Abstract
PURPOSE
A great deal of information about functionally significant domains of a protein may be obtained by comparison of primary sequences of gene homologues over a broad phylogenetic base. This study was designed to identify evolutionarily conserved domains of the photoreceptor disc membrane protein peripherin/rds by analysis of the homologue in a primitive vertebrate, the skate.
METHODS
A skate retinal cDNA library was screened using a mouse peripherin/rds clone. The 5′ and 3′ untranslated regions of the skate peripherin/rds (srds) cDNA were isolated by the rapid amplification of cDNA ends (RACE) approach. The gene structure was characterized by PCR amplification and sequencing of genomic fragments. Northern and Western blot analyses were used to identify srds transcript and protein, respectively.
RESULTS
A new homologue of peripherin/rds was identified from the skate retinal cDNA library. SRDS is a glycoprotein with a predicted molecular mass of 40.2 kDa. The srds gene consists of two exons and one small intron and transcribes into a single 6-kb message. Phylogenetic analysis places SRDS at the base of peripherin/rds family and near the division of that group and the branch leading to rds-like and rom-1 genes. SRDS protein is 54.5% identical with peripherin/rds across species. Identity is significantly higher (73%) in the intradiscal domains. Sequence comparison revealed the conservation of all residues that have been shown, on mutation, to associate with retinitis pigmentosa and showed conservation of most residues associated with macular dystrophies. Comparison with ROM-1 and other rds-like proteins revealed the presence of a highly conserved domain in the large intradiscal loop.
CONCLUSIONS
Srds represents the skate orthologue of mammalian peripherin/rds genes. Conservation of most of the residues associated with human retinal diseases indicates that these residues serve important functional roles. The high degree of conservation of a short stretch within the large intradiscal loop also suggests an important function for this domain.
The outer segments of vertebrate rod and cone photoreceptors are composed of an ordered array of membranous discs that serve as the site for phototransduction. In rods, with the exception of a few nascent discs located at the base of the outer segment, the discs are surrounded by a separate plasma membrane.1 In cones, the discs appear as a folded system of membranes that are continuous, both with each other and with the plasma membrane. Peripherin/rds, a transmembrane glycoprotein, has been localized to the rim region of mature discs and at the basal region adjacent to the cilia of rod and cone outer segments where disc morphogenesis occurs.2 It is proposed to be involved in maintenance of the flattened disc morphology and fusion of membranes, to form isolated discs.3 This is suggested by its location on the disc membrane as well as by the phenotype of retinal degeneration slow (rds) mutants. In rds/+ mice, the phenotype is a semidominant trait characterized by haploinsufficiency of peripherin/rds arising from insertion of a 9.2-kb repetitive genomic element within exon 2 of the gene.4 It has been shown that the rds defect in rds/+ retina is more deleterious to rods than cones.5 The outer segment membrane biogenesis occurs in the rds/rds but the membrane is unable to fold into the proper disc structure.
Peripherin/rds cDNA has been isolated and sequenced from human,6 cow,7 dog,8 cat,9 rat,10 mouse,11 chicken (CRDS1 and CRDS2),12 and frog (XRDS35, XRDS36, and XRDS38)13 and found to code for proteins ranging in length from 346 to 364 residues, depending on the species. The predicted polypeptide is composed of four putative transmembrane segments, relatively small (21 residues) and large (142 residues) intradiscal loops, and a long C-terminal segment exposed to the cytoplasmic side of the disc membranes (for review, see Ref. 14). In vitro biochemical studies suggested a noncovalent association between peripherin/rds and ROM-1,14–18 a nonglycosylated transmembrane protein that shares several characteristics with peripherin/rds. These characteristics include similar hydropathy profiles, gene organization, highly conserved residues, and localization to the rim region of rod and cone outer segments.14, 15, 19 Both in vivo and in vitro studies have shown that the noncovalent interactions between peripherin/rds and ROM-1 act to form homomeric and heteromeric functional core complexes. Although proper assembly between peripherin/rds and ROM-1 is believed to play a crucial role in normal outer segment structure, the functional activities and the site of interactions between the two proteins at the molecular level are not completely understood. Goldberg et al.17 used site-directed mutagenesis to determine the role of cysteine residues of peripherin/rds in the functional core complex formation. The mouse peripherin/rds has 13 cysteine residues and 11 of them are conserved in all known peripherin/rds. Sequence comparison with ROM-1, however, shows that only seven of these cysteines located in the large intradiscal loop are conserved. Some or all these cysteine residues may form intra- or intermolecular disulfide bonds. Replacement of the noncon-served cysteines showed no apparent effect on dimer formation, folding, or subunit assembly. In contrast, replacement of any of the seven conserved cysteine residues within the large intradiscal loop significantly alters these properties,17 suggesting that these residues are crucial for proper folding and subunit assembly. Furthermore, the carboxyl terminus of pe-ripherin/rds has been shown to promote membrane fusion in vitro, signifying a possible role for this protein in outer segment renewal.3,20,21 Recently, peripherin/rds has been shown to associate with the photoreceptor cGMP-gated channel Na/Ca-K exchanger.22 It has been suggested that the glutamic acid–rich protein of the channel may act as a bridge to connect the channel-exchanger complex with peripherin/rds.22 This association may play a role in connecting the rim region of the disc to the plasma membrane and/or anchoring the cGMP-gated channel in the plasma membrane.
Interest in peripherin/rds has increased since the discovery of its association with different forms of human retinal diseases. More than 80 different pathogenic mutations have been identified that are associated with retinitis pigmentosa (RP) and several forms of macular dystrophy (MD; for review, see Refs. 23–25). These mutations include base substitutions that cause missense mutations or premature termination and in-frame insertion/deletion mutations that change the reading frame. The majority of these mutations are located in the large intradiscal loop, emphasizing the important role played by this region in the function of peripherin/rds.
Most of our knowledge of the structure and function of peripherin/rds has come from studies of mammalian genes. In the present study, we determined the structure of the skate peripherin/rds (srds) gene, the sequence of the intron/exon boundaries, and the sequence analysis of the cDNA clones. We studied the evolution and structural properties of srds by comparing the skate gene with homologues from human, cow, dog, cat, mouse, rat, chicken, and the African clawed frog. We provide evidence that there are highly conserved regions in all peripherin/rds proteins, suggesting important functional roles for these domains.
MATERIALS AND METHODS
Immunoblot Analysis
Polyclonal antiserum against the D2 loop of mouse peripherin/rds (rds-D2 Ab), generously provided by Gabriel H. Travis (Jules Stein Eye Institute, UCLA School of Medicine), was used to detect the SRDS protein. A polyclonal antibody against residues 336–351 from the carboxyl terminus of ROM-1 (ROM-1–C-term Ab) was generated by immunizing rabbits with the corresponding peptides coupled to keyhole limpet hemocyanin. Dissected retinas were homogenized on ice in solubilization buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.05% SDS, 2.5% glycerol, and 1.0 mM phenylmethylsulfonyl fluoride. After solubilization at 4°C for 1 hour, the homogenates were centrifuged at 100,000g for 30 minutes, and supernatants were collected for protein concentration and Western blot analysis as described elsewhere.26 For Western blot analysis, retinal extracts were mixed with 2× Laemmli buffer containing 100 mM Tris-HCl, 4% SDS, 200 mM dithiothreitol (DTT), and 20% glycerol; separated on 10% SDS–polyacrylamide gels; and then transferred onto polyvinylidene difluoride (PVDF) membrane in transfer buffer containing 25 mM Tris-base, 190 mM glycine, 20% methanol, and 0.03% SDS. The blots were preblocked with 10% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) at 4°C overnight before incubation with rds-D2 Ab or ROM-1–C-term antibody at a dilution ratio of 1:1000 for 2 hours at room temperature. Blots were then washed several times with TBST and incubated with the secondary antibody at a dilution ratio of 1:25,000 for 1 hour at room temperature. A chemiluminescent substrate (Super Signal; Pierce, Rockford, IL) was used for detection.
