Abstract
Metazoan transcription is controlled either through coordinated recruitment of transcription machinery to the gene promoter, or through regulated pausing of RNA polymerase II (Pol II) in early elongation. We report that a striking difference between genes that use these distinct regulatory strategies lies in the “default” chromatin architecture specified by their DNA sequences. Pol II pausing is prominent at highly-regulated genes whose sequences inherently disfavor nucleosome formation within the gene, but favor occlusion of the promoter by nucleosomes. In contrast, housekeeping genes that lack pronounced Pol II pausing show higher nucleosome occupancy downstream, but their promoters are deprived of nucleosomes regardless of polymerase binding. Our results indicate that a key role of paused Pol II is to compete with nucleosomes for occupancy of highly-regulated promoters, thereby preventing the formation of repressive chromatin architecture to facilitate further or future gene activation.
Introduction
Eukaryotic gene expression begins with recruitment of the transcription machinery to a gene promoter and formation of a pre-initiation complex comprised of Pol II and general transcription factors (Roeder, 2005). This step is highly regulated and is enhanced by DNA sequence motifs within the promoter region, which are recognized by general transcription factors to stabilize transcription complex assembly (Juven-Gershon et al., 2008). Interestingly, these core promoter motifs are more prevalent at highly-regulated genes than at constitutively active housekeeping genes, suggesting that these two classes of promoters might employ different mechanisms to attract the transcription machinery (Basehoar et al., 2004; Hendrix et al., 2008).
Chromatin structure also impacts polymerase recruitment by modulating promoter accessibility, and activation of some genes requires disassembly of promoter nucleosomes by ATP-dependent chromatin remodeling complexes (Cairns, 2009). In the yeast S. cerevisiae, highly-regulated promoters are particularly likely to be occluded by nucleosomes prior to activation, making these genes reliant on nucleosome remodeling for transcription (Tirosh and Barkai, 2008). However, global mapping of nucleosomes in yeast has revealed that most promoter regions display low nucleosome occupancy even when the gene is inactive (Yuan et al., 2005; Albert et al., 2007), suggesting that assembly of promoter nucleosomes is inherently disfavored. Indeed, yeast promoter DNA sequences often contain rigid poly (dA:dT) tracts that deter nucleosome assembly (Iyer and Struhl, 1995). Accordingly, intrinsic sequence preferences for nucleosome formation contribute significantly to accessibility of yeast promoters in vivo (Sekinger et al., 2005; Kaplan et al., 2009; Zhang et al., 2009).
Human and Drosophila promoters are also generally nucleosome-deprived in a manner that is not dependent upon gene expression (Mito et al., 2005; Ozsolak et al., 2007; Mavrich et al., 2008; Schones et al., 2008). However, the mechanisms for this nucleosome depletion appear to be different than in yeast. Metazoan genes are much more G+C-rich than their yeast counterparts and, in contrast to yeast, are reported to intrinsically favor nucleosome formation around their promoters (Kaplan et al., 2009; Tillo et al., 2010). Thus, active mechanisms must contribute to the broad nucleosome-depletion observed in metazoans, such as recruitment of chromatin-remodeling complexes or association of the transcription machinery (Kim et al., 2005; Ozsolak et al., 2007). Indeed, pausing of Pol II near promoters can affect both the positioning (Mavrich et al., 2008; Schones et al., 2008) and occupancy of nucleosomes (Gilchrist et al., 2008).
Polymerase pausing was first described at the Drosophila heat shock genes, where Pol II synthesizes 25-50 nucleotides (nt) of RNA prior to heat shock and then halts to “wait” for an activating signal (Rougvie and Lis, 1988; Lis, 1998). Heat shock immediately triggers the release of paused polymerase into the gene, allowing an extremely rapid and robust transcriptional response (Lis, 1998). Rapid activation of heat shock genes is also favored by the lack of nucleosomes within the initially transcribed region (Wu, 1980) which would otherwise present barriers to efficient elongation (Izban and Luse, 1992). Although promoter-proximal pausing was once considered a rare phenomenon, recent work has demonstrated that it is a common regulatory strategy in higher eukaryotes (Muse et al., 2007; Zeitlinger et al., 2007; Core et al., 2008; Nechaev et al., 2010; Rahl et al., 2010). However, despite the growing appreciation for the widespread nature of pausing, the functions of paused Pol II remain to be elucidated.
We investigated the relationships between pausing, gene activity and chromatin structure by performing high-resolution mapping of Pol II, pause-inducing factors and nucleosomes across the Drosophila genome. Our data reveal that Pol II pausing occurs globally and plays a decisive role in determining promoter nucleosome occupancy. Moreover, we find that genes regulated by pausing rather than Pol II recruitment have distinct “default” chromatin architectures specified by their DNA sequences. Whereas recruitment-limited genes have intrinsically nucleosome-deprived promoters, genes with paused Pol II require polymerase occupancy to prevent promoter nucleosome assembly. These findings indicate that a gene's intrinsic nucleosome occupancy in the naïve, or “default” state is instructive for gene regulation, and suggest that the interplay between static information within promoter DNA sequences and the dynamics of polymerase pausing facilitates precise control of gene expression.
Results
Pausing of Pol II is Widespread and Occurs at Highly Active Genes
Regulation of Pol II pausing involves the coordinated action of both negative and positive elongation factors (Marshall and Price, 1992). Shortly after transcription initiation, the pause-inducing factors Negative Elongation Factor (NELF) and DRB-sensitivity inducing factor (DSIF) associate with the polymerase and decrease elongation efficiency (Yamaguchi et al., 2002; Wu et al., 2003; Cheng and Price, 2007; Lee et al., 2008). To examine the prevalence of pausing during early elongation, we used genome-wide ChIP-chip on high-density tiling arrays to compare NELF and DSIF distribution in Drosophila S2 cells with that of Pol II (Figure S1A and Table S1). Heatmaps representing fold enrichment over input DNA (Figure 1A) reveal a broad colocalization of NELF, total Pol II and DSIF near promoters. In fact, the average promoter signals for these factors correspond extremely well (Figure 1B and Figure S1B), indicating that NELF and DSIF generally associate with Pol II in the promoter-proximal region. Additionally, in agreement with recent reports (Rahl et al., 2010), most genes show enrichment in Pol II signal near promoters relative to downstream regions, suggesting that recruited polymerases are generally released inefficiently into genes.
Figure 1. Pol II, NELF and DSIF Globally Co-localize Near Promoters.
