Abstract
Background
When organisms experience environmental change, how does their metabolic network reset and adapt to the new condition? Methylobacterium extorquens is a bacterium capable of growth on both multi- and single-carbon compounds. These different modes of growth utilize dramatically different central metabolic pathways with limited pathway overlap.
Methodology/Principal Findings
This study focused on the mechanisms of metabolic adaptation occurring during the transition from succinate growth (predicted to be energy-limited) to methanol growth (predicted to be reducing-power-limited), analyzing changes in carbon flux, gene expression, metabolites and enzymatic activities over time. Initially, cells experienced metabolic imbalance with excretion of metabolites, changes in nucleotide levels and cessation of cell growth. Though assimilatory pathways were induced rapidly, a transient block in carbon flow to biomass synthesis occurred, and enzymatic assays suggested methylene tetrahydrofolate dehydrogenase as one control point. This “downstream priming” mechanism ensures that significant carbon flux through these pathways does not occur until they are fully induced, precluding the buildup of toxic intermediates. Most metabolites that are required for growth on both carbon sources did not change significantly, even though transcripts and enzymatic activities required for their production changed radically, underscoring the concept of metabolic setpoints.
Conclusions/Significance
This multi-level approach has resulted in new insights into the metabolic strategies carried out to effect this shift between two dramatically different modes of growth and identified a number of potential flux control and regulatory check points as a further step toward understanding metabolic adaptation and the cellular strategies employed to maintain metabolic setpoints.
Introduction
Methylobacterium extorquens AM1 is a facultative methylotrophic bacterium capable of growth on single-carbon compounds such as methanol, methylamine and formate, and multicarbon compounds such as pyruvate and succinate [1]–[3]. In addition to their role in the global carbon cycle [4], methylotrophs are of interest for their potential in the biotechnological production of valued-added chemicals from methanol, which is an inexpensive and abundant source of carbon [5]. These organisms are ubiquitous in nature and are often associated with the leaf surfaces of plants where methanol is released when the stomata open in the morning [6]–[7]. To accommodate these bursts of methanol, methylotrophs must be poised to quickly adapt, capturing available methanol while preventing buildup of its subsequent toxic oxidation product, formaldehyde, as well as further downstream toxic metabolites such as glyoxylate and glycine [8]–[9]. Because of the high flux of toxic metabolites produced during methylotrophic growth, this metabolic mode of growth presents an interesting case study regarding the regulation of and balance between production and consumption of these toxic metabolic products.
M. extorquens AM1 has been studied for decades (reviewed in [10]) and is a model organism for understanding the metabolic components required for methylotrophic growth. The most broadly studied physiological condition has been the comparison of growth on methanol vs. succinate. When growing on multi-carbon substrates, M. extorquens AM1 uses pathways that are common to many heterotrophs, including the TCA cycle, the pentose-phosphate pathway, parts of anapleurotic pathways and gluconeogenesis, and an electron transport chain involving NADH dehydrogenase [1], [11]. In contrast, growth on single-carbon compounds involves specific metabolic pathways for both energy metabolism and assimilation, and requires over 100 additional gene products for central metabolism [10]. These two modes of growth are dramatically different, with methanol growth predicted to be limited by reducing power (NAD(P)H) while succinate growth is predicted to be limited by energy (ATP) [12]–[13].
Global approaches have been developed for M. extorquens AM1 to study multiple layers in the metabolic hierarchy including transcriptomic [14], metabolomic [15]–[17] and proteomic [18]–[19] approaches, as well as analysis of enzymes and fluxes [13], [20]–[21]. These individual studies have shown that the majority of genes known to function in C1 and multicarbon metabolism are differentially expressed and produced according to their function in these two different modes of growth. Most of the amino acids, biosynthetic precursors and intermediates involved in the serine cycle (the main methylotrophic assimilatory pathway) and the TCA cycle are at similar concentrations for methanol growth and succinate growth, while intermediates of the methylotrophy-specific ethylmalonyl-CoA (EMC) pathway are highly elevated in methanol cultures (up to 24-fold differences) [13], [17].
The studies described above have provided a valuable comparison of growth conditions, but these individually-gathered datasets have not been fully integrated to provide a systems biology view of metabolism, which is crucial to understanding methylotrophic growth and predicting metabolic manipulations for industrial benefit. In addition, the dynamics of the response during the transition between growth substrates has not been studied at a global level, and that approach has great potential to provide insights into how the metabolic network is reset to allow methylotrophic growth as well as to uncover general cellular strategies for adaption to environmental change and metabolic stress.
In this study, we address how metabolism responds and adapts under perturbation as a system of interconnected metabolic pathways by adding methanol to a succinate-limited chemostat culture and following the transition of the cells from succinate growth to methanol growth, obtaining data at multiple time points for different layers in the metabolic hierarchy: transcription, enzyme activity, metabolites and fluxes. This integrated analysis provides information about the transient imbalance and the response to carbon starvation the cells experience when they are shifted out of the multi-carbon metabolic mode, the flux redistribution that occurs while the cells are adapting to the new C1 metabolic mode, and the relationships between transcript, enzyme activity, metabolite and flux during this transition. In addition, we show that unlike in other organisms, [22]–[23], M. extorquens AM1 induces its metabolic pathways in a reverse order (biomass cycles first before the pathways that generate the biomass precursor metabolites), which may be a cellular strategy to prevent the buildup of downstream toxic metabolites. Finally, we show that for those genes/enzymes common to both modes of growth, expression and activity change greatly during the transition, yet metabolite concentrations remain relatively constant underscoring the concept of metabolic setpoints [24].
Results
Parameters of the Transition Experiment and Growth Response
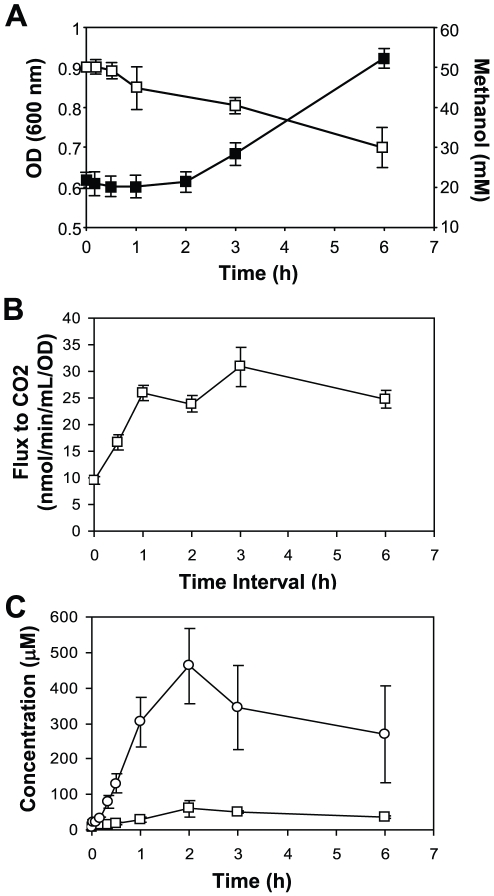
Chemostat cultures were chosen as the starting point for the substrate transition experiments due to the ability to maintain reproducible initial growth conditions. Succinate-limited chemostat cultures of M. extorquens AM1 were grown to steady state (OD ∼0.63), samples were taken as a zero time point, and then 50 mM methanol was added. Cells were harvested at multiple time points after the transition, up to 6 h, and cellular response was assessed via cell growth, carbon flux measurements, selected enzymatic activity assays, global gene expression profiling and measurement of a suite of targeted intracellular and excreted metabolites. Upon cessation of succinate addition to the growth medium and the subsequent addition of methanol, it can be estimated that the carbon flux to biomass dropped from ∼30 nmol C min−1 ml of culture−1 (calculated from the dilution rate, see Methods) to ∼0.2 nmol C min−1 ml of culture−1, which is the measured flux of methanol and CO2 carbon to biomass in succinate-grown cells exposed to methanol in batch culture [20]. This drop in flux of over two orders of magnitude would be expected to plunge the culture into carbon starvation initially, until the induction of the methylotrophic assimilatory pathways allowed increased flux to biomass from those routes. In keeping with this expectation, the culture underwent a 2 h lag phase, demonstrated by the relatively unchanged optical density (OD600) (Figure 1A), then transitioned to log phase indicating that the culture had started to actively grow on methanol. The methanol concentration in the medium began to drop between 15–30 min post methanol addition, with a concentration of ∼30 mM left at 6 h post transition. Note that since the culture was initially at steady-state under succinate limitation, this protocol effected a transition from succinate to methanol growth without the need to wash the starting culture.
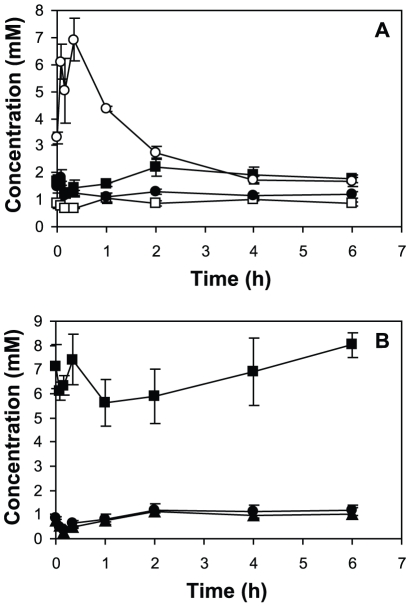
Figure 1. Optical density (OD), rates of carbon flux to CO2 and external metabolite measurements before and during the transition from succinate to methanol growth.