Enzymatic Deglycosylation of SRDS
Reduced retinal extracts (in 2% β-mercaptoethanol at 50°C for 5 minutes) were incubated with endoglycosidase F1 (Sigma-Aldrich, St. Louis, MO) at 1.0 U/150 µg of total skate retinal extract at 37°C overnight in the reaction buffer (50 mM NaH2PO4, pH 5.5).27 After deglycosylation, a 15 µg-aliquot of proteins from each sample was mixed with Laemmli sample buffer and subjected to 10% SDS-PAGE for Western blot analysis.28
Northern Blot Analyses
Total retinal RNA (5 µg) was isolated from skate (Raja erinacea), mouse, rat, and cow, with extraction reagent (Trizol; Life Technologies, Gaithersburg, MD) and run on a 1% agarose gel containing 18% formaldehyde in a buffer containing 20 mM 3-[N-morpholino]propane-sulfonic acid (MOPS; pH 7.0), 8 mM sodium acetate, and 1 mM EDTA. RNA from bovine pigment epithelium was also included as a control. The gel was stained with ethidium bromide to check the integrity of the RNA (judged by the integrity of the 28s and 18s rRNA). The RNA was then transferred to nitrocellulose membrane (Schleicher and Schuell, Keene, NH) by capillary diffusion in 20 × SSC (3 M NaCl and 0.3 M sodium citrate) and hybridized with an α-32P-labeled bovine peripherin/rds full-length cDNA probe. The probe was incubated with the blot overnight at 42°C in a solution containing 50% formamide, 2× SSPE (0.75 M NaCl, 0.06 M NaH2PO4, 0.22 M EDTA [pH7.4]), 0.5% nonfat dry milk, 1% SDS, and 0.5 mg/mL of salmon sperm DNA. The blot was washed sequentially in buffers containing 0.5% SDS plus 2× SSC for 30 minutes at 55°C, 0.5% SDS plus 0.5× SSC for 20 minutes at room temperature, and 0.5% SDS plus 0.1 × SSC for 15 minutes at room temperature. X-ray film (X-Omat AR; Eastman Kodak, Rochester, NY) was then exposed to the blot for 48 hours at −80°C with intensifying screens. Transcript sizes were determined by comparison to an RNA molecular weight ladder (Life Technologies).
Cloning and Sequence Analysis of Srds cDNA
A skate (Raja erinacea) retinal cDNA library was screened as described before.29 Approximately 1 × 106 recombinant plaques were screened, with the mouse peripherin/rds cDNA clone used as a probe. Seventeen positive clones containing different portions of peripherin/rds sequence were obtained. The clones were excised from their phagemids and subjected to digestion with EcoRI. Clones were classified according to their restriction maps and further characterized by direct DNA sequencing. A combination of subcloning and primer-directed sequencing was used to determine the DNA sequence of both strands, by the dideoxy-chain termination method in a DNA sequencing kit (Sequenase; United States Biochemical, Cleveland, OH) and [α-32P]-dATP. The complete sequence was obtained from both strands of clones within the coding region.
Transcription Initiation Site
Rapid amplification of cDNA ends (RACE)30 was used to obtain clones containing the entire 5′ end of the srds cDNA. First-strand cDNA synthesis was performed with 0.5 µg of skate retinal RNA and 35 ng of an antisense oligonucleotide complementary to exon 1 (MIN168, 5′-CAGCACCGATTTCCATCGGGCG- 3′). A dC tail was added to the 3′ end of the first strand cDNA by terminal transferase. The cDNA was then amplified with a sense primer complementary to this dC tail and an antisense nested primer directed to exon 1 (MIN163). The PCR fragment was separated on a 1% agarose gel, subcloned into a vector (pGEM-T; Promega, Madison, WI), and sequenced on both strands.
Mapping the 3′ Polyadenylation Sites
The 3′ polyadenylation sites were determined as described before.31 One microgram of skate total retinal RNA and 200 nM of an oligo(dT)-mcs primer were used to produce the first strand cDNA. This oligo (dT) primer consists of an oligo (dT17) domain and a 20-nucleotide multiple cloning site (mcs). This amplification introduced a nucleotide sequence (mcs 5′-GGCCACGCGTCGACTAGTAC-3′) into the synthesized cDNA that was used in a second PCR with an srds-specific primer located in exon 1 (MIN181). The PCR fragments were subcloned and sequenced to identify the polyadenylation sites.
Organization of the Srds Gene
Genomic DNA was isolated from the liver of a single skate according to published protocols.32 Fragments of skate genomic DNA were amplified with a PCR kit (Expand long PCR; Roche Molecular Biochemicals, Indianapolis, IN). The reactions were performed according to the manufacturer’s protocol with 250 ng genomic DNA and 200 ng of each primer in a 50-µL reaction mixture. The primers were chosen to amplify potential introns present in the coding region, because mouse, human, chicken and frog rds, as well as human and mouse rom-1 genes, harbor two introns in the coding region.4,5,33,34 The first set of primers, MIN188/MIN161, straddles exon I of mouse and human rds, whereas the second set, MIN160/MIN189, encompasses the region where introns I and II are found in mouse and human rds genes. The srds cDNA template was used as a positive control. A negative control (no template) was run with each reaction set. PCR products were cloned and sequenced.
Sequence Analysis
Sequence data obtained by direct sequencing were analyzed and edited on computer (PC/Gene software; Oxford Molecular, Ltd., Campbell, CA). The same program was used to generate a comparison of compiled DNA sequences and produce multiple alignments of predicted protein sequences from different species available in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD). The term “identity” describes residues that are completely conserved, and the term “similarity” specifies identical residues plus all conservative substitutions.
The phylogenetic relationships of all known members of the peripherin/rds and ROM-1 family of proteins were examined through further analysis of a multiple amino acid sequence alignment. The amino acid sequence data were analyzed on computer in Phylip 3.573c.35 Aligned sequence data were subjected to a distance matrix analysis using the Dayhoff PAM 280 similarity matrix.36 A neighbor-joining analysis was then used to calculate phylogenetic trees. We used the bootstrap method to assess confidence in the final tree.37 In this technique, sequence data from the aligned data set are randomly deleted with replacement from remaining aligned data. Distance matrix calculation and tree estimation procedures are performed on each of the bootstrapped data sets, followed by calculation of a consensus tree. One thousand bootstrap replicates were made of the rds data set. Branch lengths within the final tree were calculated with the neighbor-joining algorithm, using the full data set constrained to the topology of the consensus tree.
