Abstract
Synthesis of new ribosomes is an essential process upregulated during cell growth and proliferation. Here, we review our current understanding of the role that ubiquitin and ubiquitin-like proteins (UBLs) play in ribosome biogenesis, with a focus on mammalian cells. One important function of the nuclear ubiquitin-proteasome system is to control the supply of ribosomal proteins for the assembly of new ribosomal subunits in the nucleolus. Mutations in ribosomal proteins or ribosome assembly factors, stress, and many anticancer drugs have been shown to disrupt normal ribosome biogenesis, triggering a p53-dependent response. We discuss how p53 can be activated by the aberrant ribosome formation, centering on the current models of the interaction between ribosomal proteins released from the nucleolus and the ubiquitin ligase Mdm2. Recent studies also revealed multiple ubiquitin- and UBL-conjugated forms of nucleolar proteins with largely unknown functions, indicating that many new details about the role of these modifications in the nucleolus await to be discovered.
Keywords: ribosome biogenesis, ubiquitin, ribosomal proteins, nucleolar stress, p53, Mdm2
Introduction
Recent studies demonstrate that ubiquitin and ubiquitin-like proteins (UBLs)1 play important roles in ribosome biogenesis: the process of building the molecular machines that synthesize all proteins in a cell. Increased production of ribosomes is an integral part of metabolic changes that occur during cell growth,2,3 and some 105 to 106 ribosomes are typically generated prior to cell division in order to provide each daughter cell with the translation machinery.
The cytoplasmic ribosomes in eukaryotes are large ribonucleoprotein particles consisting of 2 subunits that contain the intricately folded ribosomal RNA (rRNA) and approximately 80 ribosomal proteins (RPs). Ribosome biogenesis begins with the transcription of rRNA precursors by RNA polymerase I (Pol I), followed by their processing to the mature 18S rRNA of the small (40S) subunit and 5.8S and 28S rRNAs of the large (60S) subunit. Large subunits also contain 5S rRNA, which is separately transcribed by RNA polymerase III. RPs are incorporated into the ribosomal structure concomitantly with the processing of pre-rRNA transcripts into mature rRNAs. After being assembled in the nucleus, the nascent 40S and 60S subunits are separately exported to the cytoplasm, where a few final maturation steps take place, making subunits competent for translation. Altogether, this complicated pathway requires more than 200 different protein factors and dozens of small RNAs that transiently interact with preribosomal complexes during their assembly.4
Most steps in ribosome biogenesis take place in the nucleolus, a nuclear compartment that provides an environment for the synthesis of ribosomes and several other ribonucleoprotein particles.5,6 The nucleolus is involved in a number of other cellular processes including chromosomal segregation, cell cycle control, and stress responses.7 The size of the nucleolus correlates with Pol I activity and the expression of proteins involved in ribosome biogenesis,8 reflecting the primary role of this organelle as the cellular ribosome factory. Rapidly proliferating cancer cells show upregulation of genes involved in ribosome production,9 and the presence of prominent nucleoli is one of the consistent cytological features in neoplastic cells.10
Accumulating evidence indicates that a range of events in ribosome formation and surveillance of this process utilize ubiquitin- and UBL-based regulatory circuits. In this review, we will discuss how ubiquitin and UBLs contribute to the complex milieu of the mammalian nucleolus, with the emphasis on the current understanding of the ubiquitin-dependent mechanisms involved in monitoring nucleolar function and their relation to cancerogenesis.
Ubiquitin and RPs: Balancing Supply and Demand
Biosynthesis of ribosomes is a remarkably efficient process; a growing HeLa cell is estimated to make as many as 7,500 new ribosomal subunits per minute.11 This high efficiency can only be realized through a balanced supply of hundreds of protein and RNA molecules that are necessary for manufacturing each ribosomal subunit. The RPs and accessory protein factors are synthesized in the cytoplasm and imported to the sites of ribosome assembly in the nucleolus. The rates of their synthesis are governed by growth factors and nutrients via transcriptional and translational control mechanisms.2,3,12 However, there is no mechanism that would guarantee production of dozens of individual proteins in equimolar amounts, matched precisely to the levels of rRNA transcripts. The first clues to how mammalian cells deal with this complex logistics problem came from the early observations that the newly synthesized RPs are highly unstable if rRNA transcription is inhibited,13 which suggested that cells may actively dispose of RPs that fail to assemble into ribosomes. Recent studies using quantitative mass spectrometry and live-cell imaging demonstrated that under normal growth conditions, cells import more RPs into the nucleus than export in the form of assembled ribosomal subunits, indicating rapid nuclear turnover of a fraction of RPs.14 Thus, instead of tightly controlling expression of each individual RP, mammalian cells produce these proteins in excess of what is required for ribosome synthesis and degrade those that do not become assembled with rRNA.
