Abstract
Cellular therapies have recently employed the use of small RNA molecules, particularly microRNAs (miRNAs), to regulate various cellular processes that may be altered in disease states. In this study, we examined the effect of transient muscle-specific miRNA inhibition on the function of three-dimensional skeletal muscle cultures, or bioartificial muscles (BAMs). Skeletal myoblast differentiation in vitro is enhanced by inhibiting a proliferation-promoting miRNA (miR-133) expressed in muscle tissues. As assessed by functional force measurements in response to electrical stimulation at frequencies ranging from 0 to 20 Hz, peak forces exhibited by BAMs with miR-133 inhibition (anti-miR-133) were on average 20% higher than the corresponding negative control, although dynamic responses to electrical stimulation in miRNA-transfected BAMs and negative controls were similar to nontransfected controls. Immunostaining for alpha-actinin and myosin also showed more distinct striations and myofiber organization in anti-miR-133 BAMs, and fiber diameters were significantly larger in these BAMs over both the nontransfected and negative controls. Compared to the negative control, anti-miR-133 BAMs exhibited more intense nuclear staining for Mef2, a key myogenic differentiation marker. To our knowledge, this study is the first to demonstrate that miRNA mediation has functional effects on tissue-engineered constructs.
Introduction
Alteration of microRNA (miRNA) expression represents a promising approach to promote tissue repair and regeneration.1 miRNAs are short, noncoding RNAs, ∼20–22 nucleotides in length, involved in post-transcriptional gene regulation.2,3 Many miRNAs act as repressive elements to proteins that are themselves repressors. Found either in the introns of pre-mRNAs or part of separate segments of the genome, these noncoding RNAs are processed into mature, single-stranded miRNAs that are released often after preferential incorporation into an RNA-induced silencing complex, similar in mechanism to RNA interference.4 The RNA-induced silencing complex targets sequences within the genome that are complementary, or nearly complementary, to that of the incorporated miRNA and either cleaves the mRNA target or represses translation.3,5
Myogenesis is influenced, in part, by three miRNAs, miR-1, miR-133, and miR-206.6–9 miR-1 is abundant only in cardiac and skeletal muscle and comprises nearly 50% of miRNAs found in the murine heart.8 Chen et al. showed that miR-1 and miR-133 promote skeletal muscle differentiation and proliferation, respectively.10 Specifically, miR-1 targets histone deacetylase 4, which is a transcription factor that represses Mef2, a critical component for myoblast differentiation.11 The Mef2 family of factors encode for proteins that promote differentiation at various stages in development.12,13 Similarly, miR-133 downregulates serum response factor, which blocks myoblast proliferation. Both knockdown (via antisense oligonucleotide probes) and overexpression studies confirmed that miR-1 plays a critical role in myogenesis, whereas miR-133 alone promoted proliferation and partially inhibited myoblast differentiation.10,14
Because miRNAs control critical processes in tissue development, expression of these small regulators may circumvent deleterious cell states. Care et al. found that both miR-1 and miR-133 were downregulated in cardiac hypertrophy models, and by overexpressing one or the other, a hypertrophic response was inhibited.15 In terms of cardiac function, miR-1 represses two genes, KCNJ2 and GJA1, which encode for proteins Kir2.1 and connexin 43, respectively, involved in regulating and maintaining arrhythmogenic potential; hence, miR-1 inhibition suppresses arrhythmias in infarcted hearts of rats.16 miR-133 also affected QT interval prolongation in rabbit diabetic hearts by repressing the ether-a-go-go (ERG) gene, and its overexpression in this disease state was found to be detrimental.17 Another pacing-related gene, HCN2, was a target of miR-133 and upregulated in the hypertrophic state associated with depressed miR-133.18
On the basis of the established role for miR-133 in cardiac tissue function, we hypothesized that inhibiting this miRNA increases force production by skeletal muscle cells in three-dimensional (3D) cultures by promoting differentiation. We previously showed that a collagen gel-based skeletal muscle system was successfully able to form mature, differentiated muscle in a 3D model.19
While promising, perhaps the biggest hurdle facing muscle tissue engineering is achieving contractile forces comparable to those in vivo; therefore, we examined whether modulating miRNAs in a tissue-engineered model could affect the contractile force. On the basis of preliminary results, we inhibited miR-133 and report the effect of miR-133 inhibition on skeletal muscle function in a 3D system using transient transfection of the miRNA, particularly at early time points shortly after the myogenic switch point in vitro. While modulation with viral vectors would produce a sustained response, we reasoned that for eventual clinical studies, viral vectors should be avoided, and continuous overexpression or inhibition of a miRNA may not enable the fine regulation of differentiation that is needed.