RESULTS
Identification of a Peripherin/rds-like Glycoprotein in the Skate Retina
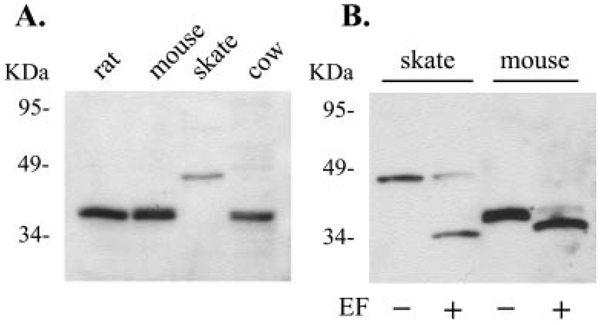
The presence of peripherin/rds-like protein in the skate retina was determined by Western blot. The blot containing extracts from retinas of skate, rat, mouse and cow was immunoreacted with anti-mouse rds-D2 Ab. This antibody recognized a single band of approximately 42 kDa in skate retina (Fig. 1A). An antibody to the mouse ROM-1 C terminus did not recognize any proteins in the skate retina (data not shown).
FIGURE 1.
Presence of a peripherin/rds-like protein in the skate retina. (A) Western blot analysis of peripherin/rds from skate, mouse, rat, and cow retinas was performed with the rds-D2 Ab. Note that the peripherin/rds detected in skate retina migrates at a position of 42 kDa, higher than the protein homologues detected in mouse, rat, and cow (approximately 36 kDa). (B) Enzymatic deglycosylation of peripherin/rds. Retinal extracts from skate and mouse were digested with endoglycosidase F (EF) before SDS-PAGE separation and Western blot analysis. Note that after deglycosylation, the migration of peripherin/rds in skate retina shifted to a position similar to that of the mouse homologue, suggesting a similar core polypeptide size for peripherin/rds in mouse and skate retinas.
The apparent molecular weight of the skate protein recognized by the rds-D2 Ab is substantially larger than the 36-kDa band detected in rat, mouse, and cow. It is possible that a different level of glycosylation of the skate protein contributes to its slow migration. To determine whether the difference in apparent size was due to glycosylation, retinal extracts from skate and mouse were treated with Endo F, a peptide-N-glycosidase that releases Asn-linked oligosaccharides from glycoproteins. This treatment produced a similar core polypeptide from mouse and skate extracts (Fig. 1B), suggesting that the proteins are similar in size.
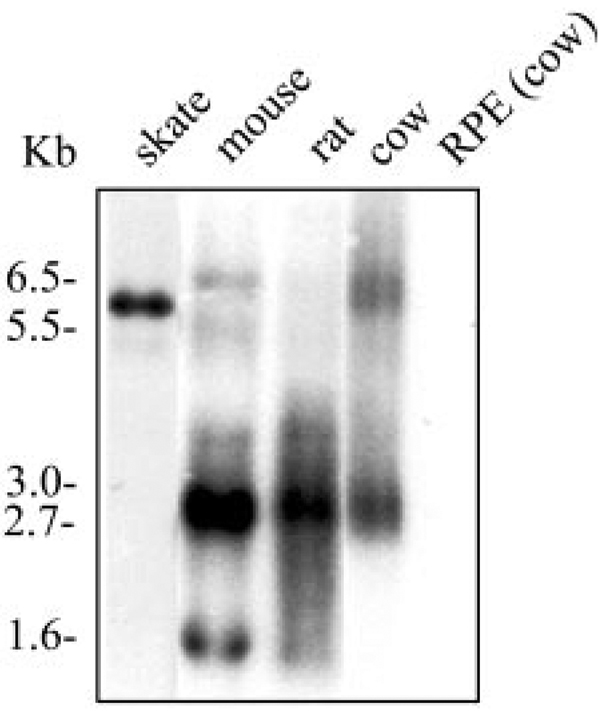
To determine whether the peripherin/rds-like protein in skate retina is related to mammalian peripherin/rds, we performed a Northern blot analysis using a cow rds cDNA probe. Figure 2 shows the result of a Northern blot containing retinal RNA isolated from skate, mouse, rat, and cow. A single 6-kb transcript labeled in skate retinal RNA indicates significant homology to mammalian peripherin/rds. Two major transcripts were detected in retinas of mouse (1.6 and 2.7 kb) and cow (2.7 and 6.5 kb), whereas one main transcript was observed in rat (2.7 kb) retinas. The 2.7-kb transcript is common in retinas of mouse, rat, and cow, while skate and cow share the 6-kb retinal transcript of rds. As expected, no rds transcripts were observed in RNA isolated from bovine pigment epithelium (Fig. 2). Northern blot analysis of RNA isolated from different skate tissues including retina, brain, heart, stomach, kidney, spleen, liver, skeletal muscle, skin, and lens indicated retina-specific expression of the this gene (data not shown), consistent with the fact that peripherin/rds is a retina-specific protein. The peripherin/rds-like glycoprotein we have identified in skate retina will be referred to as the skate RDS protein or SRDS.
FIGURE 2.
Peripherin/rds transcript in skate retina. Northern blot analysis of total RNA isolated from retinas of skate, mouse, rat, and cow and from bovine pigment epithelium was hybridized with a bovine peripherin/rds cDNA probe. Each lane contained 5 µg of total RNA. More than one transcript was observed in all the species studied except for the skate, in which a single band of ~6 kb was strongly labeled with the probe. No signal was detected in RNA isolated from the bovine pigment epithelium. RNA size standards are indicated to the left.
Isolation and Analysis of Srds cDNA
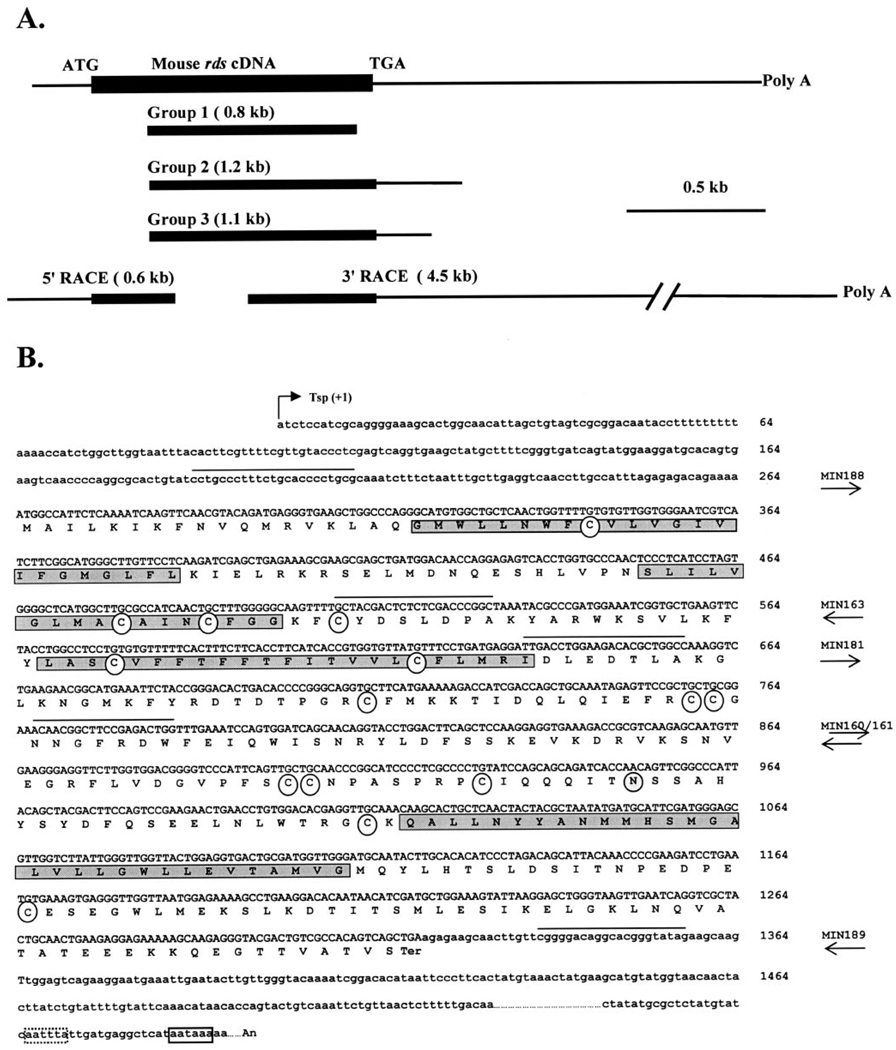
Screening of the skate retinal cDNA library with a full-length mouse rds cDNA probe yielded 17 clones, whereas screening with bovine rom-1 probes failed to yield any clones. The peripherin/rds-like clones were subjected to EcoRI digestion to determine the size of the insert and, based on the pattern of digestion, were categorized into three groups (Fig. 3A). Representative clones from each of the three groups were selected for further restriction map analysis and sequencing. Sequence comparison with mouse rds indicated that all the isolated clones were missing approximately 200 bp of the 5′ coding region, all of the 5′ untranslated region, and most of the 3′ untranslated region (Fig. 3A). To characterize the 5′ untranslated region and the 5′ end of the coding region, we performed 5′ RACE. A single fragment of approximately 600 bp was obtained. The entire coding sequence of srds was deduced from the combination of sequences obtained from the longest clone (group 2) and the PCR product from the 5′ RACE. The single open reading frame codes for a protein of 351 amino acids (Fig. 2B) with a predicted molecular mass of 40.2 kDa.