(A) Change of optical density (filled squares) and methanol concentrations (open squares). (B) Carbon flux to CO2 measured using 14C-methanol. (C) Formaldehyde (squares) and formate (circles) concentrations in the culture supernatant. Standard deviations are shown as error bars.
Global Gene Expression Response to Carbon Source Switch
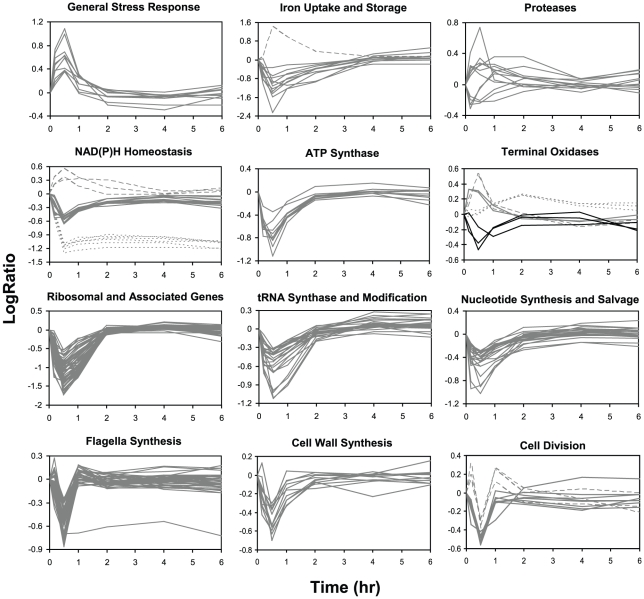
To gain insights into the global cellular response upon carbon source switch, gene expression data were analyzed to determine the cellular processes (outside of central metabolism) that showed the largest changes in their initial response to methanol addition (Figure 2) and were compared to a set of carbon-starvation response arrays as controls (Table S1). Several classes of genes responded similarly in both conditions, in keeping with the prediction that the cells would experience carbon starvation at the onset of the transition. These included genes whose expression increased in both cases, including predicted stress response genes, predicted genes involved in NADH homeostasis, cytochrome bd ubiquinol oxidase genes, and a predicted bacterioferritin (iron storage) gene. In addition, many classes of genes showed a transient decrease in expression in both conditions, either within the first 30 min or after a transient increase, and then a recovery. This pattern included genes predicted to be involved in synthesis of proteins, fatty acids, lipopolysaccharides, ATP, flagella, cell walls and nucleotide synthesis and salvage, along with cytochrome c oxidase genes and several genes predicted for cell division, chemotaxis, glycosylation, and protease activity. A similar pattern of decrease and recovery was also observed for several methylotrophy genes, discussed below. Genes encoding transhydrogenase, which interconverts NADH and NADPH [25], were significantly down-regulated (15–20 fold) in the methanol addition condition but not in the starvation control, and remain repressed during the time period of methylotrophic growth (Figure 2), consistent with previous steady state studies [14]. Cytochrome d terminal oxidase genes increased independently of the carbon starvation control while the cyotchrome o ubiquinol oxidase genes increased in opposition to the carbon starvation control, where expression decreased ∼5 fold.
Figure 2. Changes in gene expression of general cellular functions that significantly changed during the transition from succinate to methanol growth.
Graphs include genes predicted to be involved in general stress response (top left), iron uptake (top center, solid lines) and storage (top center, dashed line), peptide cleavage (top right), NAD(P)H homeostasis (upper-middle left) including NADH:ubiquinone oxidoreductase (nuo, solid lines), transhydrogenase (pnt, short dashed lines) and three additional genes (long dashed lines) predicted to encode NADH dehydrogenase 2, NADH:flavin oxidoreductase/NADH oxidase and NAD(P)H quinone oxidoreductase, ATP synthase function (upper-middle center), terminal oxidase function (upper-middle right) including cytochrome c oxidase (coxABC, solid black lines), cytochrome bd ubiquinol oxidase (qxtAB, solid gray lines), cytochrome d terminal oxidase (cydAB, long dashed lines) and cytochrome o ubiquinol oxidase (cyoABC, short dashed lines), ribosome function (lower-middle left), tRNA synthesis and modification (lower-middle center), nucleotide synthesis and salvage (lower-middle right), flagella synthesis (bottom left), cell wall synthesis (bottom middle) and cell division (bottom right). LogRatios, intensities, fold changes and p-values are shown for each gene in Table S1.
Overview of Central Metabolic Carbon Flow
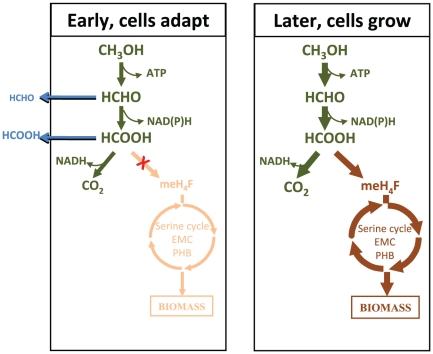
In M. extorquens AM1, methanol is first oxidized to the toxic metabolite, formaldehyde, then to formate. Formate serves as a branch point and can either be oxidized to CO2 producing reducing power, or be converted to methylene-H4F, which enters the central metabolic carbon assimilation pathways including the serine cycle, the EMC pathway, the PHB cycle and a portion of the TCA cycle, where essential intermediates are produced for cell growth [1]–[2], [21] (Figure 3). These pathways were analyzed in detail along with two core pathways involved in multi-carbon growth, the TCA cycle and the pentose-phosphate pathway. After methanol was added to succinate-grown cultures, distribution of carbon flow was assessed using 4 methods: growth curve analysis, measurement of flux to CO2 using 14C-labeled methanol, measurement of methanol, formaldehyde and formate in the culture supernatant and measurement of assimilatory pathway metabolites (described below). Growth did not occur until between 1–2 h post transition (Figure 1A), yet flux of methanol to CO2 increased significantly within the first hour, prior to growth (Figure 1B). Formaldehyde and formate concentrations in the supernatant increased until about 2 h, when levels began to decrease (Figure 1C), suggesting that before active growth occurred, both compounds were excreted and then as the cells began to divide, excretion decreased. For the first hour, approximately 1/3 of the carbon from methanol oxidation was in these two pools, the remainder in CO2. Note that the peak concentrations, 50 and 450 µM, respectively, are not toxic for M. extorquens AM1 [26]–[28]. By 2 h, the total flux to formaldehyde, formate, and CO2 of the culture (∼32 nmol min−1 [ml at 1 OD]−1) was about ¾ of the full flux measured in methanol-grown cells (41.5 nmol min−1 [ml at 1 OD]−1; [21]). These data suggest that the lag in growth was due to a block in formate assimilation, not production (summarized in Figure 3).
Figure 3. Overview of flux changes through methylotrophic central metabolism during the transition from succinate to methanol growth.
Dissimilatory steps are shown in green, assimilatory steps in brown. Methylene-H4F is abbreviated as meH4F.
Visualization of Hierarchical Changes for Central Metabolism
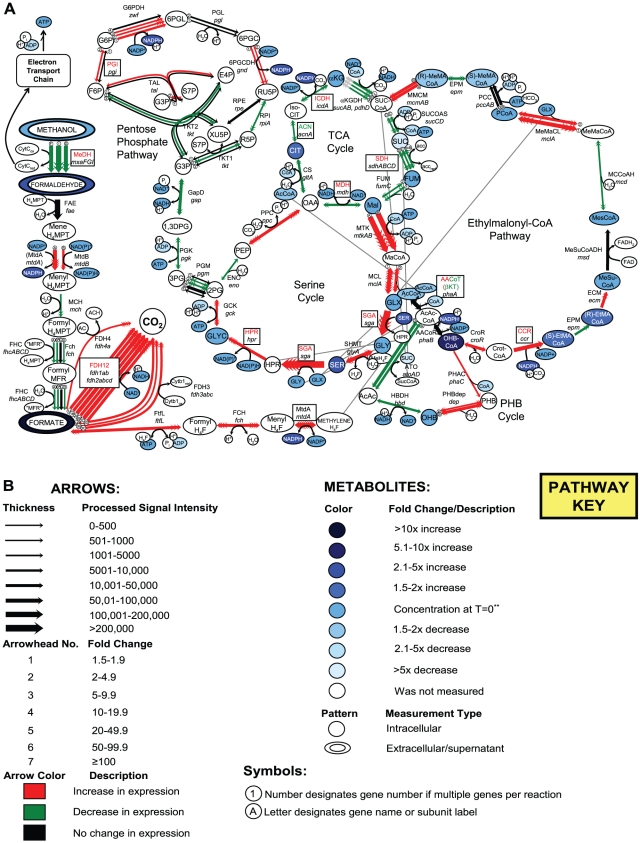
With the introduction of global “omics” level tools, it has recently become possible to investigate multiple layers of an organism's metabolic network during a condition of study. However, the power of these tools can also be a detriment, generating large amounts of data that can often be difficult to integrate and understand as a whole. To facilitate insights and infer meaning regarding the multi-leveled changes and adaptations that the metabolic network of M. extorquens AM1 undergoes during the transition from succinate- to methanol-growth, diagrams were constructed that visually compile and summarize each level of data obtained in relation to central metabolism. The initial response to methanol addition (time = 10–30 min) is shown in Figure 4A with 4B serving as a legend. Diagrams depicting the metabolic state prior to methanol addition (time = 0 min), response just prior to/at the start of cell growth (1–2 h) and during log phase cell growth (3–6 h) are included in Figure S1. These diagrams depict information about gene expression intensities (arrow thickness) and fold changes (number of arrow heads), changes in measured metabolite concentrations (color shadings), and enzymatic activities (color shadings of boxed protein names), providing insight into both the metabolic changes themselves and the level at which those changes occurred. While only semi-quantitative due to possible differing labeling and hybridization efficiencies that could occur during microarray experiments, information on gene intensities is provided since the intensity data aid in pathway interpretation of possible carbon flow. Analysis of fold changes alone can be misleading if for example, no change in expression occurs for a gene, yet expression of that gene is high, or if a gene has a significant change in expression but expression is extremely low.