Materials and Methods
Cell culture and transient transfection
Murine C2C12 myoblasts (ATCC), a subclone derived from a cell line that originated from normal adult C3H mouse leg muscle,20 were cultured on standard tissue culture flasks at passages 3–5.21 Cells were fed daily with growth medium (GM) containing high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco/Invitrogen), 8% new born calf serum (HyClone Laboratories), 8% fetal bovine serum (HyClone Laboratories), 0.5% chicken embryo extract (Accurate Chemicals), and 0.1% gentamicin (Gibco/Invitrogen) at 37°C and 5% CO2. To promote differentiation of myoblasts to fuse into myotubes, the GM was shifted to differentiation medium (DM), consisting of DMEM supplemented with 8% horse serum (HyClone Laboratories) and 0.1% gentamicin (Gibco/Invitrogen).
For inhibition studies, anti-miR antisense molecules specific for the mature miR-133a sequence, UUUGGUCCCCUUCAACCAGCUG (anti-miR miRNA Inhibitor for hsa-miR-133a, AM10413; Ambion/Applied Biosystems), were transfected with siPORT NeoFX transfection agent (Ambion) into myoblasts per manufacturer's instructions. Briefly, the transfection agent was diluted in Opti-MEM I (Gibco) and mixed with 50 nM of the anti-miR-133, single-stranded RNA or with the same concentration of the anti-miR-negative control (a random, inert nucleic acid sequence). After incubating for 10 min at room temperature, the mixture was dispensed into appropriate tissue culture flasks. A cell suspension, at subconfluent densities, was added to each flask and cultured in normal conditions for 24 h, with antibiotic-free GM. For fluorescent tracking of the transfection, a Cy-3-labeled negative control for the anti-miR was used in the same manner as described above and viewed using confocal microscopy 2 h post-transfection. This is effectively the zero time point of transfection.
Cellular proliferation assay
Cellular proliferation was assessed using an EdU assay (Click-iT(TM) EdU Imaging Kits; Invitrogen) per manufacturer's instructions. Briefly, proliferating C2C12 myoblasts were seeded in a standard six-well plate at subconfluent densities in GM. At 48 h, after reaching confluence, the cells were incubated in GM containing a 1 mM EdU solution, which only stains proliferating cells, for 2 h at 37°C, followed by a series of rinses in phosphate-buffered saline (PBS; Gibco). The cells were then fixed with 100% methanol at −20°C and rinsed several times with PBS. The cells were then incubated with the EdU reaction cocktail (prepared per manufacturer's instructions) for 30 min at room temperature with gentle rocking. The EdU-labeled cells were again rinsed with PBS, stained with Hoechst 33342, and imaged using a Nikon Eclipse TE2000-U fluorescent microscope. EdU-stained cells were counted using Image J software and assessed as a fraction of the total number of nuclei present.
Bioartificial muscle assembly
After 24 h, and before confluence was reached, myoblast cultures (nontransfected control, negative control, and miRNA-modified myoblasts) were trypsinized in a 0.05% trypsin/ethylenediaminetetraacetic acid solution (Gibco) and centrifuged at 1000 rpm for 5 min. Each cell pellet was then resuspended in a Matrigel solution (BD Biosciences) and added to a chilled solution of GM yielding final concentrations of 1.4 mg/mL type I rat tail collagen (BD Biosciences) and 3 mM NaOH to a final 3:1 ratio of collagen to Matrigel. Two million cells were suspended within this collagen gel mixture to give each bioartificial muscle (BAM) a total volume of 0.4 mL.
As previously described,22,23 BAM troughs were made using silicone tubing (Nalgene), split longitudinally with tiny sections of silicone sheets (Nalgene) that were glued to the open ends of the tubing with RTV silicone adhesive (GE). Small stainless steel minutien pins (Fine Science Tools) held Velcro tabs at the ends of the mold to act as attachment sites for the collagen/matrigel construct, and over time, to maintain passive tension. The assembled molds were glued onto the bottom of a standard tissue culture dish and gas sterilized with EtO.
After 1 h at 37°C, warm GM was added to the dishes over the gelled Matrigel construct. The BAMs were cultured for 2 days in GM and then the medium was shifted to promote differentiation. This DM consisted of high-glucose DMEM (Gibco), 8% horse serum (Hyclone Laboratories), and 0.1% gentamicin (Gibco). Cultures remained in DM for 6–8 days (shift day [SD] 6–8).
Histology
The BAMs were fixed for 2 h in a 3.7% paraformaldehyde solution at 4°C, and then rinsed and stored in PBS for histochemistry. All histochemistry procedures were performed in the Pathology Laboratory, Duke Medical Center. Briefly, samples were dehydrated in a graded series of ethanol solutions and embedded in paraffin for sectioning. Specimens were cut into 5 μm sections in both the longitudinal and cross-sectional directions and stained using hematoxylin and eosin (H&E).