How do cells degrade a surplus of RPs? Treatment of cells with proteasome inhibitors increases levels of RPs in the nucleus (and the nucleolus) but not in the cytoplasm,14,15 pointing to a role of the nuclear proteasomal pathway in their turnover. Proteasomes, which are responsible for the degradation of the bulk of proteins in eukaryotic cells, are known to operate in the nucleus.16,17 Although proteasomes were reported in the nucleolus under certain conditions,18 a systematic analysis of immunolabeling techniques suggests that they are normally present in the nucleoplasm and excluded from the nucleolus.19 Consistent with the notion that RPs are degraded outside of the nucleolus, time-lapse fluorescence imaging showed that upon inactivation of proteasomes, the lack of rRNA transcripts leads to RP accumulation in the nucleoplasm.14
The identity of ubiquitin ligases involved in targeting RPs for proteasomal degradation is unclear at the moment. RPs that are unassembled into ribosomal subunits can rapidly move between the nucleolus and the nucleoplasm.14 Does their mobility lead to random encounters with the generic protein degradation machinery in the nucleus, or are there specific enzymatic systems that track free RPs and mark them for degradation? As a hint to another layer of complexity, proteomic studies demonstrated numerous instances of modification of RPs not only by ubiquitin20,21 but also UBLs22,23 (see also below).
The load on the nuclear ubiquitin-proteasome system (UPS) from unassembled RPs may substantially increase during stress. For instance, inhibition of Pol I transcription with actinomycin D in HeLa cells does not lead to an immediate shutdown of RP synthesis.13 As a result, a large quantity of RPs that cannot be used in ribosome assembly must be turned over. The degradation of RPs does occur with a fairly efficient rate, as half-lives of most RPs under these conditions are less than 30 to 90 minutes.13 As will be discussed below, other stress conditions may create a similar situation in which massive amounts of RPs become “unemployed” and thus become potential substrates for the nuclear UPS.
Other Functions of Ubiquitin in Ribosome Biogenesis
There is growing evidence that ubiquitin is involved in diverse aspects of eukaryotic ribosome biogenesis. One of the early observations was that the small ribosomal subunit protein S27a (corresponding to Rps31 in budding yeast) and the large subunit protein L40 (Rpl40A/B in yeast) are expressed as fusions with an N-terminal ubiquitin moiety.24-26 Before assembly into ribosomes, ubiquitin is released from these RPs by proteolytic processing. In budding yeast, the new ubiquitin derived from RP fusions is sufficient for growth under normal conditions.27 This arrangement is not universal, however, because some eukaryotes express S27a as tails of polypeptides that resemble ubiquitin but have diverged amino acid sequences.28 In another variation, an additional ubiquitin-like sequence is fused N-terminally to the small subunit protein S30 in animals.29 The ubiquitin extension is required for the optimal expression of Rps31, but it needs to be removed for the efficient assembly of this RP into 40S subunits,30 implying that it facilitates folding and/or transport of Rps31 prior to assembly into ribosomes.
Treatment of human cells in culture with proteasome inhibitors alters nucleolar morphology, inhibits pre-rRNA transcription and processing, and causes accumulation of immature preribosomal particles.31,32 Given that many ribosome synthesis factors are ubiquitinated,20,21 these changes could conceivably occur as a result of a “multiple system failure” in the nucleolus. Ubiquitination has been reported to regulate turnover of several nucleolar proteins, including the Pol I transcription factor TIF-IA,32 fibrillarin,33 and the nucleolar protein N4BP1.34 However, more studies are needed to understand the exact mechanism of how this affects nucleolar function, as well as the enzymes that ubiquitinate and deubiquitinate nucleolar substrates. A precedent can be found in the recently characterized mammalian ubiquitin-specific protease USP36 that localizes to the nucleolus. Knockdown of USP36 in HeLa cells was shown to affect ubiquitination of nucleolar proteins and inhibit transcription of pre-rRNAs and their processing, leading to nucleolar fragmentation.35
Like any other complex assembly process, biosynthesis of ribosomes is bound to generate a certain amount of defective products. Misassembled ribosomal precursors are destroyed in the nucleus by the mechanisms that are still poorly understood. In addition, ribosomes with mutations in rRNAs that impair their function in translation rather than assembly become targets for degradation in the cytoplasm by the surveillance mechanism termed nonfunctional rRNA decay (NRD), recently discovered in yeast.36 Interestingly, degradation of the mutant rRNA is paralleled by ubiquitination of the defective ribosomes by the E3 enzymatic complex Mms1/Rtt101.37 It is currently unknown whether ubiquitination of RPs in the defective ribosomes initiates their destruction by the NRD machinery. Nevertheless, these observations suggest an intriguing possibility that besides tagging protein substrates for degradation, ubiquitin may also be involved in the regulation of degradation of RNA-protein complexes.
UBLs: Going with the Flow?
UBLs comprise a family of small proteins that are related to ubiquitin and share the same overall tertiary structure.38 Like ubiquitination, modification of protein substrates with UBLs is a multistep process mediated by distinct E1, E2, and E3 enzymatic activities.1 Modifications with UBLs modulate target protein interactions and function but are not commonly associated with proteasome binding and degradation.