Figure 4. Pathway schematic depicting changes that occurred in measured metabolites, gene expression, and enzymatic activities for central metabolism during the transition from succinate to methanol growth.
A boxed gene/protein name indicates the activity of this enzyme was measured. Red lettering for the protein designation indicates an increase in activity; green, decrease; black, no change. Metabolites appearing more than once are connected by gray lines. Changes are shown for (A) the initial response, time = 10–30 min with (B) serving as a legend. Graphs depicting changes for pre-methanol addition, time = 0 min; just prior to/at the start of cell growth, time = 1–2 h; and exponential cell growth, time = 3–6 h are included in Figure S1. Reaction descriptions are included in Table S2 along with gene expression intensities, LogRatios, fold changes and p-values. Mesaconyl-CoA, ethylmalonyl-CoA, methylsuccinyl-CoA were measured as free acids. **Color at T = 0 represents the concentration before methanol was added for all metabolites except for methanol which was calculated as 50 mM for the initial T = 0 value.
Response of Measured Nucleotide Pools
The adenine and pyridine nucleotide pools reflect the metabolic state of the cell. As expected for a growth downshift experiment, these compounds all showed significant changes, with most values changing by 50–100% during the time course (Figure 5). Consistent with the prediction that methanol growth is reducing power limited and succinate growth is energy limited [12]–[13], NADPH increased immediately, in keeping with an initial major downshift in biomass synthesis, peaked at the 30 min timepoint, then decreased by 6 h to a lower value than the initial value. ATP decreased immediately, in keeping with the downshift in carbon flow through the TCA cycle, then increased, then decreased, and after 1 h, slowly increased to a final value greater than that for succinate growth. The other nucleotides all decreased immediately, then rose and either stayed constant or decreased slightly, a pattern reminiscent of the starvation-induced gene expression data.
Figure 5. Changes in nucleotide concentrations before and during the transition from succinate to methanol growth.
(A) Changes of pyridine nucleotides: NAD (filled squares), NADH (open squares), NADP (filled circles), NADPH (open circles), (B) changes of adenine nucleotides: ATP (filled squares), ADP, (filled circles), AMP (filled triangles). Standard deviations are shown as error bars.
System Response for Oxidation of Methanol to Formaldehyde
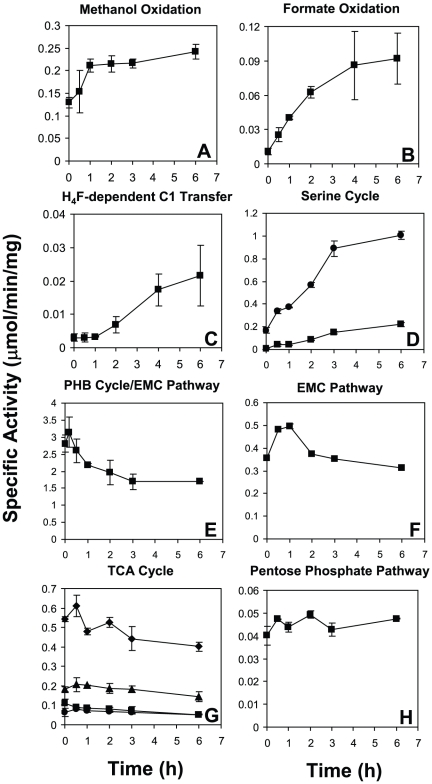
Methanol is oxidized to formaldehyde by methanol dehydrogenase. In the succinate-limited cultures, the transcripts for the genes encoding the methanol dehydrogenase subunits (mxaF, mxaI) and its cytochrome (mxaG) were very high (Figure S1A, expression intensity values are given in Table S2). Upon methanol addition, these transcripts initially decreased 2–3 fold (Figure 6), then increased and had recovered to near starting levels by 2 h, when the cells began to grow, a pattern similar to the carbon starvation response noted earlier. The other genes involved in generating the active form of methanol dehydrogenase (mxaACDEHJKLRS) showed a similar pattern (see Table S2). However, enzymatic activity of methanol dehydrogenase did not mirror transcriptional changes. Activity in the succinate-grown cells was 129 nmol min−1 mg protein−1 initially and increased over the 6 h of the experiment after methanol was added, to 242 nmol min−1 mg protein−1 (Figure 7A).
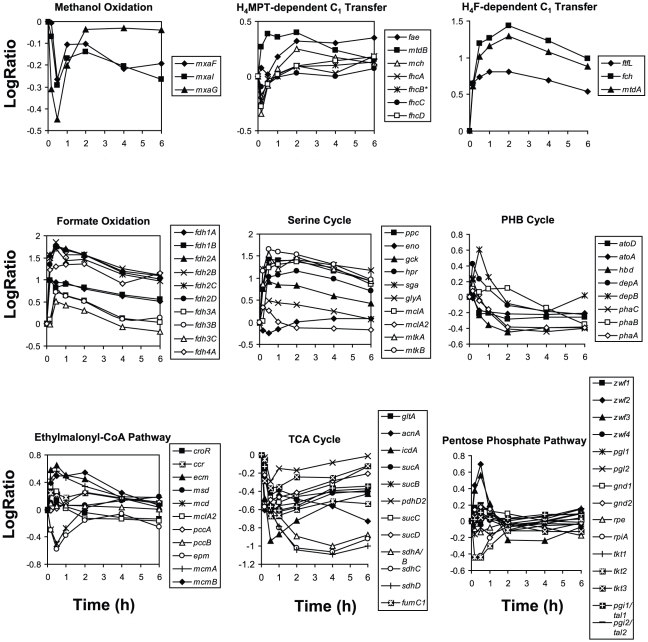
Figure 6. Changes in gene expression for central metabolic pathways during the transition from succinate to methanol growth.
Graph include genes encoding methanol dehydrogenase (mxaFI) and cytochrome c L (mxaG) (top left), H4MPT-dependent (top center) and H4F-dependent (top left) C1 transfer genes, formate oxidation genes (middle left), serine cycle genes (middle center), PHB cycle genes (middle left), EMC pathway genes (bottom left), TCA cycle genes (bottom center) and pentose phosphate pathway genes (bottom right) after addition of methanol (50 mM, final concentration) to succinate-limited chemostat-grown cells.
Figure 7. Changes in enzymatic activity before and during the transition from succinate to methanol growth.
Measured activities include (A) methanol dehydrogenase activity, (B) NAD-dependent formate dehydrogenase activity (Fdh1,2) (C) methylene-H4F dehydrogenase activity (MtdA), (D) serine-glyoxylate aminotransferase (Sga, squares) and hydroxypyruvate reductase (Hpr, circles), (E) β-ketothiolase (βKT, encoded by phaA), (F) crotonyl-CoA reductase (Ccr), (G) malate dehydrogenase (Mdh, diamonds), isocitrate dehyhdrogenase (Icd, triangles), aconitase (Acn, squares) and succinate dehydrogenase (Sdh, circles). Standard deviations are shown as error bars.