FIGURE 3.
Isolation and sequence analysis of the srds cDNA clones. (A) Positional comparison of the srds cDNA clones to the mouse cDNA sequence. The three groups of the original 17 clones and the two RACE clones are shown. Filled boxes: translated sequences; horizontal lines: untranslated sequences. (B) Compiled nucleotide and predicted amino acid sequence of the srds cDNA. The four predicted transmembrane domains are highlighted with shaded boxes in the polypeptide sequence. Arrow: the putative transcription start point (Tsp) used as +1. Open circles: conserved cysteine residues within the second intradiscal loop, which are predicted to be involved in the secondary structure of peripherin/rds and its association with other proteins, are present in the skate homologue. The conserved site for N-linked glycosylation (N229) is located in the second intradiscal loop (shaded circle). Right: names of the primers used; arrows: direction of the primer. Sequence of the primers is marked by a solid line above the nucleotide sequence. Solid- and dotted-line boxes: potential polyadenylation signals. The srds cDNA sequence has been entered into the GenBank database under accession number AF162436.
To identify the potential polyadenylation signal present in the srds transcript, a 3′ RACE experiment was performed, and the site of the poly(A) tail addition was determined by direct sequencing of the subcloned PCR product. Because the 3′ RACE product was approximately 4.5 kb in size, only approximately 400 bp of sequences were obtained from each end. Sequence analysis indicated the presence of a polyadenylation signal (AATAAA) immediately before the poly(A) tail (Fig. 2B, open solid-line box). Although the typical consensus sequence AATAAA is usually situated 10 to 30 nucleotides upstream of the poly(A) addition site, variations in the distance of these sites have been described in other genes.38 A polyadenylation-like signal (AATTTA) was identified 17 bp upstream of the poly(A) tail (Fig. 3B, dotted-line box). It is not clear which of these potential polyadenylation signals is actually used by the srds gene. The combined size of the entire coding region (1056 bp), the 5′ (264 bp), and the 3′ (~ 4500 bp) untranslated portions of the transcript was approximately 6.0 kb, in agreement with the size of the unique srds transcript identified by Northern blot (Fig. 2).
Organization of the Srds Gene
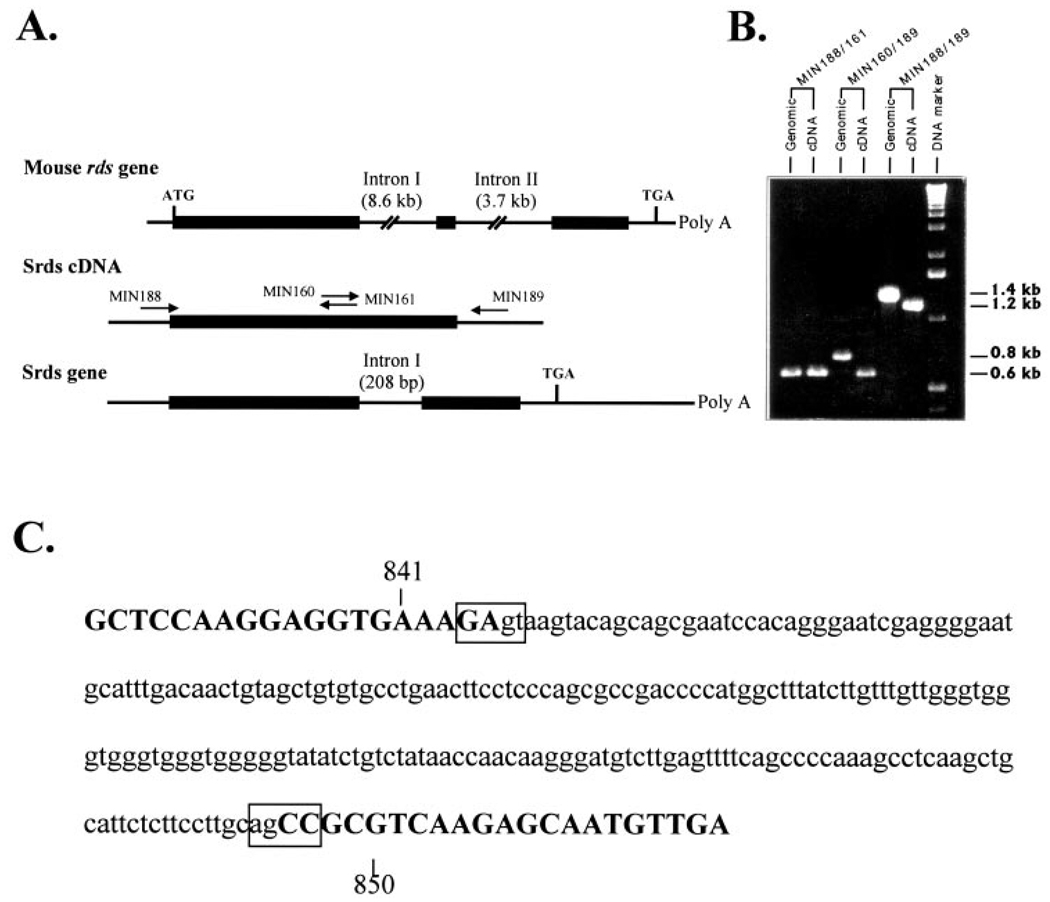
Analysis of the structure of mouse, human, frog, and chicken rds as well as mouse and human rom-1 genes revealed the presence of two introns.4,5,33,34 Although the sizes of these introns are different, their locations are the same in mouse, human, frog, and chicken. To determine whether any introns were present in the srds gene, we amplified across the entire coding region using skate genomic DNA as a template. One set of primers (Fig. 3B, MIN188/MIN161) was made against sequences of the skate gene equivalent to those within exon I of the mouse gene (Fig. 4A). The PCR yielded similar products (0.6 kb) from both the skate genomic DNA and the cDNA used as a control (Fig. 4B). A second set of primers (MIN160/MIN189, Fig. 3B) was used to amplify around the area of putative introns I and II. When genomic DNA was used as a template, the PCR product was 208 bp longer than when the cDNA was used (Fig. 4B). Sequencing of the PCR products revealed the presence of only one intron at the same location as intron I in mouse and human (Fig. 4C). We did not find intron II in the srds gene. The absence of intron II was further confirmed by amplifying across the entire coding region using a third set of primers (MIN188/MIN189, Fig. 3B). Again, the PCR product obtained with genomic DNA as a template was 208 bp longer than that obtained with the cDNA (Fig. 4B). Exon–intron boundaries were noted by divergence of the genomic sequence from that of the cDNA. The sequence of the exon–intron junction (Fig. 4C) is in agreement with both the 5′ and 3′ consensus splice site sequences.