In a recent study, multiple RPs were found to be substrates for modification by the ubiquitin-like NEDD8 polypeptide in vivo.22 The same study showed that inhibition of the proteasome increased the amount of both ubiquitinated and neddylated RPs in cellular extracts. Overexpression of the NEDD8-specific protease NEDP1 was found to decrease the half-life of one of the studied RPs, L11, but whether neddylation is a general mechanism regulating RP stability is not yet clear. Photobleaching experiments also showed changes in the nucleolar recovery rates of L11-EGFP upon knockdown of NEDD8,39 suggesting that this modification may be involved in trafficking RPs to the nucleolus.
The small ubiquitin-related modifier (SUMO) is another UBL that modifies multiple proteins involved in ribosome biogenesis. Studies in yeast found that despite the relatively low abundance of the sumoylated forms of ribosome assembly factors (0.01%-0.5% relative to nonmodified forms), blocking the SUMO pathway had strong effects on the maturation and export of ribosomal subunits from the nucleus.40 Mammalian nucleolar proteins including RPs and ribosome synthesis factors were also identified as substrates for sumoylation.23,34 Two mammalian SUMO proteases, Senp341 and Senp5,42,43 show a predominantly nucleolar localization and bind to B23/nucleophosmin, a highly abundant phosphorylated protein that concentrates in the nucleolus.44 Moreover, depletion of these proteases in mammalian cells caused accumulation of sumoylated proteins in the nucleolus and inhibited ribosome biogenesis.44,45
The great variety of nucleolar proteins identified as targets of sumoylation suggests diversity of SUMO functions in this organelle. The importance of SUMO for the nucleolar function is also underscored by the severe fragmentation of nucleoli observed in mouse cells deficient for the SUMO-conjugating E2 enzyme Ubc9.46 Apart from its major biosynthetic activities, the nucleolus is used as a storage compartment for certain regulatory proteins, which can be released in a regulated manner to exert their function elsewhere.47 The SUMO protease Senp5, which is found predominantly in the nucleolus in the interphase cells, relocalizes to the cytoplasm at G2/M, where it regulates mitochondrial fission.48 Whether the function inside and outside of the nucleolus reflects the involvement of SUMO in cross-talk between ribosome biogenesis and other cellular processes remains to be seen.
Activation of p53 by Impairment of Ribosome Biogenesis
Extensive evidence shows that perturbations in ribosome biogenesis, often referred to collectively as “nucleolar” or “ribosomal” stress, can trigger an alert in the p53 tumor suppressor system, halting cell cycle progression in some cell types and inducing apoptosis in others. In one of the first examples, conditional deletion of the small ribosomal subunit protein S6 in regenerating mouse liver was found to activate a cell cycle checkpoint.49 Studies of the ribosome assembly factor Bop1 suggested that the response to aberrant ribosome synthesis involves p53.50 A p53-dependent cell cycle arrest and apoptosis were observed after blocking rRNA transcription by a conditional knockout of the Pol I transcription factor TIF-IA,51 microinjection of specific antibodies against another transcription factor, UBF, and after treating cells with drugs that affected nucleolar integrity.52 Many of these treatments induce disintegration of nucleoli, although a recent study indicated that this occurs only when Pol I transcription or early pre-rRNA processing steps are blocked.53 When using mutants, or selective depletion, of RPs and assembly factors required for later stages in ribosome biogenesis, p53 activation was observed in cells without a visible nucleolar disruption.54-57
What is the biological function of the surveillance of ribosome assembly? p53 acts as a tumor suppressor in part by coordinating expression of genes that control cell cycle progression, DNA repair, and apoptosis in response to various types of stress.58,59 It is well established that DNA damage leads to p53 activation. By contrast, perturbation in ribosome synthesis may be caused by conditions that do not appear immediately genotoxic. However, cells with disbalanced ribosome production have misregulated translational controls, which is a potential cancerogenic factor,9 and therefore eliminating such cells could bring advantages for long-lived organisms. In addition, the activation of a p53 response by defective ribosome synthesis can trigger early embryonic lethality in vertebrates through extensive apoptosis.60-62 Surveillance of ribosome biogenesis was also proposed to integrate a diverse array of metabolic inputs into a common signal to the p53 system.50 In other words, the nucleolus may act as a sensor,52 capable of detecting a broad range of metabolic anomalies and mutations that could lead to erratic translation and thus impair the fidelity of gene expression.
Studies of the mechanisms responsible for the activation of p53 by signals emanating from the nucleolus have revealed a large (and still growing) network of interactions (Fig. 1), many of which were discussed in recent reviews.63,64 We will therefore focus here on the mechanistic aspects of the interaction between RPs and Mdm2, the ubiquitin ligase with a key role in p53 regulation, and discuss some new implications of recent studies for the existing models.
Figure 1.