System Response for Oxidation of Formaldehyde to Formate
To oxidize formaldehyde, M. extorquens AM1 uses the cofactor, tetrahydromethanopterin (H4MPT) as a carrier of one-carbon units, converting formaldehyde to formate in 5 enzymatic steps as shown in Figure 4A. Due to the unavailability of H4MPT and its conjugates, enzyme activities in this pathway were not measured. The transcript of the first enzyme required for the oxidation of formaldehyde (Fae, formaldehyde activating enzyme) was high in the succinate grown cells (Figure S1A, Table S2) and increased 2.2-fold over 6 h (Figure 6). This enzyme is key to preventing formaldehyde from building up to toxic levels inside the cell [29]. The next step in the pathway involves MtdB, which generates NAD(P)H. In addition, a second enzyme with activity towards H4MPT and NADP, MtdA, is present in M. extorquens AM1, but the main role of this enzyme appears to be in the H4F-dependent C1 transfer pathway [21], [30], so it will be discussed below, in that section. The gene encoding MtdB increased in expression immediately, leveled out at 30 min, and then decreased slowly over the rest of the time course. However, expression of the remaining genes of the formaldehyde oxidation pathway (H4MPT-dependent C1 transfer pathway) decreased initially (mch, fhcABCD, Figure 6) similar to the genes for methanol dehydrogenase, and similar to the carbon starvation response. In general, expression of these genes returned to starting levels between 1–2 hours, increasing slightly or remaining constant through the remaining 6 hrs.
System Response for Oxidation of Formate to CO2
Oxidation of formate to CO2 is carried out by formate dehydrogenase (Fdh). M. extorquens AM1 contains 4 known Fdhs, and transcripts for the genes encoding those Fdhs increased immediately and considerably, peaking in expression levels between 10 and 30 min post transition, then decreased over time (Figure 6). It has not yet been possible to assay the activities of Fdh3 and Fdh4 [26]–[27], but the combined activities of the NAD-dependent enzymes, Fdh1 and Fdh2, increased immediately and continued to increase over the 6 h time period (Figure 7B), correlating with the increase in transcripts and the flux to CO2.
Response of the H4F-dependent C1 Transfer Pathway for Conversion of Formate to Methylene-H4F
Three enzymes are required to convert formate to methylene-H4F: formyl-H4F ligase (FtfL), methenyl-H4F cyclohydrolase (Fch) and methylene-H4F dehydrogenase (MtdA). Transcripts for each of these genes responded in a similar manner, increasing immediately until 2 h, when the cells began to grow (Figure 6). Then the transcripts decreased but remained high over the course of the experiment. MtdA activity was assayed and showed a 1 h lag in activity (Figure 7C), though expression of this gene increased immediately and rose to levels 20-fold above the starting point.
Response of the Carbon Assimilation Pathways
The serine cycle and EMC pathway are the main pathways by which M. extorquens AM1 incorporates C1 compounds from methanol into biomass. The EMC pathway overlaps with the PHB, Serine and TCA cycles (Figure 4A).
Serine cycle
With the exception of enolase, expression of the serine cycle genes increased immediately and peaked at 10 min post switch (Figure 6). As is well-known [31]–[32], hydroxypyruvate reductase (Hpr) and serine-glyoxylate aminotransferase (Sga) had low activities in cells growing on succinate compared to methanol. After methanol addition, their activities increased considerably (6-fold and 44-fold, respectively) and continuously over the next 6 hours (Figure 7D), as previously reported for batch culture experiments [33]. Expression of the enolase gene decreased initially and then increased back to its original pre-transition level (Figure 6), similar to the carbon starvation response. While expression of enolase stands apart from the other serine cycle genes, unlike the other serine cycle genes, it is essential for growth on both succinate and methanol [10]. With the exception of serine, which responded in a manner similar to other amino acids (alanine, threonine, aspartate and phenylalanine; Table 1), measured serine cycle intermediates (glycerate, malate, glyoxylate and glycine) did not significantly change between the time of methanol addition and cell growth (Table 1, Figure 8). Most of the amino acids measured showed an increase within the first 30 min, then decreased to their approximate initial concentrations, which correlated with an initial increase in transcription of a number of putative protease genes (Figure 2), suggesting this response may have been due to protein turnover early in the transition.
Table 1. Intracellular concentrations of target metabolites from cells during transition from succinate growth to methanol growth.
| zero | 10 min | 20 min | 30 min | 1 h | 2 h | 3 h | ||
| Pathway | Metabolite | Concentration (mM) | ||||||
| TCA | Suc | 12.2±0.7 | 3.7±2.5 | 2.1±0.7 | 2.9±0.8 | 5.0±0.3 | 2.2±0.3 | 2.3±0.9 |
| Fum | 0.5±0.1 | 0.6±0.1 | 0.5±0.0 | 0.5±0.1 | 0.5±0.1 | 0.5±0.1 | 0.5±0.1 | |
| Mal | 1.6±0.4 | 1.7±0.4 | 1.2±0.2 | 1.3±0.4 | 1.0±0.4 | 0.9±0.1 | 1.2±0.4 | |
| 2KG | 1.4±0.3 | 1.5±0.2 | 1.7±0.3 | 1.4±0.4 | 1.5±0.3 | 2.3±0.3 | 2.5±0.4 | |
| Cit | 1.8±0.5 | 2.0±0.4 | 3.3±0.4 | 1.8±0.4 | 2.2±0.6 | 2.1±0.4 | 2.4±0.5 | |
| Pyr | 3.7±0.4 | 4.4±1.2 | 3.9±0.3 | 4.6±0.5 | 5.1±0.7 | 4.8±0.4 | 4.9±0.3 | |
| Serine | Glyc | 1.7±0.3 | 2.5±0.6 | 2.0±0.5 | 2.1±0.3 | 2.0±0.4 | 1.8±0.2 | 2.2±0.7 |
| Ser | 4.0±1.5 | 4.5±1.1 | 4.2±1.0 | 16.7±10.4 | 10.1±6.8 | 3.3±0.8 | 2.9±1.7 | |
| Gly | 0.8±0.1 | 0.8±0.1 | 0.8±0.1 | 0.9±0.2 | 1.0±0.1 | 1.0±0.0 | 1.2±0.1 | |
| PHB | OHB | 3.0±0.2 | 3.3±0.4 | 3.4±0.2 | 3.3±0.5 | 4.4±0.7 | 4.0±0.5 | 4.3±0.9 |
| OHB-CoA | 0.1±0.1 | 0.2±0.1 | NMa | 0.4±0.1 | 0.3±0.1 | NM | 0.3±0.1 | |
| EMC | EtMA | 0.6±0.1 | 0.8±0.1 | 1.1±0.1 | 1.0±0.1 | 1.2±0.1 | 2.1±0.2 | 3.1±0.4 |
| MeSuc | 0.6±0.0 | 0.7±0.1 | 0.8±0.0 | 0.7±0.1 | 0.8±0.0 | 1.7±0.1 | 4.3±0.8 | |
| P-CoA | 0.1±0.1 | 0.1±0.1 | NMa | 0.1±0.1 | 0.2±0.1 | NM | 0.3±0.1 | |
| MeMA | 0.3±0.0 | 0.1±0.1 | NM | 0.1±0.1 | 0.2±0.1 | NM | 0.3±0.1 | |
| Otherb | Ac-CoA | 0.3±0.1 | 0.2±0.1 | NM | 0.2±0.1 | 0.2±0.1 | NM | 0.3±0.1 |
| M-CoA | 0.1±0.1 | 0.3±0.1 | NM | 0.1±0.1 | 0.2±0.1 | NM | 0.2±0.1 | |
| CoA | 2.6±0.2 | 0.7±0.5 | NM | 1.5±0.2 | 1.2±0.3 | NM | 1.4±0.5 | |
| Ala | 7.6±0.7 | 9.4±2.0 | 9.4±1.1 | 14.0±1.5 | 12.9±4.1 | 9.7±0.3 | 10.0±0.6 | |
| Val | 1.0±0.1 | 0.7±0.2 | 0.9±0.1 | 1.4±0.4 | 1.5±0.6 | 1.2±0.1 | 1.3±0.2 | |
| Leu | 0.6±0.0 | 0.6±0.2 | 0.8±0.0 | 0.9±0.1 | 0.9±0.4 | 1.1±0.1 | 1.4±0.3 | |
| Ile | 0.4±0.1 | 0.5±0.1 | 0.6±0.0 | 0.7±0.1 | 0.7±0.1 | 0.5±0.0 | 0.6±0.0 | |
| Pro | 1.3±0.2 | 1.3±0.3 | 1.6±0.1 | 1.6±0.3 | 1.4±0.4 | 1.1±0.2 | 1.0±0.4 | |
| Thr | 3.1±0.4 | 3.4±0.6 | 3.4±0.6 | 6.3±3.4 | 5.2±1.1 | 3.0±0.5 | 3.3±0.7 | |
| Asp | 4.9±1.5 | 4.9±1.3 | 4.9±0.5 | 5.9±1.1 | 6.2±3.1 | 2.2±2.0 | 2.3±2.4 | |
| Phe | 0.3±0.0 | 0.4±0.1 | 0.7±0.1 | 0.5±0.2 | 0.4±0.2 | 0.2±0.1 | 0.2±0.1 | |
Numbers are averages of nine replicates, and standard deviations are listed as ± sd. The abbreviations used in the table are defined as follows: Suc = succinate; Fum = fumarate; Mal = malate; 2KG = 2-ketoglutarate; Cit = citrate; Pyr = pyruvate; Glyc = glycerate; OHB = 3-hydroxybutyrate; OHB-CoA = 3-hydroxybutyryl-CoA; EtMA = ethylmalonate; MeSuc = methylsuccinate; P-CoA = propionyl-CoA; MeMA = methylmalonate; Ac-CoA = acetyl-CoA; M-CoA = malonyl-CoA.