FIGURE 4.
Organization of the srds gene. (A) Map shows the introns in the mouse peripherin/rds cDNA and the positions of the primers used to identify the presence of introns in the skate homologue. (B) PCR amplification and analysis of DNA fragments from skate genomic DNA reveals the presence of a 208-bp intervening sequence in fragments amplified by primer pairs 160/189 and 188/189, but not in the fragment amplified by primer pair 188/161. (C) Sequence of the intron-exon junctions of the srds gene. Uppercase letters: exon sequences; lowercase letters: intron sequences.
Characteristics of the SRDS Protein
Hydrophobicity analysis of the predicted sequence of the SRDS protein revealed four transmembrane domains similar in length and location to those observed in other species (Fig. 3B, shaded boxes). The sequence contains 14 cysteine residues, five of which are located in the transmembrane domains, one in the small and seven in the large intradiscal loops, and one in the C-terminal region (Fig. 3B, open circles).
Consensus sequences for N-linked glycosylation were found in a number of locations in peripherin/rds from different species. SRDS has only one potential glycosylation site, located at position N229 (Fig. 3B, shaded circle). Notably, this site is completely conserved in all species examined. It is likely that this site, located within the large intradiscal loop, is the site used for glycosylation in all these species. Enzymatic deglycosylation of the SRDS produced a much larger mobility shift than similar treatment of the mouse homologue (Fig. 1B), suggesting that SRDS is more extensively glycosylated at this site.
Comparison of SRDS with Other Known Peripherin/rds and ROM-1
The SRDS protein shares 63% identity with all its known mammalian counterparts, 69% identity with XRDS38, and 72% identity with CRDS1. The total shared identity for all these proteins is 54.5%, although different domains share different levels of identity. For example, the four transmembrane domains and the C-terminal region retain approximately 38% identity, whereas the large hydrophilic loop between the third and the fourth transmembrane domains shares 73% identity and 93% similarity with peripherin/rds from all known species. Although C termini comparison shows lower sequence identity, the overall structural and functional homology may be retained. This can be assessed by comparisons of specific domains. A stretch of 15 residues has been identified in the C terminus of bovine peripherin/rds to possess membrane fusogenic activity.3,20,21 Therefore, we compared the equivalent domain at the C terminus of SRDS to the same region of other known peripherin/rds (see Fig. 6, solid line below the amino acid sequence at the C terminus). This area shows 44% identity and 73% similarities among all peripherin/rds. The relatively lower level of identity at this region does not suggest close structural conservation of this domain. Biochemical studies would have to be performed to assess whether the domain is conserved functionally. In contrast, the N terminus and the small intradiscal loop share 61% and 76% identity, respectively, suggesting greater conservation throughout these domains.
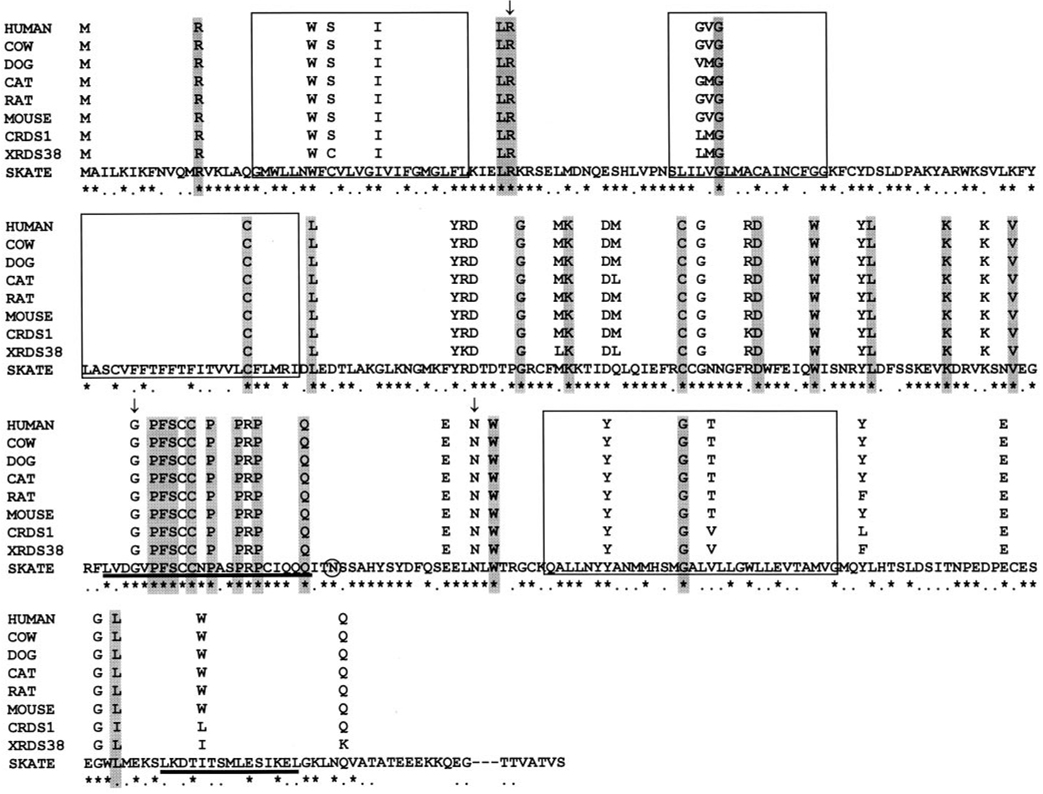
FIGURE 6.
Conservation of residues associated with human retinal diseases. The protein sequences from nine known peripherin/rds homologues are aligned. For clarity, the full amino acid sequences of the nonskate proteins have not been shown, with the exception of residues that align with positions of the human peripherin/rds that are mutated in certain retinal degenerative diseases. (⋆) Residues identical in all homologues with those of SRDS; (…) conserved residues. The unmarked residues are not conserved. Boxes: the putative positions of transmembrane domains. Shaded residues in all nine homologues are those associated with RP; unshaded residues are associated with macular dystrophies in humans. Vertical arrows: mutations that cause variable phenotypes among family members. Mutations reported here are base substitutions causing missense mutations or in-frame deletions. Shaded circle: N229 potential glycosylation site. Underscored sequences: a highly conserved region in the large intradiscal loop between the third and fourth transmembrane domains and a region of the C terminus that has been found to have membrane fusogenic properties in mammals.