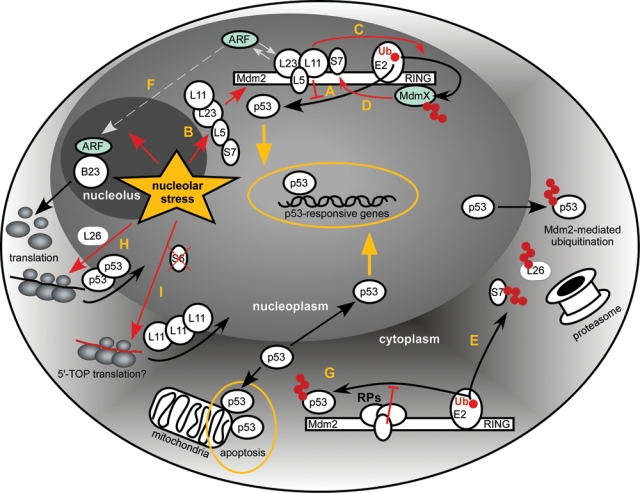
Schematic of the proposed interactions between RPs, Mdm2, and p53 resulting from perturbations in ribosome biogenesis (nucleolar stress). See text for details. Ub = ubiquitin.
Role of RPs in Modulating Mdm2 Activity
The oncoprotein Mdm2 (murine double minute 2; also referred to as HDM2 in humans) plays a pivotal role in regulating p53 levels and activity.65,66 Mdm2 is an E3 ubiquitin ligase that monoubiquitinates and polyubiquitinates p53, and the latter modification leads to the proteasomal degradation of p53.67,68 The levels of Mdm2, in turn, are regulated by p53, thus forming a feedback loop in which p53 induces Mdm2 transcription while Mdm2 targets p53 for degradation.69,70 Mdm2-knockout mice die during early stages of embryonic development, and their lethality can be rescued by deletion of TP53,71,72 providing a strong argument that one of the main functions of Mdm2 is to keep p53 activity in check.
The conserved N-terminal domain of Mdm2 is involved in binding p53, while the C-terminally located RING domain is responsible for the E3 ligase activity. The centrally located acidic domain contains a zinc finger motif of the C4 type, which binds a zinc ion via 4 cysteine residues C305, C308, C319, and C322 (numbers based on the human sequence). The crucial finding that forms the cornerstone of our current understanding of the nucleolar stress response is that several RPs, when they are not assembled into the ribosome, are capable of binding to the central acidic domain of Mdm2, resulting in the suppression of its ubiquitin ligase activity (Fig. 1A). The RPs involved include mostly those from the large ribosomal subunit, L5, L11, and L23,73-80 but 2 of the small subunit proteins, S7 and S3, were also reported to interact with Mdm2.81-83
Conditions that block transcription of rRNA or stall ribosome assembly in the nucleolus are postulated to create a pool of unused RPs that are free to interact with other molecules including Mdm2 (Fig. 1B). In this way, rather than acting as structural components of the ribosome, the RPs perform the role of “sentinels for the self-evaluation of cellular health.”64 This model is supported by the observations that forced overexpression of certain RPs in cells activates p53.74-79 Conversely, knockdown of the RPs was observed to attenuate p53 increase in response to other nucleolar stress–inducing conditions.76,77,84,85 One caveat of RP knockdowns, however, is that deficiency in any integral ribosome component affects ribosome maturation and may induce nucleolar stress. This was indeed noted in several studies.62,79
In addition to genetic inactivation of RPs or factors required for ribosome assembly, a remarkable array of chemical inhibitors is capable of blocking ribosome synthesis. A recent survey of 36 clinically relevant anticancer drugs demonstrated that 21 of them inhibited, often severely, pre-rRNA transcription and processing.53 The inhibitors most often used in a laboratory setting include actinomycin D,74-76,78 5-fluorouracil,84,86 and mycophenolic acid.85 Although not strictly proven in all cases, it is thought that the release of RPs from the nucleolus occurs when ribosome formation is disrupted by the drug treatments.
Because RPs interact with Mdm2 through sites other than the catalytic RING domain, binding of the RPs is thought to impose a conformational block preventing the transfer of ubiquitin from the RING-bound E2 enzyme to the N-terminally bound p53. In support of this, Mdm2-L11-L23 and Mdm2-L5-L11-L23 complexes were observed to retain the ability to interact with p53.78-80 It is still uncertain whether multiple RPs can bind to an Mdm2 molecule independently or if they interact with each other and dock to Mdm2 as a single multimeric complex. The interactions between RPs may account for the synergism in binding to Mdm2 observed in some studies. For instance, L11 binding to Mdm2 is increased when L11 is associated with L5 and 5S rRNA.87 The binding interface between the RPs and Mdm2 also appears complicated. Thus, it was reported that the C305F mutation in a structurally important zinc finger cystein abolished Mdm2 interaction with L5 and L11, while allowing binding to L23.80 In another study, a different amino acid substitution of the same cystein (C305S) abolished binding to L11 but not to overexpressed L5 and L23.86 These and other data raise the possibility that in addition to the zinc finger domain of Mdm2, flanking regions also contribute to the binding of individual RPs. Structural analysis of the Mdm2-RP complexes should aid in clarifying these issues.