Immunostaining and fiber diameter measurements
The BAMs were fixed for 2 h in a 3.7% paraformaldehyde solution at 4°C, rinsed in PBS, and then permeabilized with 0.05% Triton-X for 2 h at 37°C. Afterward, the samples were thoroughly rinsed and stored in PBS. For immunohistochemical staining, primary antibodies against alpha-actinin (Sigma) and myosin MF20 (Developmental Studies Hybridoma Bank) were diluted 1:500 and 1:50, respectively, in a 10% goat serum solution and incubated with the BAMs overnight at 4°C. After thorough rinsing with PBS, the construct was incubated at room temperature with a secondary Alexa Fluor-488 or -546 goat anti-mouse antibody (Invitrogen) for 2 h. In most cases, a nuclear stain, such as Sytox Green (Invitrogen), 1 μM, was added and incubated at room temperature for 1 h. Samples were then viewed via confocal microscopy.
To detect Mef2, the BAMS were fixed with 100% methanol for 5 min at −20°C. The methanol was removed, and the cells were rinsed three times with PBS. A rabbit polyclonal antibody to Mef2 (sc313; Santa Cruz Biotechnology) was diluted to 1:500 in 10% goat serum and incubated in the same manner as stated above. This antibody recognizes Mef2a and to a lesser extent Mef2c and Mef2d in human and mouse cells.
Using several sets of confocal images, fiber diameter measurements were completed for clearly visible fibers within a given field of view. Five independent sets of BAMs (n = 5) were analyzed with an average of 3–5 images per each BAM (negative control and anti-miR-133). For each of the images, ∼5–10 measurements per image were taken using Image J (NIH). The averages from all images linked to a given BAM set were then gathered for statistical analysis.
Functional force testing
BAM force measurements were gathered using a custom apparatus from Aurora Scientific comprised of a force transducer (Aurora Scientific, Model 403A) with an upper limit specification of 5 mN and a benchtop unit (Aurora Scientific, Model 801B). Controlled with a Labview software program package, electrical stimulation was applied at frequencies in a range of 0–40 Hz at 40 V, 10 ms duration, 0.03 ms delay. Data were gathered as a text file and analyzed using a custom Matlab program (The Mathworks, Inc.). For each experiment, two BAMs (for each treatment condition: nontransfected, negative control, and anti-miR-133) were analyzed and averaged. Replicate measurements on the same BAM were virtually identical. Stress measurements were based on the force output of the machine, and BAM cross-sectional area measurements were taken from histology slides. Cross-sectional areas were determined using ImageJ (NIH).
Statistical analysis
Data are presented as a mean ± standard error of the mean (SEM) unless otherwise noted. A two-factor analysis of variance or Student's t-tests were performed using the Statview 5.0 statistical analysis package. When applicable, the Tukey–Kramer post-hoc test was performed as part of the analysis of variance. A value of p < 0.05 was considered significant.
Results
Transient transfection of anti-miR-133 in BAMs
Transfection occurred while cells were still in the tissue culture flasks and immediately before cells were cast into the collagen gel. Cy3-labeling of anti-miR negative control for tracking purposes verified the successful incorporation of anti-miR-133 in differentiating myoblasts 2 h after transfection. Transfection efficiency at 2 h post-transfection was 78.9% ± 2.9% (mean ± SEM, n = 3). Although the intensity was reduced, some of the Cy3-labeled miRNA was still visible in the 3D system at SD 0, or 3 days after transfection.
Inhibition of miR-133 affects proliferation
Since DM rapidly inhibits proliferation, we measured the effect of inhibiting miR-133 upon cell proliferation 48 h after cell confluence had been reached in GM. There was no statistical difference between cells that were not transfected and those that received a scrambled sequence (negative control). Therefore, the nontransfected samples and the negative control samples were pooled into a general control category. Compared to the control, cells that were transfected with anti-miR-133 reduced proliferation by 30% (p < 0.001; n = 10 for control and n = 6 for anti-miR-133).
Inhibition of miR-133 in BAMs affects differentiation
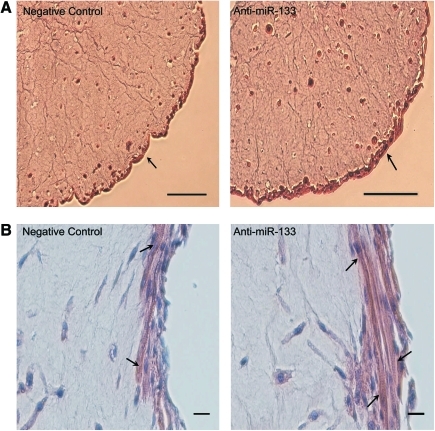
Since miR-133 promotes proliferation, we hypothesized that inhibiting miR-133 promotes differentiation. Histological analysis via H&E staining and immunostaining for various muscle-specific proteins showed organizational differences between negative control and anti-miR-133 BAMs. Transverse and longitudinal cross sections of the BAMs showed morphologic differences in BAMs with and without miR-133 inhibition, as shown in Figure 1. Areas of thick myofiber densities were more apparent in the anti-miR-133 BAMs (see arrows); however, striations were visible in both sets of BAMs showing that the structural proteins were organizing, regardless of miRNA modulation. Similar to previous findings,19 myotubes were concentrated in the outer 100 μm of the BAM periphery.