NM = not measured.
Other refers to measured metabolites that are not depicted in the central metabolic diagrams (Figure 4) or are components of multiple central metabolic pathways.
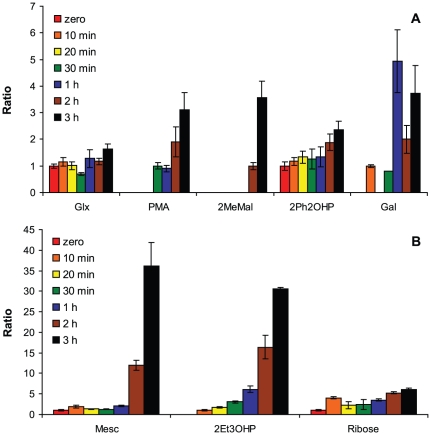
Figure 8. Metabolite ratios for selected metabolites for which concentrations are not available measured before and during the transition from succinate to methanol growth.
Ratios for metabolites that changed up to 5-fold are graphed in (A) and those that increased >5-fold are depicted in (B). Ratios are set to 1 for initial detection. Abbreviations are as follows: Glx, glyoxylate; PMA, propylmalonate; 2-MeMal, 2-methylmalate; 2-Ph2OHP, 2-phenyl-2-hydroxypropionate; Gal, galactose; Mesc, mesaconic acid; 2Et3OHP, 2-ethyl-3-hydroxypropionate. Standard deviations are shown as error bars.
EMC pathway
The majority of the EMC pathway genes were immediately and highly induced, peaking at 10–30 min and then returning to just above pre-transition levels (Figure 6). In contrast, metabolites of the EMC pathway (measured as their non-CoA derivatives) hydroxybutyrate, ethylmalonate, methylsuccinate, mesaconate, 2-methylmalate, did not show significant increases until the 2 or 3 h timepoint, while methylmalonyl-CoA and propionyl-CoA did not show significant increases until the 1 or 2 h timepoint (Table 1, Figure 8). Expression of mcd and epm encoding, respectively, mesaconyl-CoA hydratase and epimerase, mirrored each other and did not follow the pattern of the other EMC genes, decreasing immediately and returning to starting levels or higher by 4 h post transition (Figure 6), reflecting the carbon starvation response genes. Comparisons of expression changes of mcd and epm in steady-state succinate and methanol grown cells were consistent with these data, being similar in both growth conditions [14]. Between 1–2 h, carbon flux occurred via the EMC pathway, evidenced by the accumulation of mesaconate, methylsuccinate, ethylmalonate, methylmalonate, methylmalonyl-CoA and propionyl-CoA (Figure 8, Table 1, summarized in Figure S1C), and cell growth occurred. Expression of msd, encoding methylsuccinyl-CoA dehydrogenase, which is known to be required for growth on C1 compounds [34], did not significantly change, yet expression intensities suggested that this gene might be highly expressed under conditions of both methanol and succinate growth. With few exceptions, both array data sets (biological replicates) were in agreement regarding range of intensity and fold change. Notable exceptions were ccr and croR, encoding respectively, crotonyl-CoA reductase and crotonase, which increased by a small amount (1.8-fold for both) in the first array data set but not the second.
The activities of two enzymes of the EMC pathway, β-ketothiolase (βKT; encoded by phaA) and crotonyl-CoA reductase (Ccr; encoded by ccr) were measured. βKT activity, which overlaps with the EMC and PHB cycles, increased slightly initially then decreased to ∼60% of the original activity (Figure 7E), mirroring expression of phaA (Figure 6). 3-hydroxybutyryl-CoA, which is an intermediate of both the EMC pathway and PHB cycle, showed a similar pattern, increasing initially and then remaining relatively constant (Table 1). Ccr activity was high in succinate grown cultures (356 nmol min−1 mg protein−1, Figure 7F), increased 1.4-fold by 1 h, and then decreased to just below starting levels by 6 h, consistent with the minimal changes in transcripts noted above.
Using Fisher ratio analysis to assess metabolite differences between two samples, a number of metabolites were previously identified (2-methylmalate, propylmalonate, 2-phenyl-2-hydroxy propionate, 2-ethyl-3-hydroxypropionate, ribose and galactose) as being significantly higher in methanol-grown cells than succinate-grown cells [15]. These metabolites responded similarly to the EMC pathway intermediates and are shown in Figure 8, but their role in C1 metabolism, if any, is unknown.
PHB cycle
Poly-β-hydroxybutyrate (PHB) accumulates in M. extorquens AM1 as a carbon and energy storage compound [35]–[36]. The phaA and phaB genes, which are required for PHB synthesis, are shared with the EMC pathway. Expression of these genes increased slightly initially, then decreased over the course of the experiment with phaA decreasing between 30 min–1 h and phaB decreasing between 4–6 h (Figure 6). These gene products produce 3-hydroxybutyryl-CoA, which is a precursor to PHB as well as an intermediate of the EMC pathway. While expression of these genes did not change much initially and activity of βKT increased minimally (Figures 6,7E), concentration of 3-hydroxybutyryl-CoA increased 4-fold within the first 30 min after methanol addition (Table 1). Expression of the depolymerase genes (depA, depB), which hydrolyze PHB, increased 2.6- and 4.1-fold respectively at 10 min and then decreased over the time course. Genes involved in the further degradation of PHB (hbd, ato) decreased after 10 min post methanol addition and remained lower than initial levels.
Response of the TCA Cycle
Expression of all TCA cycle genes (sdhABCD, sucAB, icdA, acnA, gltA, mdh, fumC) decreased severely (Figure 6) but intensities were still well above background levels throughout the experiment (Table S2). However, aconitase (Acn) was the only measured enzyme that exhibited a significant decrease in enzyme activity (48% decrease) while other enzyme activities increased slightly and then returned to just below their starting activity (Icd, 79%; Sdh, 82%, Mdh; 74% decrease; Figure 7G). In keeping with the enzyme activity data, except for succinate, levels of the measured TCA cycle metabolites remained relatively constant, not showing distinct trends (Table 1). Succinate was high initially at time zero as expected since it was the carbon source, and decreased from 12.2 mM to 3.7 mM within 10 min of stopping medium flow (Table 1).
Response of the Pentose Phosphate Pathway
Predicted genes for the pentose phosphate pathway (zwf, pgl, gnd, fbp, pgi, tkt, fba. rpe, rpi) exist in M. extorquens AM1, often in duplicate or triplicate, yet the operation of this pathway has not been thoroughly studied. Gene expression intensities suggested that enzymes involved in this pathway are present during both succinate and methanol growth, with different portions of the pathway up-regulated during the transition. 2 of the 4 candidate zwf (glucose-6-phosphate dehydrogenase) genes were significantly up-regulated after methanol was added (4–5 fold) while 1 of 3 predicted tkt (transketolase) genes along with a predicted rpiA (ribose-5-phosphate isomerase) gene were down-regulated (∼3 fold) (Figures 4A, 6). Expression of a third putative zwf gene along with gnd1 (6-phosphogluconate dehydrogenase), gnd2 and tal2/pgi2 (transaldolase/phosphoglucose isomerase) increased slightly (1.5–1.7 fold) (Figures 4A, 6). Activity of phosphoglucose isomerase (PGI) was measured and increased minimally (Figure 7H). This result was generally consistent with the gene expression studies. Expression of pgi2 did increase but peaked only at ∼1.7-fold while pgi1 increased less than 1.3-fold (Figures 4A, 6).
Discussion
When cells are shifted from one metabolic mode to another, significant remodeling of metabolic networks occurs. With the advent of global profiling capabilities, it is now possible to study metabolic response using a systems biology approach. In this paper, we used whole genome microarrays in conjunction with targeted enzyme assays and metabolite and flux measurements to assess the dynamic response that M. extorquens AM1 undergoes when switching from multi-carbon growth to single-carbon growth. Integrating datasets targeting multiple layers in the metabolic hierarchy has generated a much more complete picture of how the cells respond and adapt to changes in their environment than any of the datasets alone, and has provided insights into the mechanisms of this response.
To switch from succinate to methanol growth, energy metabolism shifts from NADH oxidation via NADH dehydrogenase to a cytochrome-based electron transport system that accepts electrons from methanol dehydrogenase during methanol oxidation [1]. Carbon metabolism shifts from the TCA cycle and its associated pathways to a methylene H4F- and CO2-based assimilatory scheme involving the serine cycle and the EMC pathway [1], [12], [17], while NADH production shifts from the TCA cycle to the methylene H4MPT dehydrogenase and formate dehydrogenase enzymes ([1], [10]; Figures 3,4A). To accomplish this shift, over 100 genes must be up-regulated in M extorquens AM1, producing over 50 enzymes and a number of regulatory proteins [10], and the cell must accommodate high flux through toxic metabolites, such as formaldehyde, glyoxylate, and glycine.