Consistent with the important biological function of the Mdm2-RP interactions, many cancer-associated mutations of Mdm2 were found to affect the central acidic domain, including the C4 zinc finger.80 In another study, mutations in the structurally important zinc finger residues C305, C308, and C319, as well as frameshifts or nonsense mutations resulting in truncations of this domain, were detected in multiple types of human tumors.88
Expanding the p53-Mdm2 Loop
Among several additional regulators implicated in mediating stress signaling via RPs to p53, most data available to date concern MdmX, ARF, and B23/nucleophosmin.
MdmX (also called Mdm4) is a protein related to Mdm2 that regulates p53, in part by binding to Mdm2 via the RING domain and stimulating Mdm2’s E3 ubiquitin ligase activity.89,90 Treatment of cells with actinomycin D was reported to induce MdmX degradation, which was attributed to the ability of L11 to increase Mdm2 polyubiquitination of MdmX (Fig. 1C), thereby augmenting p53 response.86 Interestingly, L5 and L23 had no effect on MdmX degradation.86 MdmX was also found to cooperate with the small subunit protein S7 in inhibiting Mdm2 activity (Fig. 1D), including lower autoubiquitination in vitro.82 A possible role for MdmX may thus be to regulate activity and substrate specificity of Mdm2, thereby modulating the levels of both Mdm2 and p53. The story becomes more complicated as Mdm2 can also polyubiquitinate S7, targeting it for proteasomal degradation in the cytoplasm (Fig. 1E),82 which suggests another potential feedback mechanism affecting p53 stability.
ARF (alternative reading frame; p19ARF in mouse and p14ARF in human) is a tumor-suppressor protein that mediates p53 response to oncogene activation, viral infection, and other types of stress mainly by counteracting Mdm2 activity.91 ARF is normally found in the nucleolus, but many aspects of its nucleolar activities are not well understood. Initially, ARF was proposed to sequester Mdm2 to the nucleolus, thus inhibiting p53 degradation.92 Subsequent studies, however, suggested that ARF can interact with Mdm2 in the nucleoplasm and that the nucleolar relocalization of Mdm2 is not required for p53 activation.93,94 Interestingly, p53-independent functions of ARF have been linked to its ability to antagonize another nucleolar target, the SUMO protease Senp3.95
ARF associates with high molecular weight complexes containing RPs and other nucleolar proteins,96 binds to preribosomal particles,97 and was reported to affect rRNA processing.98,99 Although these properties seemingly put ARF in the right place to serve as a modulator of the p53 response to aberrant ribosome synthesis, p53 is activated by nucleolar stress in a variety of cell lines lacking ARF. Moreover, ARF did not compete with, or promote, binding of L5 and L11 to Mdm2, at least in immunoprecipitation experiments with ectopically expressed ARF, even though complexes between Mdm2, ARF, L11,75 and L5,80 can be detected in cells. In agreement with these data, binding sites for ARF, L5, and L11 on Mdm2 do not appear to overlap.75,80 Intriguingly, binding sites for ARF and L23 on Mdm2 do overlap, but the functional significance of this was not addressed.79 One explanation of these data is that ARF and L5/L11 act in separate pathways, which respond to different stimuli and converge only at the point of Mdm2 inactivation. Another nonexclusive possibility is that ARF provides nuances to the p53 response that are only relevant in certain cell types or in the context of an organism.
B23 (also known as nucleophosmin or NPM) is an abundant cellular protein with diverse activities, implicated in the regulation of ribosome biogenesis, embryonic development, maintaining genome stability, and tumorigenesis.100 B23 is present in large quantities in the nucleolus, but it can also shuttle between different cellular compartments.101 B23 associates with many nucleolar proteins including RPs and rRNA maturation factors102 and promotes nuclear export of ribosomes.103 B23 also binds most of the cellular ARF96,104 and is crucial for its stability.105 For its part, overexpressed ARF was reported to facilitate degradation99 and inhibit nucleocytoplasmic shuttling of B23,106 suggesting that ARF may alter the activity of a certain fraction of B23 in a cell. It is tempting to speculate that impaired ribosome assembly increases association of L23 with Mdm2, and this diverts more ARF to B23 complexes, which in turn could affect export of new ribosomes and their function in translation (Fig. 1F).
Mdm2-RP Interactions: In the Nucleus, Cytoplasm, or Both?
How does Mdm2 avoid the inhibitory influence of RPs in unstressed cells? As was discussed above, there is strong experimental evidence that mammalian cells synthesize RPs in excess of the needs of ribosome assembly.14 Besides, RPs are very abundant cellular components, and one can expect their high molar ratio relative to regulatory proteins such as Mdm2, especially in growing cells that actively produce ribosomes. One possible argument could be that unassembled RPs are normally so rapidly destroyed by the nuclear UPS that they can only effectively bind to Mdm2 when the system becomes overloaded during stress. Another untested possibility is that a crucial role might lie with the RPs that were once bound to preribosomal particles in the nucleolus but later released due to particle disassembly during stress. One key assumption in this model would be that these proteins must be somehow different from the “naive” RPs, perhaps by the presence (or absence) of some posttranslational modifications.