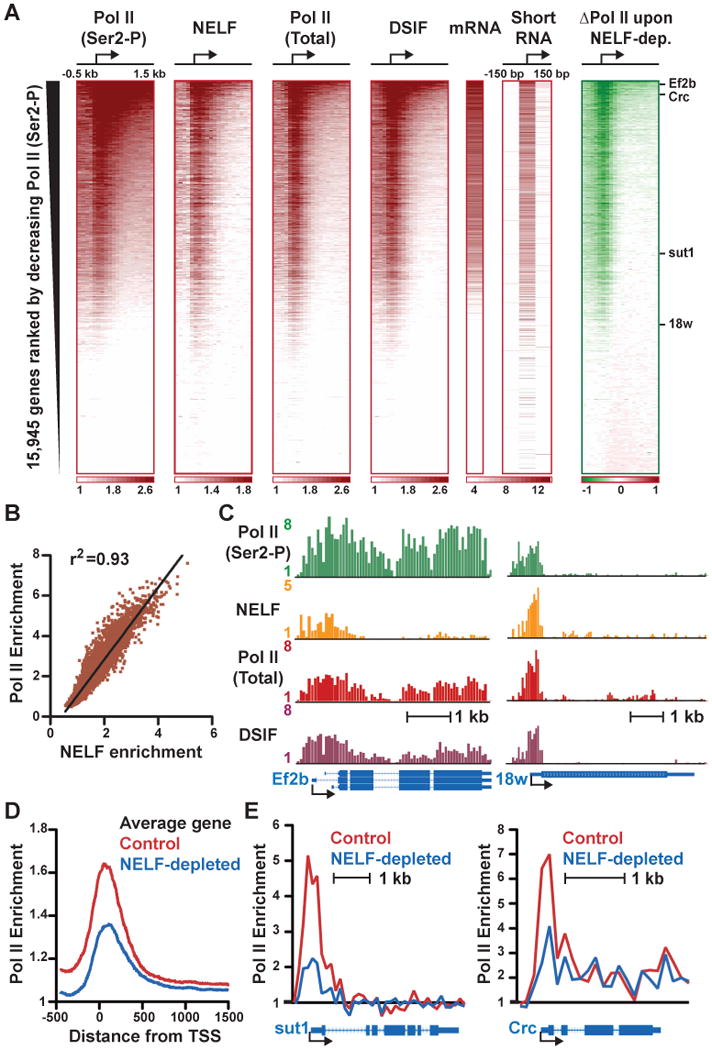
(A) Average fold enrichment over genomic DNA from ChIP-chip experiments is shown in 100 bp windows surrounding Drosophila TSSs (shown as arrows) for: actively elongating Pol II (Ser2-P), NELF (α-NELF-B), total Pol II (α-Rpb3) and DSIF (α-Spt5), with color bars at bottom indicating range. Expression levels determined by microarray (mRNA), and short RNAs derived from paused Pol II (Nechaev et al., 2010), are shown in Log2 units. The change in Pol II signal following NELF RNAi is shown at right, as compared to control samples. Range is depicted in color bar, where red signifies gain and green indicates loss in signal.
(B) The average enrichment for total Pol II and NELF around promoters (+/- 250 bp) are strongly correlated.
(C) ChIP-chip data for indicated factors displayed as fold enrichment at Ef2b (CG2238) a gene with considerable elongating Pol II (left) and 18w (CG8896) a gene with little evidence of productive elongation (right). Gene models below depict exons as boxes and introns as lines.
(D) Composite Pol II distribution profiles surrounding all promoters in control and NELF-depleted cells reveal a general decrease in promoter occupancy upon NELF RNAi.
(E) NELF-depletion affects Pol II promoter occupancy at genes with very little Pol II enrichment within the gene (sut1, CG8714), and with polymerase signal throughout the transcription unit (Crc, CG9429).
See also Figure S1.
Release of paused polymerase into productive elongation is triggered by the kinase activity of the positive transcription elongation factor b (P-TEFb) (Marshall and Price, 1995; Peterlin and Price, 2006). P-TEFb phosphorylates the Serine-2 residues on the Pol II C-terminal domain, leading to dissociation of NELF and recruitment of factors that facilitate transcription elongation and RNA processing. The tight correlation between NELF and Pol II signals near promoters suggests that each round of transcription involves NELF-mediated pausing, such that active genes should be enriched in NELF. To confirm this, we identified active genes by performing ChIP-chip with an antibody that recognizes the Serine-2 phosphorylated (Ser2-P) form of Pol II. All heatmaps shown in Figure 1A have genes rank-ordered from highest Ser2-P Pol II enrichment within the gene to lowest, clustering active genes at the top. Expression analysis confirms that genes with elevated Ser2-P Pol II signal produced significant levels of mRNA (Figure 1A, mRNA). Notably, the most active promoters were highly enriched in NELF (e.g. Ef2b, Figure 1C; Figure S1C), suggesting that NELF is universally present during early elongation, even at the most highly expressed genes.
To determine whether NELF-bound polymerases were engaged in transcription, we evaluated RNA production from each transcription start site (TSS). We found that >85% of Pol II-bound promoters generate significant short (<100 nt) transcripts (Nechaev et al., 2010), strongly supporting the idea that Pol II pauses promoter-proximally at these genes (Figure 1A, Figure S1D). Thus, the majority of Drosophila genes occupied by Pol II display the key hallmarks of polymerase pausing: i) occupancy by NELF and DSIF, ii) promoter-proximal enrichment of Pol II signal, and iii) the synthesis of short RNA transcripts. Notably, these findings suggest that it is not the initiation of NELF-mediated pausing, but rather the rate of pause release that is regulatory for transcription.
NELF Broadly Affects Promoter-proximal Pol II Occupancy
To evaluate the impact of pausing on Pol II promoter occupancy, we investigated the changes in polymerase distribution upon depletion of NELF (Figure S1E). These experiments demonstrated that NELF-depletion using RNA interference (RNAi) globally reduced promoter-proximal polymerase levels (Figure 1A, right panel; Figure S1F, P<0.0001). Composite Pol II profiles demonstrate that the average promoter signal is substantially reduced by NELF RNAi (Figure 1D), at both highly active and less active genes (Figure 1E). These results are consistent with widespread NELF-mediated pausing during early elongation, and provide further evidence that pausing is a general step in the transcription cycle. However, although NELF RNAi widely impacts Pol II promoter occupancy, polymerase loss at individual genes varies in magnitude, suggesting that some promoters are more reliant upon NELF to achieve maximal Pol II occupancy.
We investigated why genes showed differential responses to NELF RNAi, focusing on genes bound by Pol II in untreated S2 cells. Heatmaps depicting ChIP-chip signal around these promoters are shown (Figure 2A), with genes rank-ordered from most to least Pol II loss upon NELF-depletion. Consistent with NELF RNAi releasing paused polymerases (Muse et al., 2007), promoters with the highest levels of promoter-proximal Pol II and NELF enrichment in untreated cells experienced the largest losses in polymerase signal upon NELF-depletion (Figure S2A). Notably, the most NELF-affected genes (Quartile 1, Figure 2A, upper bracket) were among the most active (Figure S2B), confirming that NELF-mediated pausing plays a role at active genes.
