spreading has been correlated with propensity of MSCs to differentiate down the osteogenic lineage, hMSC spreading alone does not induce differentiation. For example, Salasznyk et al. demonstrated that hMSCs spread on collagen I, collagen IV, fibronectin, and vitronectin coated surfaces. However, significant increases osteogenic differentiations markers were only observed in the presence of pro-osteogenic supplements, which induce differentiation. Therefore, further experiments will be needed to determine whether the enhanced hMSC spreading observed here in response to FGF2 leads to enhanced propensity for osteogenic differentiation.
Hydrogel degradability promoted hMSC spreading. Three different extents of degradation were explored in these studies. Specifically, PEG hydrogel spots were formed either with no DTT “bridges” included, 2.5mM DTT “bridges”, or 5mM DTT “bridges”. In previous studies we have demonstrated that the concentration of these bridges significantly influences degradation of PEG hydrogel networks14. Specifically, in our previous studies the equilibrium swelling ratio of 8k PEG hydrogels containing 2.5mM DTT did not significantly change during the first 7 days of incubation in an aqueous solution, which indicated low levels of network degradation. Equilibrium swelling ratios of 8k PEG hydrogels containing 5mM DTT increased significantly after 7 days in aqueous solution, indicating more network degradation in hydrogel networks with 5mM DTT than networks with 2.5mM DTT. Bulk hydrogel erosion – indicated by a decrease in dry mass - was not observed in either material. Therefore, it is likely that the extent of degradation of the hydrogel spots over the culture period examined in our current study was significant, but not substantial. However, these changes in network degradability did significantly influence hMSC viability and spreading. Taken together, these results suggest that extensive network degradation is not necessary to promote hMSC spreading.
An important current limitation of this approach is the analytical technique and its throughput. Epifluorescence microscopy has several analytical limitations for observing hMSCs in 3-D hydrogels. hMSC features adjacent to each other in the same x-y plane can be resolved, but out-of-focus light may reduce the contrast between them. Also, epifluorescence limits the ability to resolve features in overlapping planes-of-focus. Confocal microscopy allows researchers to acquire images from specific planes and the reconstruct the data to form 3-D cell representations without contrast concerns48. Therefore, although this initial study did not incorporate more complex and detailed imaging techniques, ongoing studies will benefit from the use of confocal microscopy to collect more detailed information about cell morphology. In addition, the analyses described here and elsewhere rely primarily on manual analysis of fluorescence emission, which requires a fluorescent reporter, and it is not a high throughput process. Other studies have developed fluorescent markers of MSC behavior, including osteogenic49 and chondrogenic50 differentiation, and these markers may be useful in future studies using hydrogel array format. In addition, there are several high-content image analysis techniques and software tools available6,51 that may be used in conjunction with this array format in future studies to enhance analytical throughput.
Taken together, these results suggest that small changes in hydrogel network structure can significantly influence hMSC viability and spreading. Importantly, the processing approach used to generate hydrogel arrays in this study is readily adaptable. Taken together, the current study and previous studies2,20 have demonstrated the ability to generate at least 5 distinct hydrogel matrices per minute while systematically varying the hydrogel type (natural or synthetic), hydrogel degradability, cell adhesion ligand type, cell adhesion ligand density, soluble factor location, soluble factor concentration, cell type, and cell seeding density. Therefore, this general approach may be useful for screening the effects of a broad range of parameters on cell behavior.
Conclusions
Hydrogel arrays formed via an automated liquid handling approach can be used to rapidly screen for the effects of FGF2 on hMSC viability and spreading in various 3-D hydrogel matrices. The presence of soluble FGF2 significantly enhanced hMSC viability in both non-degrading and degrading PEG hydrogels devoid of cell adhesion ligands. FGF2 effects on hMSC viability were significant in a subset of conditions, but less pronounced when presented in combination with the fibronectin-derived RGDSP ligand or the laminin-derived IKVAV ligand in non-degrading PEG hydrogels. Although hMSCs typically displayed a rounded morphology in 3-D PEG hydrogels, we observed spread hMSC morphologies in degrading PEG hydrogels which contained combinations of RGDSP and 100 ng/ml FGF2. hMSCs were clearly spread in these particular environments at all time points studied. These results indicate that FGF2 may be an important component of 3-D hMSC culture environments which require high viability and cell spreading, including hydrogel matrices for tissue engineering.