FIG. 1.
Representative hematoxylin and eosin staining of negative control and anti-miR-133-transfected bioartificial muscles (BAMs), showing thicker area of myofiber density in BAMs with miR-133 inhibition. BAMs were cultured for 2 days in the growth medium and 8 days in the differentiation medium (DM) and are shown at (A) 10 × and (B) 20 × magnification. Arrows in (A) point to the mononuclear and multinucleated myotubes forming on the periphery of the BAM. Arrows in (B) point to striations observed in both sets of BAMs. Scale bars are 100 and 20 μm, respectively for (A) and (B). Color images available online at www.liebertonline.com/ten.
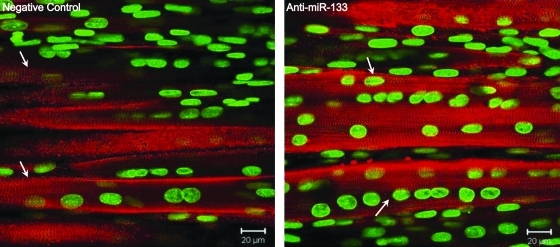
Previous work has shown that inhibiting miR-133 increases myogenin and myosin heavy chain protein expression10; therefore, we examined myofiber formation and striations with alpha-actinin and myosin antibodies. Immunostaining with alpha-actinin in BAMs at SD 6 showed visible striations in both negative control and anti-miR-133 BAMs (Fig. 2). It was difficult to assess if there was a difference in the actual degree of striation with this structural protein.
FIG. 2.
Immunostaining for alpha-actinin (red) in BAMs transfected with negative control or anti-miR-133 and cultured for 6 days in DM, with a Sytox green nuclear counterstain. Distinct striations were visible in both constructs (as shown by arrows); however, myofiber diameter appeared to be larger in the anti-miR-133 BAMs. Color images available online at www.liebertonline.com/ten.
Consistent results were obtained with the myosin immunostaining. Myosin staining with a nuclear counterstain showed that BAMs cultured until SD 6 showed the same trend as constructs that were fixed 2 days later, at SD 8. Fusion and some organization were apparent in both the negative control and anti-miR-133 BAMs, but were more pronounced in those with miRNA mediation. Similar to the alpha-actinin immunostaining, the myosin staining also revealed larger myofiber diameters in the anti-miR-133 BAMs.
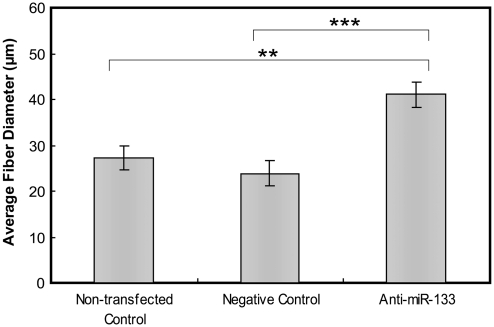
To assess the differences in fiber diameter, measurements from confocal images of five independent sets of BAMs (from SD 6 to 8) were completed. Depending on the z-stack image, each clearly visible fiber diameter was measured, and the results are shown in Figure 3. The BAMs cultured with miR-133 inhibition had, on average, a significantly larger fiber diameter. While there was no significant difference in fiber diameter between the nontransfected and negative control BAMs (27.2 ± 2.7 vs. 23.9 ± 2.6 μm), both of these sets were significantly different from the anti-miR-133 BAMs, which had an average fiber diameter of 41.1 ± 2.7 μm.
FIG. 3.
Average fiber diameter of nontransfected, negative control, and anti-miR-133 BAMs. BAMs cultured with miR-133-inhibited myoblasts cultured for 6 or 8 days had, on average, a large fiber diameter than either the corresponding nontransfected control or the negative control. Mean ± standard error of the mean, n = 5 (independent experiments), and **p < 0.005 compared to nontransfected control, ***p < 0.001 compared to negative control BAMs.
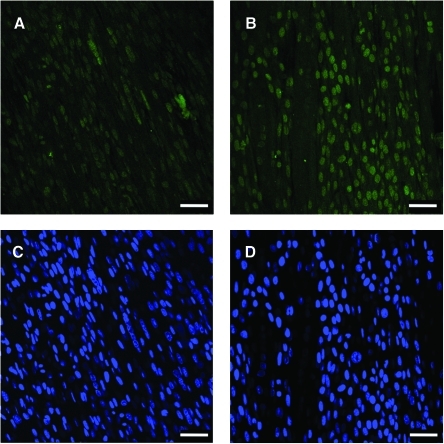
Myogenic differentiation was further assessed with Mef2 immunostaining in the negative control and anti-miR-133 BAMs. As a family of transcription factors that are involved in inducing differentiation, Mef2 is regulated by miR-1 and miR-133.10 Since miR-133 inhibited differentiation, we reasoned that blocking miR-133 would enhance the level of Mef2. Immunofluorescence was used to detect Mef2, and Figure 4 shows Mef2 immunostaining in negative control (panel A) and anti-miR-133 BAMs (panel B). Mef2 expression was nuclear in the BAMs transfected with anti-miR-133 and also in negative and nontransfected controls. While similar fractions of cells showed Mef2 nuclear staining in both anti-miR-133-treated and control BAMs, cells transfected with anti-miR-133 showed more intense staining than the controls. This suggests that the level of differentiation in the anti-miR-133 BAMs is greater compared to the negative control. However, while staining can be useful to grossly assess various states and levels of differentiation, muscle force generation is a more quantitative and functional measure of differentiation.