Figure 2. Genes with Prominent NELF-mediated Pausing Have Strong Promoters and More Focused Transcription Initiation.
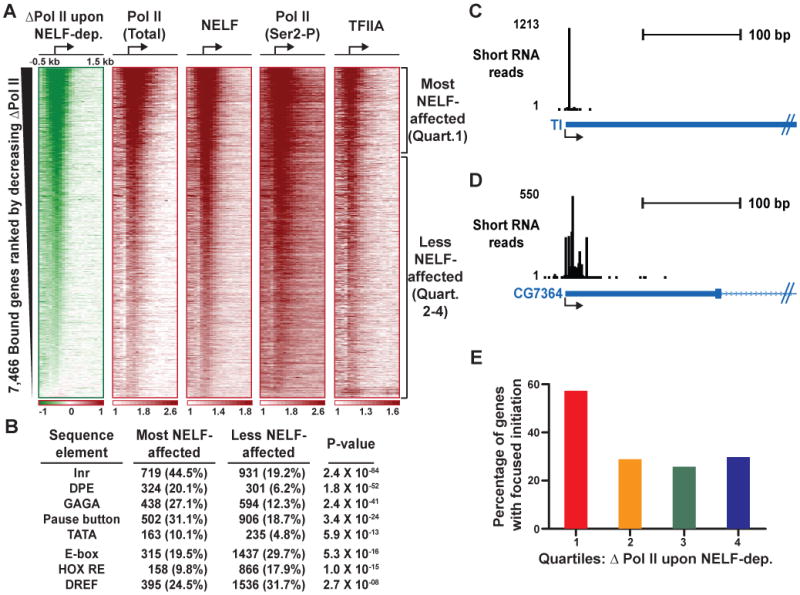
(A) Heatmaps depict loss of Pol II signal upon NELF-depletion or fold enrichment for the factors indicated at genes bound by Pol II in untreated cells. The rank order places promoter regions (+/- 250 bp) that lose the most Pol II signal upon NELF depletion at the top, and those least affected at bottom.
(B) Promoter motifs are enriched among the most NELF-affected genes (Quartile 1), whereas less NELF-affected genes (Quartiles 2-4) are more likely to possess activator binding sites, such as the E-box, homeo domain response element (Hox RE) or DNA-replication-related element binding factor (DREF). Pol II-bound genes with high confidence TSS annotation were analyzed (n=6,461; including 1,615 of the most- and 4,846 of the less NELF-affected genes), and the number and percentage of genes that possess each motif are shown, along with P-value (Fisher's exact test).
(C and D) Examples of genes that display highly focused transcription initiation (Tl, CG5490) or more dispersed initiation patterns (CG7364). Shown are the number of short RNA 5′-ends, at single-nucleotide resolution, that map near each TSS.
(E) The most NELF-affected genes (Quartile 1) have more focused initiation than genes less affected by NELF RNAi (Quartiles 2-4). Initiation was considered focused when ≥50% of total promoter-proximal reads (+/- 50 bp from TSS) mapped to a single location.
See also Figures S2 and S3.
Gene Ontology analysis of the most NELF-affected genes (Quartile 1) supports the idea that pausing is a favored regulatory mechanism at genes that require synchronous, precise control of expression (Muse et al., 2007; Gilchrist et al., 2008; Hendrix et al., 2008; Boettiger and Levine, 2009): these genes tend to encode highly-regulated components of developmental and stimulus-responsive pathways (Figure S2C). In contrast, genes less affected by NELF (Quartiles 2-4) include housekeeping genes involved in basic cellular processes (Figure S2C).
Genes with Paused Pol II Show High Levels of Pre-Initiation Complex Formation and Focused Initiation
The most NELF-affected genes also displayed a distinct sequence composition near their promoters. In agreement with recent reports (Hendrix et al., 2008; Lee et al., 2008), these genes were enriched in binding sites for GAGA factor, a protein important for pausing at the heat shock genes (Shopland et al., 1995), as well as a number of well-defined promoter motifs, such as the TATA box, Initiator (Inr) and Downstream Promoter Element (DPE). Interestingly, we found that two G+C-rich motifs that were over-represented at the most NELF-affected genes, the DPE and Pause Button, were both located between positions +26 and +33 at these genes (Figure S3A and Juven-Gershon et al., 2008). The precise coincidence of these sequence motifs with the peak of paused Pol II supports the idea that G+C-richness within the initially transcribed region influences elongation efficiency (Hendrix et al., 2008; Nechaev et al., 2010).
We found that 60% of the most NELF-affected genes possess at least one of the three core promoter motifs (TATA, Inr, DPE), compared to only 27% of the less NELF-affected genes. Strong core promoters are thought to direct transcription initiation that is focused around a single nucleotide position (e.g. Figure 2C), whereas the absence of such motifs leads to more dispersed initiation (e.g. Figure 2D; Juven-Gershon et al., 2008). Thus, we probed whether the observed enrichment in core promoter sequences at the most NELF-affected genes impacted the mode of transcription initiation at these genes. Mapping the 5′-ends of short capped RNAs (Nechaev et al., 2010) around the promoters of the most NELF-affected genes (Quartile 1) revealed that they experienced much more focused initiation than did less NELF-affected genes (Quartiles 2-4, Figure 2E and Figure S3B).
In agreement with the idea that highly NELF-affected genes contain intrinsically stronger promoters, we find that the general transcription factor TFIIA is significantly enriched at the most NELF-affected genes (Figure 2A and Figure S3C). Moreover, the general correspondence between occupancy by TFIIA and paused Pol II suggests that pausing may stabilize binding of general transcription factors, facilitating subsequent rounds of re-initiation at these promoters.
Conversely, weaker promoters with fewer core motifs were observed at genes that were less affected by NELF-mediated pausing, consistent with polymerase recruitment being inefficient, and likely rate-limiting at these genes. Moreover, the less NELF-affected genes were enriched in binding sites for transcription activators (Figure 2B), suggesting a greater reliance on extrinsic factors for recruitment of the transcription machinery. Thus, these findings point to a relationship between promoter strength and the rate-limiting step of transcription: genes where pause release is rate-limiting have strong promoters that drive efficient recruitment of Pol II, whereas recruitment-limited genes have weaker promoters and depend on additional factors for their activation.
Pol II Pausing is Linked to Nucleosome-deprivation Downstream of the TSS
We next mapped nucleosomes across the Drosophila genome using MNase-digestion of chromatin followed by high-throughput paired-end sequencing. We achieved >30-fold coverage of the genome (assuming one nucleosome every 200 bp), with 32.5 million reads that mapped uniquely to the 170 megabase genome. The distribution of these reads around TSSs of Pol II-bound genes is shown in Figure 3A as the number of read centers that mapped to each 50-bp bin. These data confirm that Pol II-occupied Drosophila promoters display a nucleosome-deprived region around the TSS (Mavrich et al., 2008). However, the most and least NELF-affected genes differed substantially in their nucleosome distributions downstream of the TSS.