Supplementary Material
Protein diffusion out of hydrogel array spots. To mimic FGF2 diffusion out of hydrogel array spots, lysozyme was fluorescently labeled with NHS-rhodamine using the manufacturer’s protocol (Pierce, Rockford, IL). Rhodamine-labeled lysozyme was placed in 10,000 molecular weight cutoff dialysis tubing (Pierce, Rockford, IL) and dialyzed against ultrapure H2O (18 MΩ resistivity). 100μM Rhodamine-Lysozyme was loaded into hydrogel spots (n=4), photocrosslinked, and placed in phosphate buffered saline. Fluorescent micrographs were acquired using an inverted, compound fluorescence microscope (IX51, Olympus) at 40X magnification at predetermined times and the buffer was replaced after every image acquisition. The fluorescence intensity was measured using ImageJ software (Freeware, NIH, Bethesda, MD).
Toxicity studies with PD 173074 FGF2 inhibitor. To determine the toxicity of the PD 173074 small molecule FGFR inhibitor (Sigma, St. Louis, MO), hMSC’s were plated in 96 well plates and allowed to attach for 1 hour. After two hours, additional hMSC media was added so that there were final concentrations of 0, 25, 50, 75, 100, and 200nM PD173074. Cell viability was measured using the CellTiter Blue Assay (Promega, Madison, WI) three days after the addition of the PD 173074. Results indicate that there is a significant decrease in cell number in solutions containing 50-100 nM inhibitor, and a more substantial decrease in cell number in the presence of 200 nM inhibitor. The decrease in cell number in the presence of 50-100 nM inhibitor is likely due to inhibition of serum-derived FGF2 signaling, as the magnitude of the decrease is not consistent with toxicity. * Denotes significant difference from 0nM condition, ANOVA p<0.05. + Denotes significant difference from 0nM condition, ANOVA p<0.001.
Acknowledgments
The authors acknowledge financial support from the National Institutes of Health (R21EB005374 and R21HL084547 to W.L.M), and the National Science Foundation (CAREER award 0745563 to W.L.M., and graduate fellowship to W.J.K.). We also acknowledge Jay Warrick and Dr. David Beebe at the University of Wisconsin for assistance with automated liquid handling.
References
- 1.Nuttelman CR, Tripodi MC, Anseth KS. Dexamethasone-functionalized gels induce osteogenic differentiation of encapsulated hMSCs. J Biomed Mater Res A. 2006;76(1):183–95. doi: 10.1002/jbm.a.30537. [DOI] [PubMed] [Google Scholar]
- 2.Jongpaiboonkit L, King WJ, Murphy WL. Screening for 3D Environments That Support Human Mesenchymal Stem Cell Viability Using Hydrogel Arrays. Tissue Eng Part A. 2008 doi: 10.1089/ten.tea.2008.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26(30):5991–8. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Benoit DS, Anseth KS. Heparin functionalized PEG gels that modulate protein adsorption for hMSC adhesion and differentiation. Acta Biomater. 2005;1(4):461–70. doi: 10.1016/j.actbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 6.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol. 2005;24(3):208–18. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Drury JL, Mooney DJ. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24(24):4337–51. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 8.Uludag H, De Vos P, Tresco PA. Technology of mammalian cell encapsulation. Adv Drug Deliv Rev. 2000;42(1-2):29–64. doi: 10.1016/s0169-409x(00)00053-3. [DOI] [PubMed] [Google Scholar]
- 9.Patel PN, Gobin AS, West JL, Patrick CW., Jr Poly(ethylene glycol) hydrogel system supports preadipocyte viability, adhesion, and proliferation. Tissue Eng. 2005;11(9-10):1498–505. doi: 10.1089/ten.2005.11.1498. [DOI] [PubMed] [Google Scholar]
- 10.Sharma B, Williams CG, Khan M, Manson P, Elisseeff JH. In vivo chondrogenesis of mesenchymal stem cells in a photopolymerized hydrogel. Plast Reconstr Surg. 2007;119(1):112–20. doi: 10.1097/01.prs.0000236896.22479.52. [DOI] [PubMed] [Google Scholar]