One other hypothesis that was not given enough consideration is that the Mdm2-RP interaction may take place in the cytoplasm. Both p53 and Mdm2 are present in the cytoplasm of unstressed cells,107 and the bulk of cellular p53 is thought to be degraded by cytoplasmic proteasomes.108-110 There is also strong evidence for the cytoplasm-specific functions of p53 including induction of apoptosis111,112 and regulation of autophagy.113 Interestingly, Mdm2 is a major, if not exclusive, factor for p53 translocation from the nucleus to the cytoplasm.109,114-116 Could it be that binding of RPs is part of a mechanism to attenuate the cytoplasmic activity of Mdm2 and protect p53 from polyubiquitination and premature proteolysis (Fig. 1G)? This mechanism would ensure that p53 cycles through the nucleus and could also serve to fine-tune p53 functions, for instance, when programmed cell death induced by cytoplasmic p53 is needed. Interestingly, while most studies on the Mdm2-RP interaction have been performed with total cellular extracts, early work indicated that RPs indeed interact with Mdm2 in the context of cytoplasmic complexes.77
RPs, Mdm2, and p53: New Twists to the Story
Recent studies uncovered more complexity in the interplay between proteins involved in ribosome formation, Mdm2, and p53. The large subunit protein L26 was shown to stimulate p53 translation (Fig. 1H) via binding to the 5′ untranslated region of the p53 mRNA.117 Further studies demonstrated that Mdm2 can directly target L26 and abolish its ability to upregulate p53 translation.118 Two separate mechanisms seem to be involved. First, Mdm2 sequesters L26 from the p53 mRNA through its acidic domain. Second, Mdm2 polyubiquitinates L26 and targets it for degradation, which occurs predominantly in the cytoplasm (Fig. 1E). Thus, L26 does not behave like other RPs that bind to Mdm2 but rather seems to be integrated in the p53-Mdm2 autoregulatory loop as an inducer of p53 and target for Mdm2.
Another recent study suggested a surprising new mechanism by which RPs from large and small ribosomal subunits cooperate in shifting the balance in the p53-Mdm2 circuit.55 Depletion of S6 in a cell culture setting was found to increase the ribosome-free L11 protein levels due to enhanced translation of the L11 mRNA (Fig 1I), leading to the inhibition of Mdm2 and activation of p53. Translational upregulation of L11 was not observed in L7a-depleted cells, even though the levels of ribosome-free L11 were also increased in this case due to impaired biogenesis of large ribosomal subunits. mRNAs that encode proteins involved in ribosome biogenesis, most notably RPs, have a common 5′ terminal oligopyrimidine (5′TOP) motif responsible for their coordinated regulation.12 At this point, it is still unclear whether L11 is the only RP upregulated in response to a deficiency in 40S subunits or if other RPs could undergo a similar upregulation through their 5′TOP elements.
There are indications that other mechanisms may be involved in the activation of p53 by nucleolar stress. Knockdown of NEDD8, which modifies multiple RPs including L11,22 dampens the increase in p53 levels in cells treated with actinomycin D,39 suggesting that neddylation contributes to the regulation of the p53 response. It was also noted that overexpression of L11 can stabilize p53 in cells lacking Mdm2,74 indicating that RPs may work in concert with other regulators to control p53 levels. Several E3 ligases including COP1,119 Pirh2,120 ARF-BP1/HECTH9,121 and E4F1122 have been shown to target p53, but it is unknown whether the scope of their regulatory activities includes monitoring nucleolar function. Finally, simultaneous knockdown of L5, L11, and L23 in zebrafish failed to prevent p53 activation,62 raising the possibility that other RPs, or pathways that do not rely on RPs, may link the nucleolus and p53. Thus, it would be of great interest to assess the functional input of additional E3 ligases in the regulation of p53 and Mdm2 after blocking ribosome formation and explore other nucleolar-derived signals that may contribute to the stabilization and activation of p53.
Concluding Remarks
The normal operation of the nucleolar biosynthetic factory is highly dependent on the supply of numerous ribosomal components and accessory factors. Recent studies suggest that ubiquitin and UBLs are important players in controlling the flow of materials to and from the nucleolus. Even more fascinating, the flow of information from the nucleolar compartment to the p53 network also employs ubiquitin-controlled circuitry. Many molecular details of the involvement of ubiquitin and UBLs in these events still remain unknown. One important question is what ubiquitin- and UBL-specific ligases and proteases carry out the numerous modifications identified among RPs and ribosome assembly factors. The interaction network mediating the nucleolar stress response has also turned out to be far more sophisticated than originally thought. A comprehensive analysis of components of this network may provide new insights into the mechanisms of tumor suppression by p53. A particularly instructive case is “ribosomopathies”—disorders caused by mutations in ribosomes or ribosome assembly factors—that are underpinned by the activation of the p53-mediated nucleolar surveillance mechanisms.123 Another important reason is the strong inhibitory effects of many anticancer drugs on rRNA and ribosome maturation.53 A better understanding of how these effects can be modulated may help in the rational design of new treatments with improved therapeutic potential.