When the sequence comparison is extended to include ROM-1 proteins, CRDS2, and XRDS35 and -36, the overall identity among the proteins is reduced to 18%, although a high degree of similarity is apparent in the large intradiscal loop. Fifty-four (80%) of the 65 identical residues in peripherin/rds and ROM-1 are located in the large loop, and a stretch of 22 residues (L205–Q226) displays nearly complete identity (see Fig. 6, solid line below the amino acid sequence at the large intradiscal loop). SRDS is five amino acids longer than peripherin/rds from human,6 cow,7 dog,8 cat,9 rat,10 mouse,11 and XRDS38,13 and six residues longer than XRDS35 and −36.13 However, it is 3 and 13 residues shorter than CRDS1 and −2, respectively.12
It has been proposed that some or all the cysteine residues in the large intradiscal loop may form intra- or intermolecular disulfide bonds.14,17 Seven cysteine residues located in the large intradiscal loop are completely conserved in all the peripherin/rds and ROM-1 proteins studied to date (Fig. 3B, open circles).
The amino acid sequences of all 15 known members of the rds and rom-1 gene families were subjected to a neighbor-joining analysis and a tree was constructed (Fig. 5). SRDS is homologous to peripherin/rds and is positioned near the base of the group including XRDS38, CRDS1, and the tight cluster of mammalian peripherin/rds. The order within this group is generally consistent with the evolutionary relationships expected among vertebrates. CRDS2, XRDS35 and -36, and mammalian ROM-1s fall outside the group, but show a clear pattern of relationship. Whereas CRDS2 is relatively closely related, the other sequences diverge more widely than do the peripherin/rds group, and do not form a distinct gene family.
FIGURE 5.
Phylogenetic analysis of the peripherin/rds and ROM-1 family of proteins. The tree was constructed using a neighbor-joining analysis of the amino acid sequence of all known members of the family. Bootstrapping was performed to estimate support for each node of the tree. Bootstrap values shown at the nodes represent the percentage of 1000 bootstrap replicates in which the node occurred. Values below 50% are not included.
Association of Conserved Residues with Human Retinal Diseases
Analysis of the residues that, when mutated, have been associated with human retinal degeneration (Fig. 6) revealed that all the residues associated with mutations that cause RP were completely conserved in all species studied. Although most of the amino acids associated with mutations that cause MD were also conserved across these species, nine were not entirely conserved. Two of them were not conserved in SRDS and XRDS38, and the others were not conserved in either CRDS1 or XRDS38. The lower degree of conservation in those residues associated with human cone-related diseases suggests that their importance in the function of the protein differs among the species studied. Furthermore, three mutations showed variable phenotypes with different members of the family (Fig. 6, residues with vertical arrows). For example, R46ter causes autosomal dominant RP (adRP)39 and diffuse retinal degeneration,40 G208D causes adRP with phenotypic variation between the same family members,41 and N244L causes adRP and bull’s eye maculopathy,42 whereas N244H causes autosomal dominant cone–rod dystrophy.43
Further comparison between peripherin/rds, ROM-1, and the rds-like proteins identified a stretch of 22 residues in the large intradiscal loop, of which 19 are identical and 2 are conserved (Fig. 6, solid line beneath the sequence). Eleven mutations in this region have been shown to associate with human retinal diseases, eight of which cause RP (P210S, F211L, S212G, C214S, P216S, P216L, P221del, Q226D), one causes MD (C213Y), one causes a mixed phenotype of RP and MD (G208D), and one causes Pattern Dystrophy when mutated to R220W or R220Q (see RetNet: http://www.sph.uth.tmc.edu/ RetNet; provided in the public domain by the University of Texas-Houston Health Science Center, Houston, TX). These data suggest a significant role for this region of the large loop in the overall function of peripherin/rds in rods, probably through its association with rod-specific proteins.
DISCUSSION
This study was undertaken to map the highly conserved residues that may play important role in peripherin/rds function by examining the evolutionary conservation of this protein among vertebrates. The class Chondrichthyes, to which skate belongs, diverged earlier from the ancestral vertebrate line than any other group in which peripherin/rds has been studied. Thus, the skate peripherin/rds sequence can provide information about the early evolution of the gene family. The skate retina is an all-rod retina in which many of the functions of both rod and cone photoreceptors are subserved by the rods.44–46 The outer segment organization of skate photoreceptors is similar to that of other vertebrate rods,46,47 and immunocytochemistry with anti-cow peripherin/rds antibody showed strong labeling in the skate outer segments (data not shown). Therefore, we expect the SRDS protein to perform the same function in the skate as it does in mammals.
Cloning of the srds gene has allowed us to compare members of the peripherin/rds gene family (including rom-1 and the rds-like genes) over a broad phylogenetic base and establish the most conserved elements of the sequence. To make such a comparison, it is necessary to understand the relationships within the gene family. We used a neighbor joining analysis of the amino acid sequences of all known members of this gene family to study these relationships. The analysis revealed a group of highly homologous rds genes, within which the expected phylogenetic relationships among vertebrates was observed. Srds gene falls near the base of this group and diverges from a point close to that of the divergence of xrds38. Bootstrap analysis lends relatively low support for this node, indicating uncertainty about the exact branching order at the most basal node of the peripherin/rds group; however, separation of this group from the remaining rds-like and rom-1 genes is well supported. This suggests that srds is an orthologue of the mammalian rds and is likely to be a functional homologue. Mammalian rom-1 and both chicken and Xenopus rds-like genes are more divergent members of the gene family, apparently paralogues to rds. These could represent functional homologues of rom-1.
The organization of the gene may offer some insights into how this gene family arose. The gene structure determined for mammalian rds, crds1 and 2, xrds38, −36, and −35 and mammalian rom-1 indicates the presence of three exons and two introns with conserved boundaries.4,5,33,34 Srds, in contrast, has only two exons and one intron in the same location as intron I in other species. There are two possibilities that could account for the relationship of srds gene to other members of the family. First, a gene duplication leading to the evolution of the rds-like and ROM-1 proteins may have occurred after the divergence of Chondrichthyes from the ancestral vertebrate line. In addition, intron II must have been acquired between the divergence of the skate and this gene duplication. Alternatively, a gene duplication leading to rom-1 and rds-like genes may have preceded the divergence of the Chondrichthyes. In this case, both introns are presumed to be present in the ancestral gene, with intron II lost from the srds gene. Previous studies have shown that the presence or absence of introns in the progenote to have profound consequences for the origin and evolution of the genes.48 We did not find an rom-1 gene homologue in the skate, but have not screened exhaustively for such genes. Further work is needed to clarify the gene complement in the skate and to resolve the question of when gene duplication may have occurred in this family.
With the described relationships in mind, it is possible to identify several conserved features of the proteins. For example, the predicted N-linked glycosylation site at N229 is completely conserved in the peripherin/rds of all species examined. Indeed, this is the only glycosylation site found in the skate sequence that contributes to the higher molecular mass seen on SDS-PAGE (Fig. 1). Although transgenic mouse study has shown that glycosylation of the mouse homologue is not critical for the normal function of peripherin/rds or its association with ROM-1, it is not known whether this is the case for SRDS.27
Information about the importance of cysteine residues in the large intradiscal loop in the formation of intra- or intermolecular disulfide bonds17 may also be gleaned from this comparison. The positions of eleven cysteine residues are conserved in all known forms of peripherin/rds (Figs. 3, 6). Sequence comparison with ROM-1, however, shows that only seven of the cysteines located in the large intradiscal loop are conserved. Studies by Goldberg et al.17 have addressed the importance of these cysteine residues. Replacement of these seven cysteines resulted in defects in dimer formation, folding, and subunit assembly of cow peripherin/rds.