- 11.Williams CG, Kim TK, Taboas A, Malik A, Manson P, Elisseeff J. In vitro chondrogenesis of bone marrow-derived mesenchymal stem cells in a photopolymerizing hydrogel. Tissue Eng. 2003;9(4):679–88. doi: 10.1089/107632703768247377. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, Kim IS, Cho TH, Lee KB, Hwang SJ, Tae G, Noh I, Lee SH, Park Y, Sun K. Bone regeneration using hyaluronic acid-based hydrogel with bone morphogenic protein-2 and human mesenchymal stem cells. Biomaterials. 2007;28(10):1830–7. doi: 10.1016/j.biomaterials.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Wang DA, Williams CG, Yang F, Cher N, Lee H, Elisseeff JH. Bioresponsive phosphoester hydrogels for bone tissue engineering. Tissue Eng. 2005;11(1-2):201–13. doi: 10.1089/ten.2005.11.201. [DOI] [PubMed] [Google Scholar]
- 14.Hudalla GA, Eng TS, Murphy WL. An approach to modulate degradation and mesenchymal stem cell behavior in poly(ethylene glycol) networks. Biomacromolecules. 2008;9(3):842–9. doi: 10.1021/bm701179s. [DOI] [PubMed] [Google Scholar]
- 15.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Photo- and electropatterning of hydrogel-encapsulated living cell arrays. Lab Chip. 2005;5(1):111–8. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 17.Koh WG, Itle LJ, Pishko MV. Molding of hydrogel microstructures to create multiphenotype cell microarrays. Anal Chem. 2003;75(21):5783–9. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- 18.Koh WG, Revzin A, Pishko MV. Poly(ethylene glycol) hydrogel microstructures encapsulating living cells. Langmuir. 2002;18(7):2459–62. doi: 10.1021/la0115740. [DOI] [PubMed] [Google Scholar]
- 19.Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A. A cell-laden microfluidic hydrogel. Lab Chip. 2007;7(6):756–62. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- 20.Jongpaiboonkit L, King WJ, Lyons GE, Paguirigan AL, Warrick JW, Beebe DJ, Murphy WL. An adaptable hydrogel array format for 3-dimensional cell culture and analysis. Biomaterials. 2008;29(23):3346–56. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianchi G, Banfi A, Mastrogiacomo M, Notaro R, Luzzatto L, Cancedda R, Quarto R. Ex vivo enrichment of mesenchymal cell progenitors by fibroblast growth factor 2. Exp Cell Res. 2003;287(1):98–105. doi: 10.1016/s0014-4827(03)00138-1. [DOI] [PubMed] [Google Scholar]
- 22.Go MJ, Takenaka C, Ohgushi H. Effect of forced expression of basic fibroblast growth factor in human bone marrow-derived mesenchymal stromal cells. J Biochem. 2007;142(6):741–8. doi: 10.1093/jb/mvm189. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Sawada R, Fujiwara Y, Seyama Y, Tsuchiya T. FGF-2 suppresses cellular senescence of human mesenchymal stem cells by down-regulation of TGF-beta2. Biochem Biophys Res Commun. 2007;359(1):108–14. doi: 10.1016/j.bbrc.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 24.Sotiropoulou PA, Perez SA, Salagianni M, Baxevanis CN, Papamichail M. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2006;24(2):462–71. doi: 10.1634/stemcells.2004-0331. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt A, Ladage D, Schinkothe T, Klausmann U, Ulrichs C, Klinz FJ, Brixius K, Arnhold S, Desai B, Mehlhorn U, et al. Basic fibroblast growth factor controls migration in human mesenchymal stem cells. Stem Cells. 2006;24(7):1750–8. doi: 10.1634/stemcells.2005-0191. [DOI] [PubMed] [Google Scholar]
- 26.Martin I, Muraglia A, Campanile G, Cancedda R, Quarto R. Fibroblast growth factor-2 supports ex vivo expansion and maintenance of osteogenic precursors from human bone marrow. Endocrinology. 1997;138(10):4456–62. doi: 10.1210/endo.138.10.5425. [DOI] [PubMed] [Google Scholar]