Figure 3. Nucleosomes are Depleted Downstream of Promoters with Highly Paused Pol II.
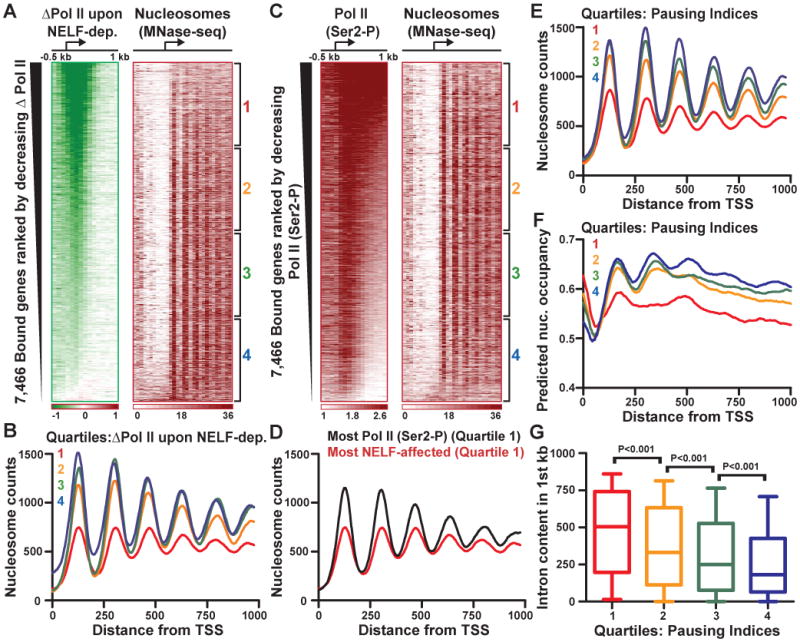
(A) The most NELF-affected genes are preferentially depleted of downstream nucleosomes. Heatmaps show the change in Pol II signal upon NELF depletion for Pol II-bound genes (as in Figure 1A) and nucleosome occupancy determined by paired-end MNase-seq (color intensity indicates the number of read centers that lie in each 50 bp bin).
(B) Pol II-bound genes were divided into quartiles based on the effect of NELF-depletion on Pol II promoter occupancy, from most affected (Quartile 1) to least (Quartile 4). Nucleosome distribution at genes in each quartile was determined by summing the number of nucleosome centers mapping to each position from the TSS to +1 kb.
(C) Transcription elongation modestly disrupts chromatin architecture. Heatmaps show Ser2-P Pol II signal and nucleosome distribution at genes rank ordered by levels of Ser2-P enrichment within the gene.
(D) Nucleosome occupancy is lower downstream of the most NELF-affected genes than at genes with the most active elongation (panel C, Quartile 1).
(E) Nucleosome occupancy at genes separated into quartiles by Pausing indices, where Quartile 1 represents genes with the most pausing.
(F) Predicted nucleosome occupancy at genes in each quartile of Pausing indices, based on intrinsic DNA sequence preferences of nucleosome formation (Kaplan et al., 2009).
(G) Intron content is shown for genes in each quartile of Pausing indices, revealing significantly elevated intron levels at genes that are highly affected by NELF-depletion (Kruskal-Wallis test, boxes depict 25-75th percentiles, whiskers show 10-90th).
See also Figure S4.
Genes that were less affected by NELF-depletion (Quartiles 2-4) exhibit a canonical, well-organized nucleosome architecture with a clear periodicity (Figure 3A, ∼170 bp inter-nucleosomal spacing). In contrast, the most NELF-affected genes (Quartile 1) show lower nucleosome occupancy and less organized chromatin structure. Composite metagene analysis of nucleosome distribution showed that the most NELF-affected genes contain far fewer nucleosomes within the initially transcribed region than genes less impacted by NELF RNAi (Figure 3B).
Pol II disrupts nucleosomes as it transcribes, and the considerable levels of Ser2-P Pol II detected at the most NELF-affected genes raised the possibility that the observed nucleosome deprivation could result from polymerase elongation. To address this issue, we analyzed nucleosome occupancy at Pol II-bound genes when ordered by descending levels of active elongation (Ser2-P Pol II signal; Figure 3C and Figure S4A). If polymerase elongation were largely responsible for low nucleosome occupancy, then the most actively transcribed genes should be particularly depleted of nucleosomes. In contrast, despite having much higher levels of Ser2-P Pol II signal (Figure S4B), genes with the most active elongation exhibit higher nucleosome density than the most NELF-affected genes (Figure 3D), indicating that Pol II elongation is not the dominant cause of nucleosome disruption within NELF-affected genes.
Nucleosome Depletion at Paused Genes Argues Against a Role for Nucleosomes in Establishing Paused Pol II
To further probe the link between paused Pol II and promoter-proximal nucleosome organization, we investigated nucleosome distributions at genes with varying levels of pausing, as judged by their “Pausing index” (calculated as the ratio of the Pol II signal near promoters (TSS +/- 250 bp) to the downstream region (+500 bp to the end of the gene), as described in Muse et al., 2007). Higher ratios reflect greater promoter-proximal enrichment of polymerase, and thus genes with the highest Pausing indices (Quartile 1) display the most paused Pol II. Consistent with our analysis of NELF-affected genes, the most paused genes show the lowest nucleosome occupancy within the initially transcribed region (Figure 3E, Figure S4C).
Pol II-bound genes containing TATA, Inr, or PB/DPE motifs also show reduced nucleosome density downstream of the TSSs relative to the average bound gene (Figure S4D), consistent with recent reports suggesting that these motifs are associated with diminished nucleosome organization (Albert et al., 2007; Mavrich et al., 2008). However, genes with GAGA elements showed the lowest average nucleosome occupancy, similar to that at the most NELF-affected genes (Figure S4D). These data are consistent with the known role of GAGA factor in recruiting chromatin remodeling complexes (Tsukiyama et al., 1994) and suggest that GAGA binding broadly leads to histone eviction. Notably, the presence of GAGA-binding sites at the most NELF-affected genes corresponded to a dramatic depletion of promoter-proximal nucleosomes (Figure S4E), indicating that many of these genes, like the Drosophila heat shock genes (Wu, 1980), are effectively nucleosome-free within the initially transcribed region. This finding argues strongly against recent suggestions that nucleosomes cause pausing by imposing a stable barrier to elongation (Schones et al., 2008; Mavrich et al., 2008). In contrast, we find higher promoter-proximal nucleosome occupancy at genes that display less pausing, implying that the presence of nucleosomes is unlikely to establish paused Pol II.