Carbon Starvation
Our time course experiments demonstrated that when the cells were transitioned from a state of succinate growth to methanol growth, they underwent a period of transient carbon starvation followed by periods of recovery and growth. During the starvation phase, the cells were thrown out of metabolic balance and growth ceased, and during the recovery period, all of the methylotrophic metabolic pathways rapidly changed to allow the cells to shift metabolic modes.
During the starvation period (0–30 min), gene expression patterns largely reflected those for a starvation control. In both the starvation control and the methanol switch experiment, expression of genes predicted to be involved in many core cellular processes such as cell structure synthesis, ribosome synthesis and protein production, flagellar synthesis, and iron uptake were down-regulated, conserving energy. Stress response genes were up-regulated along with several protease genes, which correlated with the increase in free amino acid pools detected in cell extracts. Expression of cytochrome c oxidase genes decreased by 30 min and then rose by 2 h mirroring expression of methanol dehydrogenase genes and cytochrome c (encoded by mxaG) used during methanol metabolism suggesting the possibility of a coordinated response. Immediately after the transition, ATP dropped, suggesting that energy metabolism was curtailed, while NADPH rose transiently, in keeping with a drop in biosynthetic consumption, and then dropped, consistent with a drop in production.
Transient imbalance
Cells growing on succinate maintain significant capacity to oxidize methanol to formaldehyde and then to formate. For the first h after the switch from succinate to methanol, the cells excreted about 1/3 of the total production of these compounds into the medium, mainly as formate. This result suggests that the capacity to consume formate was initially less than the formate production capacity, but by converting most of the formaldehyde to formate and excreting the excess, the cells could survive the transition without buildup of formaldehyde to toxic levels. It is likely that excretion serves as a buffer for flux capacity to these intermediates, allowing the cell to adjust formate consumption capacity according to need, rather than maintaining a high constitutive capacity. During this time period, the formate dehydrogenases rapidly induced and the flux of methanol to CO2 reached nearly full capacity by the time the culture started growing, at about 2 h. By maintaining significant methanol oxidation capacity in the absence of substrate, the cells can take advantage of transient methanol sources to obtain energy, without committing to the considerable effort of inducing the entire system of methylotrophic metabolism.
Post-transcriptional regulation
After the first 30 min, virtually all of the genes involved in methylotrophy began to induce, as expected. However, a number of examples were identified in which enzyme activities did not reflect transcript changes, suggesting post-transcriptional regulation. These included: 1) methanol dehydrogenase, for which enzyme activity increased rapidly during the time that transcripts dropped, suggesting a mechanism of enzyme activation; 2) methylene H4F dehydrogenase, for which transcripts increased 14-fold but enzyme activity stayed constant for the first hour, suggesting a mechanism of post-transcriptional inhibition, either of translation or of enzyme activity, and 3) TCA cycle enzymes (Icd, Sdh, and Mdh), for which the activities stayed relatively constant while transcripts dropped, even after cell division, suggesting either a mechanism of enzyme stabilization or activation.
Flux redistribution
Surprisingly, during the time period from 30 min to 2 h while the flux capacity from methanol to CO2 was increasing, net flux through methylotrophy assimilatory pathways to biomass did not occur, even though both transcripts and enzyme activities increased significantly. Since intermediates of the EMC pathway did not accumulate, the assimilatory block must have occurred before that pathway. A candidate step for this block was identified, the conversion of methenyl H4F to methylene H4F by methylene H4F dehydrogenase, since as noted above, this enzyme did not increase in activity during this time period. Once this activity increased, flux to biomass occurred.
These results suggest a metabolic strategy in which the assimilatory pathways are primed to receive assimilatory carbon, but carbon flux occurs only after they are poised and ready, and only after the dissimilatory flux has increased substantially. Such a strategy might decrease the likelihood of accumulation of intermediates produced in the biomass cycles that could inhibit growth, such as glyoxylate and glycine [8]–[9], ensuring a smooth transition between these two metabolic modes. Our measurements showed that glyoxylate and glycine did not show major changes during this transition.
Another hypothesis generated from these data was that M. extorquens AM1 may fine-tune the levels of these toxic metabolites through the Mcd step (mesaconyl-CoA hydratase) in the EMC pathway, which produces/regenerates the glyoxylate required to drive the serine cycle. The expression pattern of mcd was notable, since it decreased significantly initially, and only slowly returned to a level slightly above the succinate level. Once intermediates of the EMC pathway began to accumulate, those generated before the Mcd step accumulated to a much higher level than those generated after the Mcd step, consistent with a flux bottleneck at this step. Because glyoxylate is toxic at high levels, it would be advantageous for the cell to regulate the production and consumption of this metabolite. Since glyoxylate is the precursor to glycine during methylotrophic growth, by controlling glyoxylate levels, both of these potentially toxic metabolites are managed. This potential flux bottleneck at the Mcd step would allow the cells to control the balance between need and toxicity for glyoxylate and is an interesting candidate for further study.
Another possible metabolic strategy for carbon partitioning during this transition suggested by these results involved 3-hydroxybutyryl-CoA. This metabolite is an intermediate for both PHB synthesis (carbon storage) and the EMC pathway (carbon assimilation), and is thus a key branchpoint. Succinate-grown cells contain higher levels of PHB than methanol-grown cells [13], and so during the transition, a shift must occur that redirects flux from PHB synthesis to the EMC pathway. Our results showed that 3-hydroxybutyryl-CoA increased almost 6-fold in the first 30 min, before significant increases were observed for the EMC pathway intermediates, at the time when PHB synthesis gene expression was beginning to decrease. In other organisms, the partitioning of acetyl-CoA flux between PHB synthesis and assimilation is controlled by levels of NADPH or the ratio of NADPH/NADP [37]–[38]. The accumulation of 3-hydroxybutyryl-CoA correlated with the increase in NADPH levels and the NADPH/NADP ratio seen during the first 30 min after methanol addition, suggesting that this mechanism of control may also be present in M. extorquens AM1. Later, when flux began to increase through the EMC pathway, both 3-hydroxybutyryl-CoA and NADPH levels dropped. By decreasing flux into the PHB cycle first, then increasing the enzymes of the EMC pathway, the cells could effectively shunt 3-hydroxybutyryl-CoA into primary assimilation and away from synthesis of the storage product, PHB.
Modular response
The results presented here showed that in general, each pathway responded as a separate module, with gene expression changes and enzymatic activities responding as a group. One notable exception was the EMC pathway, in which the majority of the genes increased during the transition yet the patterns of this increase differed (Figure 6).
Another example was the H4MPT-dependent C1 transfer pathway in which expression of mtdB and fae stood out as different from the other genes involved in the oxidation of formaldehyde to formate. Fae is present in the cell at very high levels [18]–[19] and likely is involved in keeping intracellular levels of free formaldehyde low. The rapid increase of fae transcription may reflect the need to respond to a sudden increase in flux from methanol to formaldehyde. Likewise, MtdB is the first NAD(P)H-generating enzyme in the oxidative pathway, and it responded more similarly to the other NADH-generating step, the formate oxidation pathway. It may be advantageous to the cell to control reduced pyridine nucleotide-generating capacity separately from the other oxidative steps, and act to increase that capacity immediately upon downshift.
Metabolic set points
These datasets also revealed another major feature, which is the prevalence of metabolic set points. During the transition from succinate to methanol growth, central metabolic pathways change significantly, with correspondingly large changes in flux through these pathways, on the order of 10-fold [20]. Such changes might be expected to reflect transient swings in metabolite concentrations as the enzyme requirements and activities change. However, the concentration of those metabolites that are involved in both succinate and methanol growth such as fumarate, malate, α-ketoglutarate, citrate, pyruvate and glycerate stayed remarkably constant, with less than 2-fold changes during the transition. A similar phenomenon has been reported for E. coli wild-type and mutant strains in steady-state cultures under different growth conditions, and these authors suggested that metabolic setpoints may be a mechanism to maintain metabolic robustness in a changing environment [24]. Metabolites that were required for growth on both succinate and methanol but did not appear as metabolic setpoints in this study include succinate and serine. Succinate decreased from 12.2 mM to 3.7 mM within 10 min of stopping medium flow, consistent with the termination of succinate as the carbon source. Serine responded similarly to other amino acids which correlated with an increase in protease gene expression (Table 1, Figure 2).
Summary
In summary, the multi-level datasets we have generated in this study coupled to our pathway-level analysis have resulted in new insights into the metabolic strategies carried out to effect this shift between two dramatically different modes of growth, one energy-limited (succinate), and the other reducing-power limited (methanol). Analysis of the dynamic response of modular sections of the central metabolic network is an important approach to determine how cells respond and adapt to carbon substrate perturbation. Our work showed that the cells use a “down-stream priming” strategy, inducing biomass cycles before formate production to carry out this transition in a way that does not accumulate toxic intermediates to inhibitory levels. In addition, this work has uncovered key candidates for regulatory control points, as a further step towards understanding metabolic adaptation and the cellular strategies employed to maintain metabolic setpoints.