- 27.Solchaga LA, Penick K, Porter JD, Goldberg VM, Caplan AI, Welter JF. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203(2):398–409. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- 28.Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem Biophys Res Commun. 2001;288(2):413–9. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- 29.Cruise GM, Scharp DS, Hubbell JA. Characterization of permeability and network structure of interfacially photopolymerized poly(ethylene glycol) diacrylate hydrogels. Biomaterials. 1998;19(14):1287–94. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 30.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Systematic modulation of Michael-type reactivity of thiols through the use of charged amino acids. Bioconjug Chem. 2001;12(6):1051–6. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 31.Skaper SD, Kee WJ, Facci L, Macdonald G, Doherty P, Walsh FS. The FGFR1 inhibitor PD 173074 selectively and potently antagonizes FGF-2 neurotrophic and neurotropic effects. J Neurochem. 2000;75(4):1520–7. doi: 10.1046/j.1471-4159.2000.0751520.x. [DOI] [PubMed] [Google Scholar]
- 32.Trudel S, Ely S, Farooqi Y, Affer M, Robbiani DF, Chesi M, Bergsagel PL. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103(9):3521–8. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- 33.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, Green D, Schlessinger J, Hubbard SR. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. Embo J. 1998;17(20):5896–904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bansal R, Magge S, Winkler S. Specific inhibitor of FGF receptor signaling: FGF-2-mediated effects on proliferation, differentiation, and MAPK activation are inhibited by PD173074 in oligodendrocyte-lineage cells. J Neurosci Res. 2003;74(4):486–93. doi: 10.1002/jnr.10773. [DOI] [PubMed] [Google Scholar]
- 35.Zaragosi LE, Ailhaud G, Dani C. Autocrine fibroblast growth factor 2 signaling is critical for self-renewal of human multipotent adipose-derived stem cells. Stem Cells. 2006;24(11):2412–9. doi: 10.1634/stemcells.2006-0006. [DOI] [PubMed] [Google Scholar]
- 36.Khetan SK, Joshua S, Burdick Jason A. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5:1601–1606. [Google Scholar]
- 37.Shihabuddin LS, Ray J, Gage FH. FGF-2 is sufficient to isolate progenitors found in the adult mammalian spinal cord. Exp Neurol. 1997;148(2):577–86. doi: 10.1006/exnr.1997.6697. [DOI] [PubMed] [Google Scholar]
- 38.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2(3):185–90. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 39.Hentges S, Boyadjieva N, Sarkar DK. Transforming growth factor-beta 3 stimulates lactotrope cell growth by increasing basic fibroblast growth factor from folliculo-stellate cells. Endocrinology. 2000;141(3):859–867. doi: 10.1210/endo.141.3.7382. [DOI] [PubMed] [Google Scholar]
- 40.Batas B, Jones HR, Chaudhuri JB. Studies of the hydrodynamic volume changes that occur during refolding of lysozyme using size-exclusion chromatography. Journal of Chromatography A. 1997;766(1-2):109–119. doi: 10.1016/s0021-9673(96)01020-5. [DOI] [PubMed] [Google Scholar]
- 41.Onuma K, Kanzaki N, Kobayashi N. Association of calcium phosphate and fibroblast growth factor-2: a dynamic light scattering study. Macromol Biosci. 2004;4(1):39–46. doi: 10.1002/mabi.200350037. [DOI] [PubMed] [Google Scholar]
- 42.Tanihara M, Suzuki Y, Yamamoto E, Noguchi A, Mizushima Y. Sustained release of basic fibroblast growth factor and angiogenesis in a novel covalently crosslinked gel of heparin and alginate. J Biomed Mater Res. 2001;56(2):216–21. doi: 10.1002/1097-4636(200108)56:2<216::aid-jbm1086>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 43.Moscatelli D. Metabolism of receptor-bound and matrix-bound basic fibroblast growth factor by bovine capillary endothelial cells. J Cell Biol. 1988;107(2):753–9. doi: 10.1083/jcb.107.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusnati M, Tanghetti E, DellEra P, Gualandris A, Presta M. alpha(v)beta(3) integrin mediates the cell-adhesive capacity and biological activity of basic fibroblast growth factor (FGF-2) in cultured endothelial cells. Molecular Biology of the Cell. 1997;8(12):2449–2461. doi: 10.1091/mbc.8.12.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varas L, Ohlsson LB, Honeth G, Olsson A, Bengtsson T, Wiberg C, Bockermann R, Jarnum S, Richter J, Pennington D, et al. alpha 10 integrin expression is up-regulated on fibroblast growth factor-2-treated mesenchymal stem cells with improved chondrogenic differentiation potential. Stem Cells and Development. 2007;16(6):965–978. doi: 10.1089/scd.2007.0049. [DOI] [PubMed] [Google Scholar]