FIG. 4.
Immunostaining for Mef2 (green) in BAMs transfected with (A) negative control or (B) anti-miR-133 and cultured for 6 days in DM. The Hoechst nuclear counterstain (blue) is also shown for the (C) negative control and (D) anti-miR-133 BAMs. All scale bars are 50 μm. Color images available online at www.liebertonline.com/ten.
Functional force response in nontransfected BAMs
One of the most definitive tests for tissue-engineered constructs is its downstream function. For skeletal muscle constructs, spontaneous twitching and excitability, and contractions in response to electrical stimulation are two ways to determine function in vitro. BAMs cultured with nontransfected myoblasts were used as a baseline to assess overall construct health. Although the direct effect of miRNA mediation on BAM function was compared to the negative control (inert nucleic acid sequence), we first examined the nontransfected BAMs to verify that there were no functional differences by using a transfection agent.
The excitability of nontransfected BAMs to electrical stimulation frequencies ranging from 0 to 20 Hz was measured. Constructs displayed consistent peaks in force in response to a given frequency of stimulation, which was applied for 500 ms from a time of 1 to 1.5 s. However, no tetanus was achieved with these BAMs, and responses degraded when stimulation frequencies were >20 Hz. Average peak forces for nontransfected BAMs cultured from SD 6 to 8 were 0.88 ± 0.18 mN at a 1 Hz stimulation frequency, 1.03 ± 0.21 mN at a 5 Hz frequency, 1.02 ± 0.18 mN at a 10 Hz frequency, and 1.11 ± 0.21 mN at a 20 Hz frequency (mean ±SEM, n = 5). There was evidence that suggests a slight increase in average peak force with increasing stimulation frequency from 1 to 5 Hz. The peak forces were fairly consistent at frequencies >5 Hz. There was no statistical difference in peak force values with different frequencies of stimulation, and data were gathered over five independent experiments.
Functional force response in anti-miR-133-treated BAMs
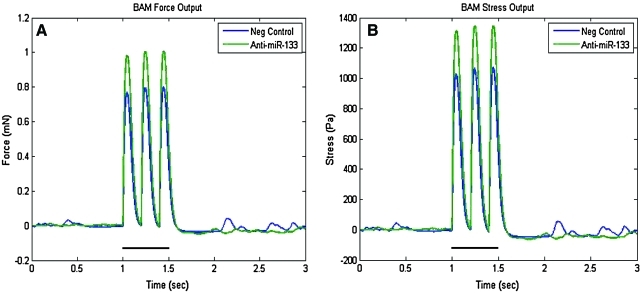
BAMs cultured with myoblasts that were transiently transfected with a miR-133a inhibitor (anti-miR-133) exhibited maximum forces that were on average 20% greater in magnitude than those cultured with the negative control cells. The response to electrical stimulation in BAMs cultured with transfected (both negative control and anti-miR-133) myoblasts was similar to that observed with the nontransfected constructs. A comparative set of peaks at a 5 Hz stimulation frequency (applied stimulation at 1–1.5 s) is shown in Figure 5. In this particular example, the spontaneous twitch force of the anti-miR-133 BAMs was higher than the negative control constructs. The overall spontaneous twitch force (with no electrical stimulation) varied greatly between independent runs with a mean ± SEM as follows: for the nontransfected BAMs, 0.33 ±0.12 mN; negative control BAMs, 0.49 ± 0.20 mN; anti-miR-133 BAMs, 0.57 ± 0.19 mN. There was no significant difference in the passive forces between any of the sets of BAMs.
FIG. 5.
Representative functional force measurements at a stimulation frequency of 5 Hz for BAMs cultured with negative control and anti-miR-133a myoblasts. BAMs were cultured for 6 days in DM. Data are presented as (A) force output, mN, and (B) stress output, Pa, where cross-sectional area was considered. The bold black line segment signifies the time period of electrical stimulation. Color images available online at www.liebertonline.com/ten.