Materials and Methods
Materials
All chemicals, enzymes and chemical standards were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. Analytical- reagent grade potassium dihydrogen phosphate and sodium dihydrogen phosphate were obtained from Fisher Scientific (Hampton, NH). Absolute ethanol and HPLC-grade methanol (Sigma, St. Louis, MO, USA) were used for extraction procedures and preparation of buffer for HPLC analysis. Nanopure (Barnstead Life Sciences model, Thermo Fisher Scientific, Waltham, MA) water was used in the preparation of media, buffer, standard and sample solutions. To prepare formaldehyde, 0.15 gm of paraformaldehyde was resuspended in 10 ml nanopure water in a glass vial, which was then stoppered, crimp sealed and autoclaved for 5 min.
Chemostat cultivation
For a starting point (time = 0 in), the carbon-shift experiment utilized cells from a steady-state chemostat culture of M. extorquens AM1 growing on limiting succinate. M. extorquens AM1 was cultivated in a minimal medium [14] containing 3.7 mM succinate as a growth-limiting nutrient in a 2.2-liter bench-top BioFlo 110 Modular Fermentor (New Brunswick Scientific) with a 1 liter working volume, resulting in an OD600 of ∼0.63. Dilution rate was maintained at 0.163 h−1. Other parameters of the chemostat operation were the same as described previously [13]. After the culture reached steady-state, the cells were manipulated in two ways: for enzyme assays, flux measurements and nucleotide determinations, the flows were stopped and methanol was added to the succinate-limited steady-state culture to a final concentration of 50 mM. For RNA isolation, steady-state cells were transferred to flasks before 50 mM methanol addition and incubated at 28°C. For each approach, cells were harvested at different time points as described in the text.
Transcriptomics
Expression studies were carried out using 60-mer oligonucleotide custom designed Agilent arrays (Santa Clara, CA) as previously described [14]. RNA was extracted immediately and digested with DNase I as described previously [14]. cDNA production and labeling, hybridization and array scanning were carried out by MOgene (St. Louis, MO), using the DNase I-digested RNA. Data extraction and initial processing were as previously described [14]. For the first biological replicate, cells were transferred to a flask, methanol was added (50 mM) and time points were harvested at 0, 10, 30, 60, 120, 240 and 360 min post methanol addition, and each time point included a technical duplicate (dye-swap). Most genes in this study peaked in their induction or repression by 2 h. To confirm the array data expression trends and peak times, a second biological replicate (second chemostat culture) was repeated for the 10, 30, 60 and 120 min time points, each also with a technical duplicate (dye-swap). For carbon starvation arrays, cells were transferred to a flask with no additional carbon and incubated for 30 min prior to harvest. For convenience, time point RNA samples were compared against RNA prepared prior to the transition, directly harvested from the chemostat (time zero, not transferred to flask). LogRatios were adjusted using a time zero flask transfer comparison to remove affects from flask transfer thus setting the time zero expression ratio at 1. Both raw and adjusted data are included in Tables S1, S2. Intensity and LogRatio data were used to determine peak intensities and fold changes to generate the metabolic diagrams. A p-value cutoff of 0.001 (Log p-value ≤−3) was used for validity of expression fold changes. Raw and processed gene expression data are available at the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE22031.
Measurement of methanol in culture supernatant
Methanol concentration in the growth medium was measured according to Mangos and Haas [39] with a minimum of 3 replicates. The method used alcohol oxidase (EC 1.1.3.13), peroxidase (EC 1.11.1.7) and 2,2′-azinobis-(3-ethylbenzthiazoline-6- sulfonic acid) (ABTS) (Calbiochem, EMD Biosciences, Gibbstown, NJ) to determine methanol concentrations.
Measurement of formaldehyde and formate in culture supernatant
Formate measurements
Formate was measured as previously described for 3 biological replicates [27].
Formaldehyde measurements
Formaldehyde was measured for 3 biological replicates using the Nash method [40] in which formaldehyde concentration is proportional to the absorbance at 410 nm.
Enzyme assays
Cells for enzyme activities (50 ml) were harvested by centrifugation at 4500× g using a Sorvall RC-5B centrifuge at 4°C for 10 min, washed in 100 mM potassium phosphate buffer, pH 7.5, pelletted, overlaid with 1 ml of buffer and frozen at −80°C. Cell pellets were thawed and resuspended in 1–5 ml of 100 mM potassium phosphate buffer, pH 7.5 and passed through a French press. Cell extracts were centrifuged at 14,000 rpm for 1–10 min to remove cell debris, depending on the potential oxygen sensitivity of the enzyme to be assayed. A minimum of 2 biological replicates were assayed for the following enzymes: methanol dehydrogenase (MeDH), formate dehydrogenase (Fdh), methylene-H4F dehydrogenase (MtdA), β-ketothiolase (βKT), crotonyl-CoA reductase (Ccr), hydroxypyruvate reductase (Hpr), serine-glyoxylate aminotransferase (Sga), malate dehydrogenase (Mdh), aconitase (Acn), succinate dehydrogenase (Sdh), isocitrate dehydrogenase (Icd) and phosphoglucose isomerase (Pgi). βKT, Hpr, Sga and Mdh were measured as previously described [13]. The remaining enzyme activities were measured in a final volume of 200 µL in a Spectramax 190 plate reader (Molecular Devices, Sunnyvale, CA) at room temperature and contained ∼25–100 µg protein. Absorbance data were converted to a 1 cm pathlength by dividing the slope by the conversion factor, 0.541 and protein concentrations were determined spectrophotometrically [41]. With the exception of the 200 µL volume, assays for Fdh [26], MtdA [42], Acn [43], were as described previously. MeDH activity was measured as described [1] except the assay contained 0.4 mM KCN. Ccr activity was measured as described [44] except that 2 mM NADPH was used. Sdh activity was measured as described previously [45] with modifications as described [46]. Icd activity was measured by following the production of NADPH at 340 nm using an extinction coefficient of 6.2 mM−1 cm−1. The reaction contained 100 mM potassium phosphate buffer, pH 7.5, 1 mM MgCl2, 2 mM NADPH, cell extract (∼25–50 µg protein) and was initiated by addition of 20 mM isocitrate. Pgi activity was measured by modification of a coupled assay [47] and followed the production of NADPH at 340 nM in a quartz microtiter plate using an extinction coefficient of 6.2 mM−1 cm−1. The reaction contained 100 mM potassium phosphate buffer, pH 7.5, 1 mM MgCl2, 2 mM NADPH, glucose-6-phosphate dehydrogenase (5 U/ml), cell extract (∼25–50 µg protein) and was initiated by addition of 4 mM fructose-6-phosphate.
Nucleotide measurements by LC-MS/MS
5 ml of the cell culture was carefully and rapidly pipetted into the center of 25 ml 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-buffered (70 mM, pH 6.8) aqueous 60% methanol (v/v) solution (−40°C). The quenched biomass was precipitated in a refrigerated centrifuge (6 min, 10,000 rpm, −20°C; Dupont Sorvall RC5B, Waltham, MA, USA). The supernatant was removed and the cell pellets were resuspended in 5 ml of the same methanol solution and centrifuged for 6 min at 10,000 rpm. For ATP, ADP, AMP, NAD and NADP extraction, 1 ml of boiling HEPES buffered ethanol solution (75% v/v ethanol/water, pH 5.2) was added to a given cell pellet and incubated at 100°C for 3 min [13]. The extracted cell suspension was cooled on ice for 3 min and the cell debris was removed by centrifugation at 5000 rpm for 5 min. The cell-free metabolite extract was again centrifuged at 14,000 rpm for 8 min. The supernatant was transferred into a 2 ml glass vial and dried in a vacuum centrifuge (CentriVap Concentrator System, Labconco, MO, USA). The dried sample, prior to analysis using LC-MS/MS, was re-dissolved in 100 µL purified water for analysis. NADH and NADPH were extracted by a hot alkaline extraction method [13]. 0.8 ml of 0.8 N KOH at 85°C with 1% (v/v) Triton X-100 was added to a given cell pellet, and the mixture was incubated for 3 min at 85°C. After cooling on ice for 3 min, the mixture was centrifuged at 5,000 rpm at 4°C for 5 min. The supernatant was partially neutralized with HClO4 to pH 10 and centrifuged at 14,000 rpm for 8 min. The supernatant was analyzed by LC-MS/MS immediately. LC-MS/MS experiments were carried out on a Waters (Milford, MA, USA) LC-MS system consisting of a 1525 µ binary HPLC pump with a 2777C autosampler coupled to a Quattro Micro API triple-quadrupole mass spectrometer (Micromass, Manchester, UK) as described previously [48]. A minimum of two biological replicates with two technical duplicates were measured for each time point.
CO2 assimilation flux
Carbon fluxes for steady-state succinate-limited chemostat cultures were estimated using the dilution rate at steady state (0.163 h−1), the dry weight of cells per ml of culture at an OD600 of 1.0 (0.278 mg; [21]), and the percentage of total biomass, by weight, that is carbon (47%; [49]). Production of CO2 was measured for 2 biological replicates using 14C-labeled methanol essentially as described previously [50]; net CO2 production was added to CO2 assimilation flux estimated at 50% of the carbon in biomass as indicated by the fact that ∼50% of carbon labeling can be eliminated in the presence of high unlabeled CO2 as compared with a CO2-free environment ([51]; G.J. Crowther, unpublished data). Carbon assimilation into biomass via methylene H4F was calculated from changes in OD600 using the conversion factors noted above (0.278 mg dry weight per ml of culture at an OD600 of 1.0; 47% of biomass is carbon) and also the estimate that 50% of the carbon in biomass comes from assimilated CO2.