- 46.Connelly JT, Garcia AJ, Levenston ME. Inhibition of in vitro chondrogenesis in RGD-modified three-dimensional alginate gels. Biomaterials. 2007;28(6):1071–83. doi: 10.1016/j.biomaterials.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Connelly JT, Garcia AJ, Levenston ME. Interactions between integrin ligand density and cytoskeletal integrity regulate BMSC chondrogenesis. J Cell Physiol. 2008;217(1):145–54. doi: 10.1002/jcp.21484. [DOI] [PubMed] [Google Scholar]
- 48.Rizzuto R, Carrington W, Tuft RA. Digital imaging microscopy of living cells. Trends in Cell Biology. 1998;8(7):288–292. doi: 10.1016/s0962-8924(98)01301-4. [DOI] [PubMed] [Google Scholar]
- 49.Tai G, Christodoulou I, Bishop AE, Polak JM. Use of green fluorescent fusion protein to track activation of the transcription factor osterix during early osteoblast differentiation. Biochem Biophys Res Commun. 2005;333(4):1116–22. doi: 10.1016/j.bbrc.2005.05.195. [DOI] [PubMed] [Google Scholar]
- 50.Osaki M, Tan L, Choy BK, Yoshida Y, Cheah KS, Auron PE, Goldring MB. The TATA-containing core promoter of the type II collagen gene (COL2A1) is the target of interferon-gamma-mediated inhibition in human chondrocytes: requirement for Stat1 alpha, Jak1 and Jak2. Biochem J. 2003;369(Pt 1):103–15. doi: 10.1042/BJ20020928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abraham VC, Taylor DL, Haskins JR. High content screening applied to large-scale cell biology. Trends Biotechnol. 2004;22(1):15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein diffusion out of hydrogel array spots. To mimic FGF2 diffusion out of hydrogel array spots, lysozyme was fluorescently labeled with NHS-rhodamine using the manufacturer’s protocol (Pierce, Rockford, IL). Rhodamine-labeled lysozyme was placed in 10,000 molecular weight cutoff dialysis tubing (Pierce, Rockford, IL) and dialyzed against ultrapure H2O (18 MΩ resistivity). 100μM Rhodamine-Lysozyme was loaded into hydrogel spots (n=4), photocrosslinked, and placed in phosphate buffered saline. Fluorescent micrographs were acquired using an inverted, compound fluorescence microscope (IX51, Olympus) at 40X magnification at predetermined times and the buffer was replaced after every image acquisition. The fluorescence intensity was measured using ImageJ software (Freeware, NIH, Bethesda, MD).
Toxicity studies with PD 173074 FGF2 inhibitor. To determine the toxicity of the PD 173074 small molecule FGFR inhibitor (Sigma, St. Louis, MO), hMSC’s were plated in 96 well plates and allowed to attach for 1 hour. After two hours, additional hMSC media was added so that there were final concentrations of 0, 25, 50, 75, 100, and 200nM PD173074. Cell viability was measured using the CellTiter Blue Assay (Promega, Madison, WI) three days after the addition of the PD 173074. Results indicate that there is a significant decrease in cell number in solutions containing 50-100 nM inhibitor, and a more substantial decrease in cell number in the presence of 200 nM inhibitor. The decrease in cell number in the presence of 50-100 nM inhibitor is likely due to inhibition of serum-derived FGF2 signaling, as the magnitude of the decrease is not consistent with toxicity. * Denotes significant difference from 0nM condition, ANOVA p<0.05. + Denotes significant difference from 0nM condition, ANOVA p<0.001.