Sequence alignment of the SRDS with other members of the peripherin/rds family revealed a striking conservation of residues that have been linked to human retinal diseases. All the residues that, when mutated, cause RP are conserved in all species. There is less, though still significant, conservation of residues that cause cone dystrophies when mutated. Both XRDS38 and CRDS1 do not conserve some residues that have been associated with cone dystrophies. This suggests that the requirements for proper function in cones may differ among species. However, the high conservation of residues essential for rod function suggests that the requirements for this function are stricter.
One stretch of the large intradiscal loop is particularly highly conserved, not only among peripherin/rds homologues, but also among the rds-like and ROM-1 proteins. There is an exceptionally high density of sites associated with retinal diseases in this region (11/22 residues). These data suggest a significant role for this section of the large loop in the overall function of peripherin/rds, ROM-1, and the rds-like proteins in establishing and maintaining the integrity of photoreceptor outer segments.
In addition to the importance of D2 loop in peripherin/rds function, the membrane fusogenic activity of the C terminus of the bovine homologue has been localized to a 15-amino acid amphophilic α-helix domain.3,20,21 Although this fusion domain is highly conserved among all mammalian homologues, the corresponding region in SRDS shares less homology than the protein as a whole. Thus, the functional conservation of this domain will remain in question until biochemical experiments are performed.
Peripherin/rds and ROM-1 normally assemble as a heterotet-rameric complex at the photoreceptor disc rims.16 In contrast to peripherin/rds, no mutations in ROM-1 alone have been associated with retinal disease. Despite repeated attempts in screening the skate cDNA library with reduced stringency with a bovine rom-1 cDNA probe, we were unable to isolate a skate rom-1 clone. Furthermore, we were unable to detect the presence of ROM-1 on Western blot analysis, using antibody specific to the mouse ROM-1 C-terminal region. It is possible that rom-1 like genes are not present in the skate retina.
In summary, we have identified a new member of the peripherin/rds family in the skate retina. The predicted structure of SRDS is similar to that of other members of the family with the C terminus being five amino acids longer in the skate. Sequence comparisons suggest that mammalian ROM-1 and rds-like proteins may have evolved from the rds gene, with the rds-like gene serving as an ancestral gene for rom-1.
Acknowledgments
Supported by National Eye Institute Grant EY10609 (MIN) and Core Grant for Vision Research EY12190; the Foundation Fighting Blindness, Baltimore, Maryland (MIN, MRA); and the Knights Templar Eye Foundation, Illinois. MIN is a recipient of the Research to Prevent Blindness James S. Adams Scholar Award.
Footnotes
Disclosure: C. Li, None; X.-Q. Ding, None; J. O’Brien, None; M.R. Al-Ubaidi, None; M.I. Naash, None
References
- 1.Steinberg RH, Fisher SK, Anderson DH. Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol. 1980;190:501–518. doi: 10.1002/cne.901900307. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa K, Molday LL, Molday RS, Williams DS. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J Cell Biol. 1992;116:659–667. doi: 10.1083/jcb.116.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boesze-Battaglia K, Lamba OP, Napoli AA, Jr, Sinha S, Guo Y. Fusion between retinal rod outer segment membranes and model membranes: a role for photoreceptor peripherin/rds. Biochemistry. 1998;37:9477–9487. doi: 10.1021/bi980173p. [DOI] [PubMed] [Google Scholar]
- 4.Ma J, Norton JC, Allen AC, et al. Retinal degeneration slow (rds) in mouse results from simple insertion of a t haplotype-specific element into protein-coding exon II. Genomics. 1995;28:212–219. doi: 10.1006/geno.1995.1133. [DOI] [PubMed] [Google Scholar]
- 5.Cheng T, Al-Ubaidi MR, Naash MI. Structural and developmental analysis of the mouse peripherin/rds gene. Somat Cell Mol Genet. 1997;23:165–183. doi: 10.1007/BF02721369. [DOI] [PubMed] [Google Scholar]
- 6.Travis GH, Christerson L, Danielson PE, et al. The human retinal degeneration slow (RDS) gene: chromosome assignment and structure of the mRNA. Genomics. 1991;10:733–739. doi: 10.1016/0888-7543(91)90457-p. [DOI] [PubMed] [Google Scholar]
- 7.Connell GJ, Molday RS. Molecular cloning, primary structure, and orientation of the vertebrate photoreceptor cell protein peripherin in the rod outer segment disc membrane. Biochemistry. 1990;29:4691–4698. doi: 10.1021/bi00471a025. [DOI] [PubMed] [Google Scholar]
- 8.Moghrabi WN, Kedzierski W, Travis GH. Canine homolog and exclusion of retinal degeneration slow (rds) as the gene for early retinal degeneration (erd) in the dog. Exp Eye Res. 1995;61:641–643. doi: 10.1016/s0014-4835(05)80059-4. [DOI] [PubMed] [Google Scholar]
- 9.Gorin MB, Snyder S, To A, Narfstrom K, Curtis R. The cat RDS transcript: candidate gene analysis and phylogenetic sequence analysis. Mamm Genome. 1993;4:544–548. doi: 10.1007/BF00364792. [DOI] [PubMed] [Google Scholar]
- 10.Begy C, Bridges CD. Nucleotide and predicted protein sequence of rat retinal degeneration slow (rds) Nucleic Acids Res. 1990;18:3058. doi: 10.1093/nar/18.10.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Travis GH, Brennan MB, Danielson PE, Kozak CA, Sutcliffe JG. Identification of a photoreceptor-specific mRNA encoded by the gene responsible for retinal degeneration slow (rds) Nature. 1989;338:70–73. doi: 10.1038/338070a0. [DOI] [PubMed] [Google Scholar]
- 12.Weng J, Belecky-Adams T, Adler R, Travis GH. Identification of two rds/peripherin homologs in the chick retina. Invest Ophthalmol Vis Sci. 1998;39:440–443. [PubMed] [Google Scholar]
- 13.Kedzierski W, Moghrabi WN, Allen AC, et al. Three homologs of rds/peripherin in Xenopus Laevis photoreceptors that exhibit covalent and non-covalent interactions. J Cell Sci. 1996;109:2551–2560. doi: 10.1242/jcs.109.10.2551. [DOI] [PubMed] [Google Scholar]
- 14.Molday RS. Photoreceptor membrane proteins, phototransduction, and retinal degenerative. Invest Ophthalmol Vis Sci. 1998;39:2491–2513. [PubMed] [Google Scholar]
- 15.Moritz OL, Molday RS. Molecular cloning, membrane topology, and localization of bovine rom-1 in rod and cone photoreceptor cells. Invest Ophthalmol Vis Sci. 1996;37:352–362. [PubMed] [Google Scholar]