Footnotes
This work was supported by American Heart Association grant 09SDG2140065 to N.S. and by National Institutes of Health (NIH) grant GM074091 to D.P.
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin like proteins. Annu Rev Cell Dev Biol. 2006;22:159-80 [DOI] [PubMed] [Google Scholar]
- 2. Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384-91 [DOI] [PubMed] [Google Scholar]
- 3. Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855-63 [DOI] [PubMed] [Google Scholar]
- 4. Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673-83 [DOI] [PubMed] [Google Scholar]
- 5. Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177-235 [DOI] [PubMed] [Google Scholar]
- 6. Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boisvert F, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574-85 [DOI] [PubMed] [Google Scholar]
- 8. Derenzini M, Trerè D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291-7 [PMC free article] [PubMed] [Google Scholar]
- 9. Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179-92 [DOI] [PubMed] [Google Scholar]
- 10. Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385-9 [DOI] [PubMed] [Google Scholar]
- 12. Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321-30 [DOI] [PubMed] [Google Scholar]
- 13. Warner JR. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977;115:315-33 [DOI] [PubMed] [Google Scholar]
- 14. Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersen JS, Lam YW, Leung AKL, et al. Nucleolar proteome dynamics. Nature. 2005;433:77-83 [DOI] [PubMed] [Google Scholar]
- 16. Wójcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579-89 [DOI] [PubMed] [Google Scholar]
- 17. Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci. 2005;118:5231-42 [DOI] [PubMed] [Google Scholar]
- 18. Arabi A, Rustum C, Hallberg E, Wright APH. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci. 2003;116:1707-17 [DOI] [PubMed] [Google Scholar]
- 19. Scharf A, Rockel TD, von Mikecz A. Localization of proteasomes and proteasomal proteolysis in the mammalian interphase cell nucleus by systematic application of immunocytochemistry. Histochem Cell Biol. 2007;127:591-601 [DOI] [PubMed] [Google Scholar]
- 20. Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921-6 [DOI] [PubMed] [Google Scholar]
- 21. Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145-51 [DOI] [PubMed] [Google Scholar]
- 22. Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matafora V, D’Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394-401 [DOI] [PubMed] [Google Scholar]
- 25. Redman KL, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438-40 [DOI] [PubMed] [Google Scholar]
- 26. Chan YL, Suzuki K, Wool IG. The carboxyl extensions of two rat ubiquitin fusion proteins are ribosomal proteins S27a and L40. Biochem Biophys Res Commun. 1995;215:682-90 [DOI] [PubMed] [Google Scholar]
- 27. Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035-46 [DOI] [PubMed] [Google Scholar]
- 28. Catic A, Sun ZJ, Ratner DM, et al. Sequence and structure evolved separately in a ribosomal ubiquitin variant. EMBO J. 2007;26:3474-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olvera J, Wool IG. The carboxyl extension of a ubiquitin-like protein is rat ribosomal protein S30. J Biol Chem. 1993;268:17967-74 [PubMed] [Google Scholar]
- 30. Lacombe T, García-Gómez JJ, de la Cruz J, et al. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol. 2009;72:69-84 [DOI] [PubMed] [Google Scholar]
- 31. Stavreva DA, Kawasaki M, Dundr M, et al. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol Cell Biol. 2006;26:5131-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fátyol K, Grummt I. Proteasomal ATPases are associated with rDNA: the ubiquitin proteasome system plays a direct role in RNA polymerase I transcription. Biochim Biophys Acta. 2008;1779:850-9 [DOI] [PubMed] [Google Scholar]
- 33. Chen M, Rockel T, Steinweger G, Hemmerich P, Risch J, von Mikecz A. Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol Biol Cell. 2002;13:3576-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma P, Murillas R, Zhang H, Kuehn MR. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:1227-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Endo A, Matsumoto M, Inada T, et al. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci. 2009;122:678-86 [DOI] [PubMed] [Google Scholar]
- 36. LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell. 2006;24:619-26 [DOI] [PubMed] [Google Scholar]
- 37. Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009;23:963-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panse VG, Kressler D, Pauli A, et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423-7 [DOI] [PubMed] [Google Scholar]
- 42. Di Bacco A, Ouyang J, Lee H, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gong L, Yeh ETH. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869-77 [DOI] [PubMed] [Google Scholar]
- 44. Yun C, Wang Y, Mukhopadhyay D, et al. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nacerddine K, Lehembre F, Bhaumik M, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769-79 [DOI] [PubMed] [Google Scholar]
- 47. Visintin R, Amon A. The nucleolus: the magician’s hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:372-7 [DOI] [PubMed] [Google Scholar]
- 48. Zunino R, Braschi E, Xu L, McBride HM. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284:17783-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Volarevic S, Stewart MJ, Ledermann B, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045-7 [DOI] [PubMed] [Google Scholar]
- 50. Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yuan X, Zhou Y, Casanova E, et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. 2005;19:77-87 [DOI] [PubMed] [Google Scholar]