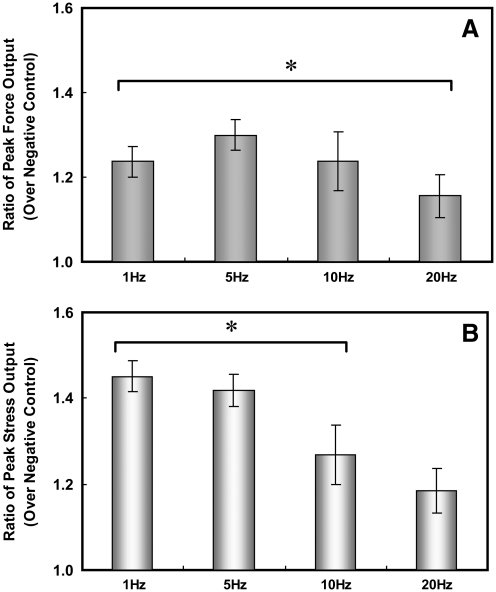
For statistical verification, forces were measured on independent BAM constructs, and the ratio of peak force output over its corresponding negative control force was analyzed. This ratio is statistically different from a ratio of 1 with forces and stresses generated by electrical stimulation at frequencies at or below 20 Hz (Fig. 6). While the ratio of peak force output of miRNA-mediated BAMs at stimulation frequencies >20 Hz was generally >1, it was not statistically significant. Although the ratio of stresses is higher than the ratio of forces, there was no trend in the cross-sectional area of the BAMs. The variability was attributed to differences in these areas over independent experiments, particularly seen at 20 Hz where there was no statistically significant difference in the ratio compared to 1. These summarized results show that inhibition of miR-133 led to increased forces and stresses exhibited by BAMs in response to electrical stimulation.
FIG. 6.
Bar graph showing ratio of peak (A) force and (B) stress of anti-miR-133 BAMs over negative control BAMs cultured for 6–8 days in DM. At stimulation frequencies of 1, 5, and 10 Hz, the peak force and stress ratios were significantly greater than a ratio of 1 (no change with ratio of 1). Mean ± standard error of the mean, n = 5 (independent experiments), *p < 0.02.
Discussion
In this study, we tested the hypothesis that transient transfection of anti-miR-133 in a BAM system promoted muscle differentiation and force production. BAMs were cultured with C2C12 myoblasts that were transiently transfected with miR-133 inhibitors (anti-miR-133). The anti-miR-133 BAMs produced more organized fibers, as assessed by H&E histological stains as well as alpha-actinin, myosin, and Mef2 immunostaining. Fiber diameters were generally larger in anti-miR-133 BAMs than in the negative controls. The presence of exogenous miRNAs (anti-miRs) was diminished by SD 0, although still present at some level; interestingly, the peak force output of the anti-miR-133 BAMs on SD 6–8 was, on average, 20% higher than that of the negative control.
Importantly, our results are the first to show that transient inhibition of miR-133 produced higher functional force. Since miR-133 is highly involved in the developmental process of skeletal muscle maturation, continuous alterations of miRNA levels or gross deviations from baseline levels may lead to detrimental results. In our system, transient transfection was used 1 day before BAMs had been cast into their collagen gels, which provided sufficient time to limit proliferation. With this, differentiation, subsequent myoblast fusion, and myofiber maturation commenced earlier in the anti-miR-133 BAMs. Previous studies have shown that continuous injection of anti-miR-133 in mice inhibited miR-133 and led to cardiac hypertrophy.15 Consistent with these results, we found that a transient block on miR-133 expression was enough to produce a hypertrophic response as larger fiber diameters were observed (Fig. 3). On the basis of our 2D work (unpublished results) and by others,10 miR-133 and miR-1 increase dramatically over time as the cell differentiates, and levels of both increase in tandem during differentiation, despite their opposing effects.
By inhibiting miR-133 in the early stages, we expected that the higher ratio of miR-1 to 133 would inhibit proliferation and promote differentiation. Others have suggested that single injections of a given antagomir are sufficient to have an effect on its downstream target.24,25 Indeed, we found that transient addition of anti-miRs in the early stages of muscle development led to reduced proliferation and a characteristically differentiated construct. In our study, the levels of anti-miR-133 nucleic acids were low by SD 0, which suggests that miR-133 acts at an early stage in the process of differentiation. Whereas a transient inhibition of miR-133 in the beginning stages of myoblast fusion and maturation may enhance the differentiation process, continuous inhibition may lead to deleterious effects as shown by the Care et al.15 study. Therefore, the approach of using nonviral and transient delivery of oligonucleotides may be a better treatment option for miRNA-dependent disease states.
In addition to functional changes, some morphological differences were noted as well. When miR-133 was inhibited, fiber diameter increased compared to the negative control. In a cardiac mouse model, hypertrophy occurred when miR-133 was inhibited continuously for 1 month.15 However, unlike cardiac tissue, skeletal muscle has satellite cells that have regenerative properties and can ultimately re-populate a damaged area. Therefore, hyperplasia cannot be excluded at this point, even though our reasonable assumption is that a small degree of hypertrophy is occurring with an initial inhibition of miR-133 since proliferation is somewhat suppressed.