- 16.Goldberg AF, Molday RS. Subunit composition of the peripherin/rds-rom-1 disk rim complex from rod photoreceptors: hydrodynamic evidence for a tetrameric quaternary structure. Biochem. 1996;35:6144–6149. doi: 10.1021/bi960259n. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg AFX, Loewen CJR, Molday RS. Cysteine residues of photoreceptor peripherin/rds: role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry. 1998;37:680–685. doi: 10.1021/bi972036i. [DOI] [PubMed] [Google Scholar]
- 18.Loewen CJ, Molday RS. Disulfide-mediated oligomerization of Peripherin/Rds and Rom-1 in photoreceptor disk membranes: implications for photoreceptor outer segment morphogenesis and degeneration. J Biol Chem. 2000;275:5370–5378. doi: 10.1074/jbc.275.8.5370. [DOI] [PubMed] [Google Scholar]
- 19.Bascom RA, Manara S, Collins L, Molday RS, Kalnins VI, McInnes RR. Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron. 1992;8:1171–1184. doi: 10.1016/0896-6273(92)90137-3. [DOI] [PubMed] [Google Scholar]
- 20.Boesze-Battaglia K, Stefano FP, Fenner M, Napoli AA., Jr A peptide analogue to a fusion domain within photoreceptor peripherin/rds promotes membrane adhesion and depolarization. Biochim Bio-phys Acta. 2000;1463:343–354. doi: 10.1016/s0005-2736(99)00226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller-Weeks S, Boesze-Battaglia K, Fitzgerald C. Deletional analysis of the rod photoreceptor cell peripherin/rds carboxy-terminal region. Exp Eye Res. 2002;75:143–154. doi: 10.1006/exer.2002.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poetsch A, Molday LL, Molday RS. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J Biol Chem. 2001;276:48009–48016. doi: 10.1074/jbc.M108941200. [DOI] [PubMed] [Google Scholar]
- 23.Kohl S, Giddings I, Besch D, Apfelstedt-Sylla E, Zrenner E, Wissinger B. The role of the peripherin/RDS gene in retinal dystrophies. Acta Anat. 1998;162:75–78. doi: 10.1159/000046471. [DOI] [PubMed] [Google Scholar]
- 24.Musarella MA. Molecular genetics of macular degeneration. Doc Ophthalmol. 2001;102:165–177. doi: 10.1023/a:1017510515893. [DOI] [PubMed] [Google Scholar]
- 25.Zack DJ, Dean M, Molday RS, et al. What can we learn about age-related macular degeneration from other retinal diseases? Mol Vis. 1999;5:30. [PubMed] [Google Scholar]
- 26.Wu T-H, Ting TD, Okajims T-I, et al. Opsin localization and rhodopsin photochemistry in a transgenic mouse model of retinitis pigmentosa. Neuroscience. 1998;87:709–717. doi: 10.1016/s0306-4522(98)00173-0. [DOI] [PubMed] [Google Scholar]
- 27.Kedzierski W, Bok D, Travis GH. Transgenic analysis of rds/peripherin N-glycosylation: effect on dimerization, interaction with rom1, and rescue of the rds null phenotype. J Neurochem. 1999;72:430–438. doi: 10.1046/j.1471-4159.1999.0720430.x. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien J, Al-Ubaidi MR, Ripps H. Connexin 35: a gap-junctional protein expressed preferentially in the skate retina. Mol Biol Cell. 1996;7:233–234. doi: 10.1091/mbc.7.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frohman MA, Dush MK, Martin GR. Rapid production of full length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Ubaidi MR, Pittler SJ, Champagne MS, Triantafyllos JT, McGinnis JF, Baehr W. Mouse opsin: gene structure and molecular basis of multiple transcripts. J Biol Chem. 1990;265:20563–20569. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Kajiwara K, Hahn LB, Mukai S, Travis GH, Berson EL, Dryja TP. Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature. 1991;354:480–483. doi: 10.1038/354480a0. [DOI] [PubMed] [Google Scholar]
- 34.Li C, O’Brien J, Al-Ubaidi MR, Naash MI. Organization of the chicken and Xenopus peripherin/rds gene. In: Anderson RE, La-Vail MM, Hollyfield JG, editors. New Insights into Retinal Degenerative Diseases. New York: Alan R. Liss; 2001. pp. 269–277. [Google Scholar]
- 35.Felsenstein J. PHYLIP: Phylogeny Inference Package (ver. 3.2) Cladistics. 1989;5:164–166. available at http://evolution.genetics.washington.edu/phylip.html/
- 36.Dayhoff MO. Atlas of Protein Sequence and Structure. Suppl. 3. Vol. 5. Washington, DC: National Biomedical Research Foundation; 1978. [Google Scholar]
- 37.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 38.Parnes JR, Robinson RR. Multiple mRNA species with distinct 3′ termini are transcribed from the β2-microglobulin gene. Nature. 1983;302:449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- 39.Lam BL, Vandenburgh K, Sheffield VC, Stone EM. Retinitis pigmentosa associated with a dominant mutation in codon 46 of the peripherin/RDS gene (arginine-46-stop) Am J Ophthalmol. 1995;119:65–71. doi: 10.1016/s0002-9394(14)73815-2. [DOI] [PubMed] [Google Scholar]
- 40.Meins M, Gruning G, Blankenagel A, et al. Heterozygous “null allele” mutation in the human peripherin/RDS gene. Hum Mol Genet. 1993;2:2181–2182. doi: 10.1093/hmg/2.12.2181. [DOI] [PubMed] [Google Scholar]
- 41.Trujillo Tiebas MJ, Gimenez Pardo A, Garcia Sandoval B, Ayuso Garcia C. Phenotypic variation in a family affected by autosomal dominant retinal dystrophy caused by the Gly208Asp mutation in the RDS peripherin gene (Letter) Med Clin. 2002;118:716. doi: 10.1016/s0025-7753(02)72505-0. [DOI] [PubMed] [Google Scholar]
- 42.Nakazawa M, Kikawa E, Kamio K, Chida Y, Shiono T, Tamai M. Ocular findings in patients with autosomal dominant retinitis pigmentosa and transversion mutation in codon 244 (Asn244Lys) of the peripherin/RDS gene. Arch Ophthalmol. 1994;112:1567–1573. doi: 10.1001/archopht.1994.01090240073028. [DOI] [PubMed] [Google Scholar]
- 43.Nakazawa M, Kikawa E, Chida Y, Wada Y, Shiono T, Tamai M. Autosomal dominant cone-rod dystrophy associated with mutations in codon 244(Asn244His) and codon 184 (Tyr184Ser) of the peripherin/RDS gene. Arch Ophthalmol. 1996;114:72–78. doi: 10.1001/archopht.1996.01100130068011. [DOI] [PubMed] [Google Scholar]
- 44.Dowling JE, Ripps H. Visual adaptation in the retina of the skate. J Gen Physiol. 1970;56:491–520. doi: 10.1085/jgp.56.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowling JE, Ripps H. On the duplex nature of the skate retina. J Exp Zool Suppl. 1991;5:55–65. doi: 10.1002/jez.1402560509. [DOI] [PubMed] [Google Scholar]
- 46.Ripps H, Dowling JE. Structural features and adaptive properties of photoreceptors in the skate retina. J Exp Zool Suppl. 1990;5:46–54. doi: 10.1002/jez.1402560508. [DOI] [PubMed] [Google Scholar]
- 47.Szamier RB, Ripps H. The visual cells of the skate retina: structure, histochemistry, and disc-shedding properties. J Comp Neurol. 1983;215:51–62. doi: 10.1002/cne.902150105. [DOI] [PubMed] [Google Scholar]
- 48.Long M, de Suoza SJ, Gilbert W. Evolution of the intron-exon structure of eukaryotic genes. Curr Opin Gen Dev. 1995;5:774–778. doi: 10.1016/0959-437x(95)80010-3. [DOI] [PubMed] [Google Scholar]