Nucleosome Occupancy is Intrinsically Disfavored Downstream of Paused Promoters
To evaluate the role of DNA sequence in establishing different chromatin structures, we determined the favored positions for nucleosome occupancy around Drosophila promoters using algorithms based on inherent sequence preferences for nucleosome formation (Kaplan et al., 2009). Surprisingly, these analyses revealed that sequences downstream of the most highly paused promoters intrinsically disfavor nucleosome occupancy (Figure 3F, Quartile 1), suggesting that the nucleosome depletion observed at these genes is specified by their DNA sequence (compare Figure 3F and Figure S4C).
Notably, introns are enriched in nucleosome-disfavoring sequences and have lower nucleosome occupancy than exons in vivo (Schwartz et al., 2009). Highly-regulated Drosophila genes, and in particular those involved in development, are known to possess long introns, leading us to investigate whether an elevated intron content downstream of highly-paused genes might contribute to their nucleosome depletion. Indeed, genes with the highest Pausing indices had significantly higher intron content within the first 1kb than did less paused genes (Figure 3G). These intriguing results suggest that introns may serve a role in deterring nucleosome formation at highly-paused genes, helping to establish distinct downstream chromatin architectures.
Promoters of Highly Paused Genes Favor Nucleosome Assembly
Consistent with prior work (Mito et al., 2005; Ozsolak et al., 2007; Mavrich et al., 2008; Schones et al., 2008), we found that Pol II-bound promoters are generally depleted of nucleosomes (Figure 4A). However, it remained unclear whether this depletion is entirely caused by the presence of Pol II, or if sequence preferences for nucleosome formation contribute as well (Tillo et al., 2010). To address this question, we determined the predicted nucleosome occupancy around promoters in each Pausing index quartile. Strikingly, genes with the highest Pausing indices (Figure 4B, Quartile 1) contain promoters that intrinsically favor assembly of a nucleosome over the TSS, whereas this tendency is diminished at genes with less pausing (Quartiles 2-4). This result suggests that highly paused promoters encode an inherently repressive chromatin structure that is counteracted by pausing of Pol II. In contrast, less paused promoters may not require Pol II pausing to deter promoter nucleosome formation, instead possessing sequences that disfavor nucleosome assembly.
Figure 4. DNA Sequences at Paused Genes Favor High Promoter Nucleosome Occupancy.
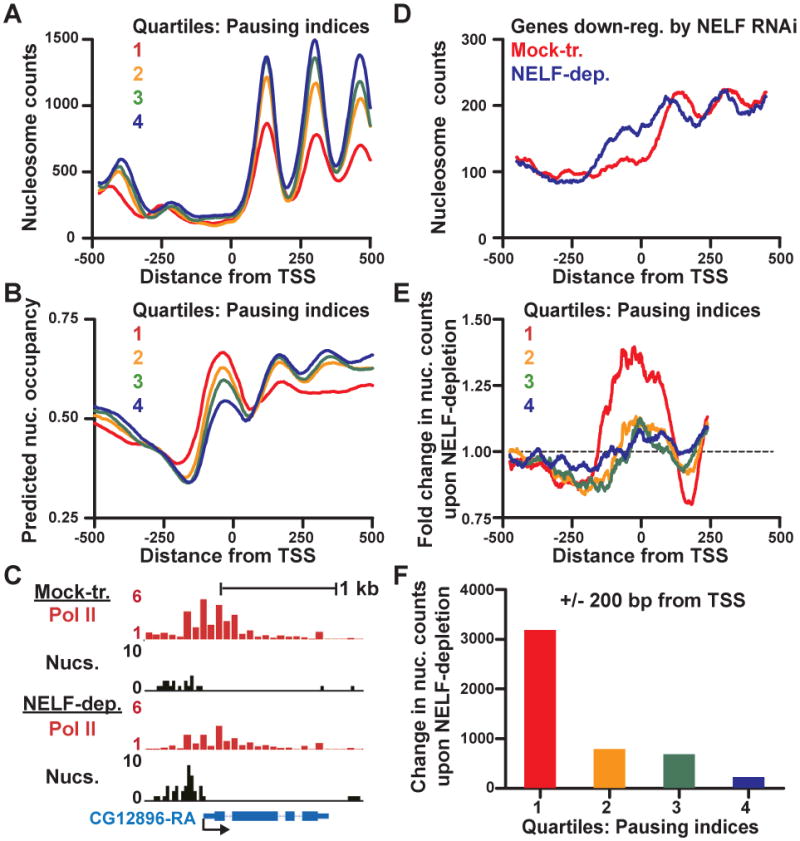
(A) Nucleosome occupancy at Pol II-bound genes (n=7,466) separated into quartiles by Pausing indices, where Quartile 1 represents the most paused genes. Nucleosome occupancy at genes in each quartile was determined by summing the number of nucleosome centers mapping to each position.
(B) Predicted nucleosome occupancy at genes in each quartile of Pausing indices, based on intrinsic DNA sequence preferences of nucleosome formation, as in (Kaplan et al., 2009).
(C) Loss of Pol II upon NELF-depletion is accompanied by increased nucleosome occupancy. Pol II ChIP-chip fold enrichment (red) and MNase-seq read distribution (black, depicts read centers in 25-bp bins) around a highly NELF-affected gene (CG12896) in mock-treated and NELF-depleted samples.
(D) Genes down-regulated by NELF-depletion show increased promoter nucleosome occupancy. Nucleosome occupancy (calculated as in A) at genes whose expression decreased >2-fold following NELF-depletion.
(E and F) NELF-depletion leads to increased nucleosome occupancy over highly paused promoters. The change in nucleosome counts upon NELF-depletion (MNase-seq reads in NELF-depleted/Mock-treated samples) is shown for genes in each quartile of Pausing indices as: fold change in read number at each position (E) or the raw increase in the number of nucleosome reads +/- 200 bp from the TSS (F).
We showed previously that loss of paused Pol II upon NELF depletion leads to increased nucleosome occupancy and down-regulation of gene expression at several highly paused promoters (Gilchrist et al., 2008). The data shown in Figure 4B suggest that increased nucleosome occupancy at these genes is driven by sequences that favor nucleosome assembly. To test this model on a global scale, we mapped nucleosomes in NELF-depleted and Mock-RNAi treated cells using MNase-seq. Figure 4C shows one example of a highly paused gene with low promoter nucleosome occupancy in mock-treated cells. Depletion of NELF results in a reduction in Pol II promoter signal, and an accompanying increase in promoter-proximal nucleosome levels. This finding can be extended broadly to genes whose expression is down-regulated following NELF RNAi (>2-fold change, see microarray expression data, Table S3), which show increased promoter nucleosome occupancy in NELF-depleted cells (Figure 4D). Interestingly, these genes also show a shift in nucleosome position, with downstream nucleosomes moving towards the promoter following NELF-depletion, implying a dynamic relationship between Pol II and nucleosome binding at these promoters.