Several different approaches have been utilized to attain a more native muscle structure in vitro and include using combinations of cells and matrices, mechanical and electrical stimulation over time, and changes to surface morphology.22,26–30 The forces observed in our BAM constructs with the C2C12 cell line were on the same order of magnitude as others have reported for primary cells,22,29,30 as there were no direct comparisons using a pure myoblast cell line. Constructs cultured with primary human myoblasts in a collagen gel had passive forces (without electrical stimulation) in the 0.5 mN range,22 which were slightly higher than our passive forces. Passive forces in myooids cultured with C2C12 myoblasts and 10T1/2 fibroblasts were found to be 0.37 ± 0.05 mN, neonatal rat constructs had forces 0.31 ±0.03 mN, and adult rat soleus myooids forces were lower at 0.12 ± 0.03 mN.28 Engineered skeletal muscle, cultured from slow soleus rat and fast tibialis anterior rat myoblasts up to 28 days with 14 days of electrical stimulation (five electrical pulses every 4 s at 20 Hz), produced higher forces in soleus muscle (0.55 vs. 0.3 mN for no electrical stimulation),29 which was in the same range as our results. Rat myoblasts cast into a fibrin gel produced constructs that exhibited maximum twitch and tetanic forces on the order of 0.3 and 0.8 mN, respectively.30
When grown on an aligned collagen gel matrix, engineered skeletal muscle strips produce similar forces, but had a lower maximal tetanic force at 0.5 mN with a 100 Hz stimulation frequency.27 Differences were also observed between unaligned and aligned matrices (0.14 ± 0.05 vs. 0.31 ± 0.09 mN average tetanic forces).26 In terms of stress, myooids grown up to 40 days with fetal and adult rat myoblasts exhibited stresses of 1–4 kPa,31 and similar stresses were observed with those cultured with a mixture of C2C12 myoblasts and 10T1/2 fibroblasts.28 The stresses seen with our constructs were on the same order of magnitude, but it is important to note that our measurements were taken at much earlier time points to assess functional force during development.
While we have established proof of principle that regulating miRNA expression has functional effects on myoblast differentiation, there are several other factors that must be addressed before the contractile force equals the value of native muscle. At its maximum contraction, native muscle exhibits forces on the order of 300–400 kPa32; however, it is also critical to note that nearly all engineered skeletal muscle contain voids of matrix or remnant gel, which reduce the overall force output. Myotube formation remained isolated to the outer periphery of the BAM, thereby making the effective cross-sectional area smaller than the entire cross-sectional area of the collagen gel. Myofiber organization was also not completely parallel and as densely packed as in native muscle.
In our case and others, we were unable to attain consistent tetanus with a pure C2C12 myoblast population as the constructs were unable to maintain constant forces when stimulation frequencies were above 20 Hz. To date, the tissue-engineered models that show tetanic forces have involved a myoblast cell line mixed with a fibroblast population or constructs cultured with primary myoblasts. Our results can be explained by the absence of mature neuromuscular junctions, which allows myofibers to respond to an electrical impulse.33 However, it is key to note that the forces of these constructs were measured at extremely early time points where the formation of myotubes and maturation of muscle fibers are just beginning.
Despite the work needed to attain native-like skeletal muscle, exogenous and transient inhibition of miRNAs may be a viable way to modulate skeletal muscle function. Future studies will aim to elucidate the mechanisms behind miRNA mediation on muscle function, with a particular focus on several regulatory gene pathways. Further, long-term studies to examine transient transfection on muscle function in vitro will be completed to understand the lasting effects of this treatment.
Conclusions
This is the first study to demonstrate that miRNA mediation has a downstream functional effect on tissue-engineered constructs. While other studies have focused on continuous or stable transfections on various tissues, our results show that differentiation of skeletal myoblasts in vitro may be enhanced by transient transfections of miRNAs. Peak forces exhibited by anti-miR-133 BAMs were on average 20% higher than its corresponding negative control to electrical stimulation (from 0 to 20 Hz), and responses to electrical stimulation in miRNA-mediated BAMs (along with negative controls) were similar to nontransfected controls. Immunostaining also showed more distinct striations and myofiber organization in BAMs with miR-133 inhibition over the negative control. Fiber diameters were also significantly larger in these BAMs over both the nontransfected and negative controls. Additionally, Mef2 immunofluorescence, a myogenic marker, was more intense in the nuclei of skeletal muscle cells in BAMs with miR-133 inhibition over the negative control. This study highlights the possibilities of nonviral, miRNA transfection to change downstream function in vitro, and eventually in vivo.
Acknowledgments
The authors would like to thank I-Chien Liao and Dr. Kam Leong for use of their force testing machine along with Matt Brown for configuring the Labview code. We would also like to thank Nima Badie for his expertise and assistance with Matlab programming for analyzing the force measurements. The authors also appreciate the help of Dr. Li Cao with additional insight into Matlab configuration for assessing staining intensities. We thank Lisa Satterwhite for assistance with the Mef2 immunofluorescence. The MF-20 mouse monoclonal antibody developed by Donald A. Fischman, M.D., was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Funding was provided by the McChesney Foundation and NIH Grant R21-AR55195, RO1-EB-002408.