- 52. Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burger K, Mühl B, Harasim T, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hölzel M, Rohrmoser M, Schlee M, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fumagalli S, Di Cara A, Neb-Gulati A, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hölzel M, Orban M, Hochstatter J, et al. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem. 2010;285:6364-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lindström MS, Nistér M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS One. 2010;5:e9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306-16 [DOI] [PubMed] [Google Scholar]
- 59. Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702-12 [DOI] [PubMed] [Google Scholar]
- 60. Panić L, Tamarut S, Sticker-Jantscheff M, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Azuma M, Toyama R, Laver E, Dawid IB. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J Biol Chem. 2006;281:13309-16 [DOI] [PubMed] [Google Scholar]
- 62. Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4:e4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: a never ending tail? DNA Repair (Amst). 2009;8:483-90 [DOI] [PubMed] [Google Scholar]
- 67. Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25-7 [DOI] [PubMed] [Google Scholar]
- 68. Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945-51 [DOI] [PubMed] [Google Scholar]
- 69. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126-32 [DOI] [PubMed] [Google Scholar]
- 70. Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899-908 [DOI] [PubMed] [Google Scholar]
- 71. Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203-6 [DOI] [PubMed] [Google Scholar]
- 72. Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206-8 [DOI] [PubMed] [Google Scholar]
- 73. Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lohrum MAE, Ludwig RL, Kubbutat MHG, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577-87 [DOI] [PubMed] [Google Scholar]
- 75. Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dai M, Zeng SX, Jin Y, Sun X, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dai M, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475-82 [DOI] [PubMed] [Google Scholar]
- 79. Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lindström MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol. 2007;27:1056-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen D, Zhang Z, Li M, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029-37 [DOI] [PubMed] [Google Scholar]
- 82. Zhu Y, Poyurovsky MV, Li Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst). 2009;8:1215-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sun X, Dai M, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052-9 [DOI] [PubMed] [Google Scholar]
- 85. Sun X, Dai M, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem. 2008;283:12387-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774-84 [DOI] [PubMed] [Google Scholar]
- 88. Schlott T, Reimer S, Jahns A, et al. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol. 1997;182:54-61 [DOI] [PubMed] [Google Scholar]
- 89. Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663-73 [DOI] [PubMed] [Google Scholar]
- 92. Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20-6 [DOI] [PubMed] [Google Scholar]
- 93. Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445-52 [DOI] [PubMed] [Google Scholar]
- 94. Korgaonkar C, Zhao L, Modestou M, Quelle DE. ARF function does not require p53 stabilization or Mdm2 relocalization. Mol Cell Biol. 2002;22:196-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kuo M, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378-87 [DOI] [PubMed] [Google Scholar]
- 96. Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rizos H, McKenzie HA, Ayub AL, et al. Physical and functional interaction of the p14ARF tumor suppressor with ribosomes. J Biol Chem. 2006;281:38080-8 [DOI] [PubMed] [Google Scholar]
- 98. Sugimoto M, Kuo M, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell. 2003;11:415-24 [DOI] [PubMed] [Google Scholar]
- 99. Itahana K, Bhat KP, Jin A, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151-64 [DOI] [PubMed] [Google Scholar]
- 100. Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147-53 [DOI] [PubMed] [Google Scholar]
- 101. Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379-90 [DOI] [PubMed] [Google Scholar]
- 102. Lindström MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283:15568-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Maggi LBJ, Kuchenruether M, Dadey DYA, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Korgaonkar C, Hagen J, Tompkins V, et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kuo M, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brady SN, Yu Y, Maggi LBJ, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2- sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang YV, Wade M, Wong E, Li Y, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci U S A. 2007;104:12365-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2- mediated degradation of p53. Proc Natl Acad Sci U S A. 1999;96:3077-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. O’Keefe K, Li H, Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol Cell Biol. 2003;23:6396-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563-8 [DOI] [PubMed] [Google Scholar]
- 115. Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569-73 [DOI] [PubMed] [Google Scholar]
- 116. Lohrum MA, Woods DB, Ludwig RL, Bálint E, Vousden KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol. 2001;21:8521-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49-63 [DOI] [PubMed] [Google Scholar]
- 118. Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86-92 [DOI] [PubMed] [Google Scholar]
- 120. Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779-91 [DOI] [PubMed] [Google Scholar]
- 121. Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071-83 [DOI] [PubMed] [Google Scholar]
- 122. Le Cam L, Linares LK, Paul C, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775-88 [DOI] [PubMed] [Google Scholar]
- 123. Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196-205 [DOI] [PMC free article] [PubMed] [Google Scholar]