Furthermore, the increase in nucleosome occupancy over the TSS following NELF RNAi is a general feature of highly paused genes. Comparing the nucleosome levels in NELF-depleted vs. mock-treated cells revealed a considerable increase in nucleosome occupancy surrounding promoters of the most paused genes (Figure 4E and 4F, Quartile 1). In contrast, NELF RNAi resulted in much smaller changes in nucleosome occupancy at genes with less paused Pol II (Figure 4F). These results demonstrate that NELF-mediated pausing inhibits nucleosome occupancy of the most highly paused promoters, and that loss of pausing allows these genes to assume the “default” nucleosome organization specified by the underlying DNA sequence.
Pol II Binding Inhibits Nucleosome Occupancy at the Most Paused Genes
We further investigated nucleosome architecture around highly paused vs. less paused promoters by comparing Pol II and nucleosome occupancy at individual genes in their repressed and activated states. To accomplish this, we took advantage of the fact that a 24-hour treatment of Drosophila cells with the steroid hormone ecdysone causes marked changes in gene expression (Dimarcq et al., 1997). Pol II ChIP-chip was performed with and without ecdysone treatment to identify genes that transitioned between Pol II-bound and unbound states (or vice versa) during this treatment. We then used quantitative PCR on MNase-digested chromatin to investigate changes in nucleosome occupancy that accompanied these Pol II transitions, focusing on genes that were highly paused (Quartile 1), or less paused (Quartiles 3 or 4) in the active state.
We found that highly paused genes had nucleosome-occluded promoters in the absence of Pol II, and polymerase binding substantially reduced nucleosome levels at these genes (Figures 5A, 5B and Figure S5). In contrast, genes lacking paused Pol II (Figures 5C and 5D) were generally depleted of nucleosomes, even in the unbound state. These data further support the notion that sequences around highly paused promoters specifically favor nucleosome assembly, and that paused Pol II prevents nucleosome formation around these promoters.
Figure 5. The Relationship Between Pol II and Nucleosome Occupancy at Highly Paused and Less Paused Genes.
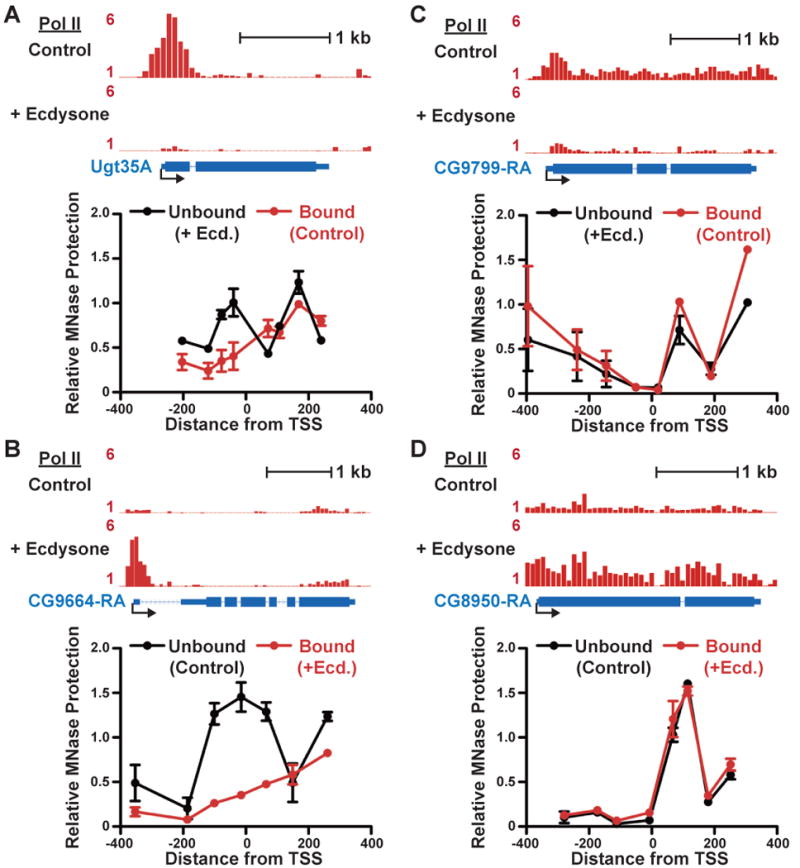
Pol II distribution in S2 cells +/- 24-hour treatment with ecdysone is depicted as fold enrichment from ChIP-chip experiments. MNase protection assays are shown below to compare nucleosome occupancy at each gene in the Pol II-bound vs. unbound state. Data points represent average qPCR signal of DNA protected against MNase digestion from two biological replicates at primer pairs centered at the indicated distance from the TSS; error bars depict range.
(A) Ugt35A (CG6644), a gene with a high Pausing index in control cells that becomes unbound by Pol II following treatment with ecdysone.
(B) CG9664, a gene unbound by Pol II in control cells that becomes highly paused in ecdysone-treated cells.
(C) Ecdysone causes CG9799, a gene with a low Pausing index in control cells, to become unbound by Pol II.
(D) CG8950, an unbound gene in control cells, has uniform Pol II distribution upon ecdysone treatment.
See also Figure S5.
Genes Adopt their Predicted Nucleosome Organization in the Absence of Pol II Binding
The above data suggests that sequences around the most highly paused genes in Drosophila S2 cells inherently favor nucleosome occupancy that is high over promoters and lower downstream. To ascertain whether these characteristics would be conserved in a different context, we determined Pol II distribution in Drosophila 0-16 hour old embryos and compared this to embryo nucleosome occupancy reported previously (Mavrich et al., 2008). Figure 6A displays these data, with genes rank-ordered by descending Pausing index in embryos. Importantly, we find that nucleosome depletion downstream of the most highly paused promoters is not limited to S2 cells, but is maintained in developing embryos (Figure 6B). Likewise, calculation of predicted nucleosome occupancies for genes in each Pausing index quartile in embryos corroborated data from S2 cells (Figure 6C; Figure S6A): the most highly paused genes in embryos exhibited higher predicted nucleosome occupancy over the promoter than within the gene (Figure 6C, Quartile 1), whereas genes with the least Pol II pausing (Quartile 4) favored higher nucleosome occupancies downstream.
Figure 6. Promoters Adopt their Predicted Nucleosome Configuration in the Absence of Paused Pol II.