Disclosure Statement
No competing financial interests exist.
References
- 1.Mack G.S. MicroRNA gets down to business. Nat Biotechnol. 2007;25:631. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005;579:5850. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez M.A. Liu J. Hannon G.J. Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Baskerville S. Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wienholds E. Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M. Rauhut R. Yalcin A. Meyer J. Lendeckel W. Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 9.Sempere L.F. Freemantle S. Pitha-Rowe I. Moss E. Dmitrovsky E. Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J.F. Mandel E.M. Thomson J.M. Wu Q. Callis T.E. Hammond S.M., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma K. Chan J.K. Zhu G. Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol. 2005;25:3575. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potthoff M.J. Arnold M.A. McAnally J. Richardson J.A. Bassel-Duby R. Olson E.N. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007;27:8143. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potthoff M.J. Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 14.Wang D.Z. Micro or mega: how important are MicroRNAs in muscle? Cell Cycle. 2006;5:1015. doi: 10.4161/cc.5.10.2742. [DOI] [PubMed] [Google Scholar]
- 15.Care A. Catalucci D. Felicetti F. Bonci D. Addario A. Gallo P., et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 16.Yang B. Lin H. Xiao J. Lu Y. Luo X. Li B., et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 17.Xiao J. Luo X. Lin H. Zhang Y. Lu Y. Wang N., et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 18.Luo X. Lin H. Pan Z. Xiao J. Zhang Y. Lu Y., et al. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008;283:20045. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- 19.Rhim C. Lowell D.A. Reedy M.C. Slentz D.H. Zhang S.J. Kraus W.E., et al. Morphology and ultrastructure of differentiating three-dimensional mammalian skeletal muscle in a collagen gel. Muscle Nerve. 2007;36:71. doi: 10.1002/mus.20788. [DOI] [PubMed] [Google Scholar]
- 20.Blau H.M. Pavlath G.K. Hardeman E.C. Chiu C.-P. Silberstein L. Webster S.G., et al. Plasticity of the differentiated state. Science. 1985;230:758. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 21.Yaffe D. Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 22.Powell C.A. Smiley B.L. Mills J. Vandenburgh H.H. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am J Physiol Cell Physiol. 2002;283:C1557. doi: 10.1152/ajpcell.00595.2001. [DOI] [PubMed] [Google Scholar]
- 23.Vandenburgh H. DelTatto M. Shansky J. Lemaire J. Chang A. Payumo F., et al. Tissue engineered skeletal muscle organoids for reversible gene therapy. Hum Gene Ther. 1996;7:2195. doi: 10.1089/hum.1996.7.17-2195. [DOI] [PubMed] [Google Scholar]
- 24.van Rooij E. Sutherland L.B. Thatcher J.E. DiMaio J.M. Naseem R.H. Marshall W.S., et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rooij E. Marshall W.S. Olson E.N. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam M.T. Huang Y.C. Birla R.K. Takayama S. Microfeature guided skeletal muscle tissue engineering for highly organized 3-dimensional free-standing constructs. Biomaterials. 2009;30:1150. doi: 10.1016/j.biomaterials.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Yan W. Fotadar U. George S. Yost M. Price R. Terracio L. Tissue engineering of skeletal muscle. Microsc Microanal. 2005;11(Suppl 2):1. doi: 10.1089/ten.2006.0408. [DOI] [PubMed] [Google Scholar]
- 28.Dennis R.G. Kosnik P.E. Gilbert M.E. Faulkner J.A. Excitability and contractility of skeletal muscle engineered from primary cultures and cell lines. Am J Physiol Cell Physiol. 2001;280:C288. doi: 10.1152/ajpcell.2001.280.2.C288. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y.C. Dennis R.G. Baar K. Cultured slow vs. fast skeletal muscle cells differ in physiology and responsiveness to stimulation. Am J Physiol Cell Physiol. 2006;291:C11. doi: 10.1152/ajpcell.00366.2005. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y.-C. Dennis R.G. Larkin L. Baar K. Rapid formation of functional muscle in vitro using fibrin gels. J Appl Phys. 2005;98:706. doi: 10.1152/japplphysiol.00273.2004. [DOI] [PubMed] [Google Scholar]
- 31.Kosnik P.E. Faulkner J.A. Dennis R.G. Functional development of engineered skeletal muscle from adult and neonatal rats. Tissue Eng. 2001;7:573. doi: 10.1089/107632701753213192. [DOI] [PubMed] [Google Scholar]
- 32.King A.M. Loiselle D.S. Kohl P. Force generation for locomotion of vertebrates: skeletal muscle overview. IEEE J Oceanic Eng. 2004;29:684. [Google Scholar]
- 33.Larkin L.M. Van der Meulen J.H. Dennis R.G. Kennedy J.B. Functional evaluation of nerve-skeletal muscle constructs engineered in vitro. In Vitro Cell Dev Biol Anim. 2006;42:75. doi: 10.1290/0509064.1. [DOI] [PubMed] [Google Scholar]