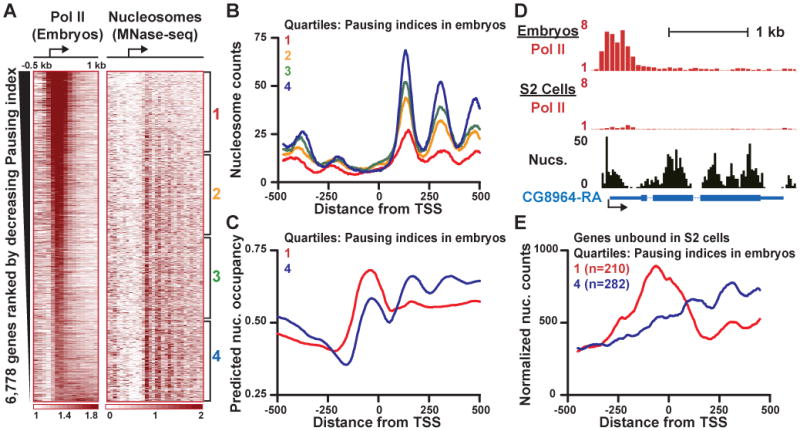
(A) Pol II distribution and nucleosome occupancy (H2A.Z nucleosomes from Mavrich et al., 2008) in Drosophila embryos. Genes are rank ordered by descending Pausing indices in embryos.
(B) The most highly paused genes in embryos are depleted of downstream nucleosomes. Pol II-bound genes in embryos were divided into quartiles based on Pausing indices and composite metagene analyses of nucleosome reads around their promoters were generated from the data in (Mavrich et al., 2008).
(C) Predicted nucleosome occupancy at genes in each quartile of Pausing indices as determined in Drosophila embryos.
(D) A promoter with a high Pausing index in embryos (top panel) that is not occupied by Pol II in S2 cells (middle panel) becomes occluded by nucleosomes in the unbound state. Bottom panel shows nucleosome occupancy at the unbound gene in S2 cells as determined by MNase-seq, with read centers displayed in 25 bp bins.
(E) In the absence of Pol II, in vivo nucleosome occupancy closely resembles predictions. Nucleosome occupancies were determined in S2 cells, where these genes are not bound by Pol II. Shown are genes that are highly paused (Quartile 1) or lacking paused Pol II (Quartile 4) in embryos.
See also Figure S6.
These experiments also identified many promoters that showed high Pausing indices in embryos but were unoccupied by Pol II in S2 cells (e.g. Figure 6D, Figure S6B). If our model is correct, then these promoters should be depleted of nucleosomes in the presence of paused Pol II, but occluded by nucleosomes in the absence of polymerase binding. To test this idea, we analyzed nucleosome distribution at genes that were highly paused in embryos (Quartile 1) but were unbound by Pol II in S2 cells. In support of our model, these genes contain a nucleosome positioned directly over the promoter in the Pol II-unbound state (S2 cells, Figure 6E), and this nucleosome is displaced in the presence of paused Pol II (embryos, Figure S6C). Notably, although Pol II-binding substantially decreases promoter-proximal nucleosome occupancy at these genes, it only modestly affects downstream nucleosome levels (Figure S6C). In contrast, promoters with the lowest Pausing indices in embryos were generally nucleosome-deprived regardless of Pol II occupancy (Figures 6E and S6D), and had higher nucleosome density within the gene, consistent with sequence-based predictions. Taken together, our data demonstrate that genes generally assume their sequence-predicted nucleosome architecture in the absence of the transcription machinery. That genes with different levels of pausing possess such distinct default states suggests that there is a fundamental relationship between intrinsic chromatin structure and gene regulatory strategies.
Discussion
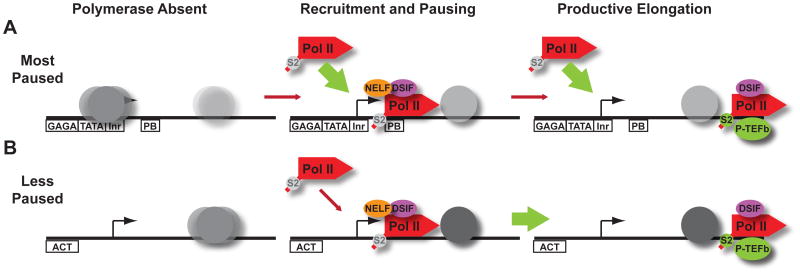
Our data support a general model for gene regulation wherein the underlying DNA sequence around promoters directly influences both chromatin architecture and the step in the transcription cycle that is rate-limiting for gene expression. We find that genes with high levels of Pol II pausing (Figure 7A) inherently favor the formation of nucleosomes over the promoter, establishing an active competition between Pol II and nucleosomes for promoter occupancy. We propose that this intrinsically repressive chromatin structure prevents aberrant expression of paused genes, which are often components of highly-regulated pathways. Nucleosome remodeling, likely initiated by proteins such as GAGA factor, would be required to disassemble nucleosomes at these promoters and allow for gene activity (small red arrow). Nucleosome removal would uncover strong promoter motifs that facilitate efficient, stable recruitment of the transcription machinery (large green arrow). Extended NELF-mediated pausing of polymerase at these promoters makes the transition to productive elongation slow (small red arrow). However, upon pause release, low levels of downstream nucleosomes would minimize barriers to transcription elongation and additional Pol II molecules would be rapidly recruited to maintain high Pol II occupancy and prevent nucleosome formation.
Figure 7. Different Default Chromatin Architectures Specify Distinct Gene Regulatory Strategies.
(A) The most paused promoters are inherently occluded by nucleosomes (shown as gray ovals, where color intensity denotes occupancy levels) prior to Pol II binding. Regulated chromatin remodeling (red arrow) can expose strong promoter motifs (shown as boxes below DNA) that allow for efficient Pol II recruitment (green arrow). Subsequent recruitment of P-TEFb and pause release are also regulated at these genes (second red arrow), providing an additional opportunity for gene regulation.
(B) Less paused genes display weaker, nucleosome-deprived promoter regions. Polymerase recruitment is rate-limiting at these genes (red arrow), and perhaps more dependent on activators (ACT, binding site shown as box). Pol II is bound by DSIF and NELF at these genes, but pausing is transient and the polymerase moves efficiently into the gene (green arrow).
In contrast, genes that lack extended pausing (Figure 7B) appear to disfavor promoter nucleosome assembly and instead harbor nucleosomes flanking the nucleosome-deprived promoter region. Localized DNA accessibility near TSSs could both help target the transcription machinery to the promoter region and diminish the requirement for nucleosome remodeling to allow gene activity. The dearth of core promoter elements could make these genes more reliant on activator binding for recruitment of the transcription machinery, and Pol II recruitment would be the rate-limiting step for expression of these genes (small red arrow). Pausing would be short-lived at these genes, and despite higher downstream nucleosome occupancy, polymerase escapes efficiently into productive synthesis.
Importantly, these two strategies present different opportunities for gene regulation. Highly paused genes present two distinct steps at which they can be regulated: promoter accessibility and release of Pol II from pausing. We propose that this two-step mechanism facilitates precise control of gene expression. We envision that the first step, nucleosome remodeling, functions as a molecular switch that relieves repression by chromatin to permit expression. This step can be temporally uncoupled from gene activation and could potentiate genes for future activation rather than prompting their immediate expression. The second step, release of paused Pol II, might be analogous to a volume dial, which permits fine-tuning of expression levels in response to changing conditions. Transcription levels could be rapidly regulated solely by manipulating the efficiency of P-TEFb recruitment through its interactions with DNA-binding transcription activators and histone modifications (Peterlin and Price, 2006; Rahl et al., 2010). This idea is supported by observations that activation of highly paused genes is both fast and synchronous (Lis, 1998; Boettiger and Levine, 2009). In contrast, genes that lack promoter-proximal pausing and nucleosome occupancy rely chiefly on a single-step mechanism to alter gene expression: regulated, step-wise recruitment of the transcription machinery. This mode of regulation has been suggested to be inherently more stochastic and prone to transcriptional noise (Boettiger and Levine, 2009), which may explain why many genes regulated by recruitment are constitutively active housekeeping genes.
We provide evidence that NELF-mediated pausing during early elongation is a general feature of the transcription cycle that is exploited at some genes to regulate transcription output. We propose that each round of transcription entails pausing, perhaps serving as an early “checkpoint” to ensure proper maturation of the elongation complex before release into productive elongation. At some genes, this halt in elongation may be transient, whereas at others it may involve a long-lived paused complex that becomes rate-limiting for gene expression. Importantly, these results imply that the release from pausing through P-TEFb recruitment is an important, regulated step that broadly impacts gene expression, in agreement with recent work (Peterlin and Price, 2006; Rahl et al., 2010). We note that general recruitment of NELF during early elongation likely explains the seemingly paradoxical observation made in several systems that NELF levels increase at activated genes that experience robust recruitment of additional Pol II.
Our data also reveal that the inherent preference towards repression of highly-regulated promoters by nucleosome occlusion is an evolutionarily conserved phenomenon (Tirosh and Barkai, 2008). Moreover, our results are in agreement with recent work in yeast which reveals that Pol II plays a role in displacing nucleosomes from promoter regions (Weiner et al., 2009). However, in yeast, nucleosome disassembly is coupled directly to gene activation, whereas in Drosophila nucleosome disassembly is coupled to Pol II pausing. Perhaps Drosophila and other metazoans have evolved promoter-proximal pausing as an additional layer of regulation to accommodate increased demands for precise and rapid gene regulation during development and organismal responses to stress. In addition, it might be beneficial to maintain highly-regulated promoters poised in an open chromatin state, to prevent their incorporation into the more inaccessible, condensed heterochromatin that exists in metazoans.
In summary, we report that a primary function of paused Pol II is to prevent promoter-proximal nucleosome formation. This represents a fundamental shift in our thinking about the role of Pol II pausing, which has long been thought to simply repress gene expression. Instead, we argue that pausing should be viewed as a mechanism to fine-tune gene expression, and to potentiate genes for further or future activation. In addition, we have shown that sequence-specified “default” nucleosome architecture instructs the regulatory properties of Drosophila promoters. We propose that metazoans have evolved a gene regulatory strategy in which nucleosomes and paused Pol II compete for promoter occupancy, affording multiple opportunities for regulation of gene expression.
Experimental Procedures
ChIP-chip Experiments
Untreated and RNAi-treated Drosophila S2 cells were cross-linked, immunoprecipitated, amplified and labeled for ChIP-chip as described previously (Gilchrist et al., 2009). NimbleGen tiling arrays that span the Drosophila genome (2.1 million probes) were probed according to manufacturer's instructions. Data shown represent average probe signals from at least two biological replicates. Antibodies and detailed methods are described in Extended Experimental Procedures.
Defining Nucleosome Positions by MNase-seq
MNase-digested chromatin from untreated, Mock-treated and NELF-depleted S2 cells was prepared as described in (Gilchrist et al., 2008) except that 200 μl chromatin was digested with 20 units MNase (Worthington) for 45 minutes at 25° C. Following gel purification, mono-nucleosome sized fragments (100-200 bp) were subjected to sequencing using the Illumina paired-end protocol. The resulting data set from untreated samples included 32.5 million unique read pairs identifying both ends of fragments ≥120 bp and ≤180 bp in length, whereas 11.2 million read pairs were obtained from each of the RNAi-treated samples.
Predictions of Nucleosome Occupancy
D. melanogaster (Fly dm3) genome-wide nucleosome positioning prediction data for average occupancy (predicted probability for each position in the genome to be covered by any nucleosome) were downloaded as described (Kaplan et al., 2009). Genomic position average occupancy values were placed in gene context relative to TSSs using custom scripts. The resulting predictions of nucleosome occupancy with respect to individual TSSs were used to generate metagene analyses of predicted nucleosome occupancy for select groups of genes as noted in the text.
Supplementary Material
Acknowledgments
We thank S. Nechaev and G. Hu for critical reading of the manuscript. We acknowledge L. Pederson for production of the NELF-B protein, J. Tucker and the NIEHS microarray core for help with arrays, and S. Dai and J. Grovenstein for computational support. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to K.A.(Z01 ES101987) and L.L.(ES 101765).
Footnotes
Author Contributions D.A.G and K.A. designed experiments, D.A.G, G.D.S., B.X. and Y.G. performed experiments, D.A.G, D.C.F, L.L. and K.A. performed data analysis and D.A.G and K.A. prepared the manuscript.
Author information Genomic data described in this work have been deposited in GEO under accession #GSE20472.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science (New York, NY. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA Polymerase II Elongation Complexes Before and After the P-TEFb-mediated Transition into Productive Elongation. The Journal of biological chemistry. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science (New York, NY. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarcq JL, Imler JL, Lanot R, Ezekowitz RA, Hoffmann JA, Janeway CA, Lagueux M. Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect biochemistry and molecular biology. 1997;27:877–886. doi: 10.1016/s0965-1748(97)00072-6. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Fargo DC, Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods (San Diego, Calif. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & development. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. The EMBO journal. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. The Journal of biological chemistry. 1992;267:13647–13655. [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Current opinion in cell biology. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Molecular and cellular biology. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor symposia on quantitative biology. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Molecular and cellular biology. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. The Journal of biological chemistry. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nature genetics. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nature genetics. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science (New York, NY. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nature biotechnology. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nature structural & molecular biology. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Molecular cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & development. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PloS one. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome research. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome research. 2009;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & development. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Molecular and cellular biology. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science (New York, NY. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nature structural & molecular biology. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.