CoA and CoA ester measurements by HPLC
CoA and short-chain CoA esters, including acetyl-CoA, malonyl-CoA, methylmalonyl-CoA, 3-hydroxybutyryl-CoA and propionyl-CoA, were measured by reversed-phase high-performance liquid chromatography.
Liquid nitrogen was used to quench metabolism. Extraction was performed by adding 1.5 ml boiling 20% ethanol in 5 mM sodium acetate/acetic acid buffer (pH 3) containing 2 mM DTT to each sample and incubating at 100°C for 3–5 min. Extracted cell suspension was cooled on ice for 3 min and the cell debris was removed by centrifugation at 5,000 rpm for 5 min. The cell-free metabolite extract was dried in a vacuum centrifuge (CentriVap Concentrator system, 79820-00, Labconco, MO, USA) to complete dryness. Deoxy-guanosine was added as an internal standard to correct for volume changes during sample preparation and injection. A Nova-Pac C18 reverse-phase column (3.9×150 mm, 60 Å, 4 µm) from Waters (Milford, MA) was used to separate CoA and CoA esters. A UV detector was used, with the wavelength set to monitor absorbance at 260 nm. Buffer A was composed of 0.1 M sodium dihydrogen phosphate and 0.075 M sodium acetate (pH 4.5); buffer B consisted of 40% buffer A and 60% methanol. The flow rate was 0.8 ml/min and the following HPLC gradient time program was used: 4 min at 100% of buffer A; linearly increase to reach 30% of buffer B at 34 min; increase to 100% buffer B in 7 min; keep at 100% buffer B for 3 min; re-equilibrate the column with buffer A for 6 min. Hydroxylamine reacts with CoA esters to form CoA and the corresponding free acids, and this reaction was used to confirm CoA compounds. Neutral hydroxylamine solution was prepared by adjusting NH2OH-HCl solution with 10 N NaOH to pH 7. To three volumes of an analyte solution, one volume of 2 M neutral NH2OH solution was added. The mixture was incubated at 30°C for approximately 1 hour to achieve complete conversion of CoA esters to CoA.
These CoA compounds were measured from two biological replicate samples taken at 0 min (before methanol addition), 10 min, 30 min, 1 h, and 3 h after methanol addition. Standard curves were generated for quantitation of the CoA compounds by adding known amounts to cell extracts (data not shown). Recoveries of CoA and CoA esters under the extraction condition were determined by spiking known amounts of standard CoA compounds to a cell culture sample before quenching and extraction. Recoveries ranged from 70–100%, depending on the compound.
GC×GC-TOFMS analysis of cell extracts
Two-dimensional gas chromatography coupled with time-of-flight mass spectrometry was used to analyze cell extract samples for the measurement of amino acids, organic acids and carbohydrates. Samples were taken at 0 min (before methanol addition), 10 min, 20 min, 30 min, 1 h, 2 h and 3 h after methanol addition, and were analyzed by GC×GC-TOFMS as described [1]. Numbers are averages of nine replicates (three extractions with 3 biological replicate injections for each extraction).
Fisher Ratio analysis of cell extracts
Fisher ratio analysis was used to assess metabolite differences between two conditions as previously described [15]. Numbers are averages of nine replicates (three extractions with 3 biological replicate injections for each extraction).
Supporting Information
Pathway schematics depicting changes that occurred in measured metabolites, gene expression, and enzymatic activities for central metabolism after cells were transitioned from succinate to methanol growth. A boxed gene/protein name indicates the activity of this enzyme was measured. Red lettering for the protein designation indicates an increase in activity; green, decrease; black, no change. Metabolites appearing more than once are connected by gray lines. Diagrams are shown for the following time periods: (A) pre-methanol addition, time = 0 min; (B) initial response, time = 10–30 min post methanol addition; (C) just prior to/at the start of cell growth, time = 1–2 h post methanol addition; and (D) exponential cell growth, time = 3–6 h post methanol addition with (E) serving as a legend. Gene expression intensities are represented by arrow thickness while gene expression fold changes are depicted by arrowhead number. Abbreviations and reaction descriptions are included in Supplementary Table II along with gene expression intensities, LogRatios, fold changes and p-values. Mesaconyl-CoA, ethylmalonyl-CoA, methylsuccinyl-CoA were measured as free acids. **Color at T = 0 represents the concentration before methanol was added for all metabolites except for methanol which was calculated as 50 mM for the initial T = 0 value.
(0.21 MB PDF)
Gene expression data for cellular processes that significantly changed during the transition from succinate to methanol growth.
(0.96 MB XLS)
Gene expression data for central metabolic pathways during the transition from succinate to methanol growth.
(0.40 MB XLS)
Acknowledgments
We thank Mila Chistoserdova, Marina Kalyuzhnaya and Norma Cecilia Martinez-Gomez for critical reading of this manuscript, Bo Hu for his aid in producing and maintaining chemostat grown cells and Julia Vorholt for initial enzyme characterization.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by grants from National Institute of General Medical Sciences (GM58933 to MEL) and the Ruth Kirschstein National Research Service Award, 08FGM78835A to ES, and 5F32GM070297 to GJC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Anthony C. New York, NY: Academic Press; 1982. The biochemistry of methylotrophs. [Google Scholar]
- 2.Green P. The genus Methylobacterium. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editors. The Prokaryotes, 2 edn. Berlin: Springer; 1991. pp. 2342–2349. [Google Scholar]
- 3.Lidstrom ME. Aerobic methylotrophic prokaryotes. In: Stackebrandt, editor. The Prokaryotes, 3 edn. New York, NY: Springer-Verlag; 2001. pp. 223–244. [Google Scholar]
- 4.Chistoserdova L, Lapidus A, Han C, Goodwin L, Saunders L, et al. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy. J Bacteriol. 2007;189:4020–7. doi: 10.1128/JB.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, et al. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009;2:107–15. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Corpe WA, Rheem S. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol. 1989;62:243–50. [Google Scholar]
- 7.Hüve K, Christ MM, Kleist E, Uerlings R, Niinemets U, et al. Simultaneous growth and emission measurements demonstrate an interactive control of methanol release by leaf expansion and stomata. J Exp Bot. 2007;58:1783–93. doi: 10.1093/jxb/erm038. [DOI] [PubMed] [Google Scholar]
- 8.Salem, AR, Hacking AJ, Quayle JR. Lack of malyl-CoA lyase in a mutant of Pseudomonas AM1. J Gen Microbiol. 1976;81:525–7. doi: 10.1099/00221287-81-2-525. [DOI] [PubMed] [Google Scholar]
- 9.Harder W, Quayle JR. The biosynthesis of serine and glycine in Pseudomonas AM1 with special reference to growth on carbon sources other than C1 compounds. Biochem J. 1971;121:753–62. doi: 10.1042/bj1210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol. 2003;185:2980–7. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Dien SJ, Okubo Y, Hough MT, Korotkova N, Taitano T, et al. Reconstruction of C3 and C4 metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology. 2003;149:601–9. doi: 10.1099/mic.0.25955-0. [DOI] [PubMed] [Google Scholar]
- 12.Van Dien SJ, Lidstrom ME. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol Bioeng. 2002;78:296–312. doi: 10.1002/bit.10200. [DOI] [PubMed] [Google Scholar]
- 13.Guo X, Lidstrom ME. Physiological analysis of M. extorquens AM1 grown in continuous and batch cultures. Arch Microbiol. 2006;186:139–49. doi: 10.1007/s00203-006-0131-7. [DOI] [PubMed] [Google Scholar]
- 14.Okubo Y, Skovran E, Guo X, Sivam D, Lidstrom ME. Implementation of microarrays for Methylobacterium extorquens AM1. Omics. 2007;11:325–40. doi: 10.1089/omi.2007.0027. [DOI] [PubMed] [Google Scholar]
- 15.Guo X, Lidstrom ME. Metabolite profiling analysis of Methylobacterium extorquens AM1 by comprehensive two-dimensional gas chromatography coupled with time-of flight mass spectrometry. Biotechnol Bioeng. 2008;99:929–40. doi: 10.1002/bit.21652. [DOI] [PubMed] [Google Scholar]
- 16.Kiefer P, Portais JC, Vorholt JA. Quantitative metabolome analysis using liquid chromatography-high-resolution mass spectrometry. Anal Biochem. 2008;382:94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Peyraud R, Kiefer P, Christen P, Massou S, Portais JC, et al. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc Natl Acad Sci U S A. 2009;106:4846–51. doi: 10.1073/pnas.0810932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laukel M, Rossignol M, Borderies G, Völker U, Vorholt JA. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and nonmethylotrophic conditions. Proteomics. 2004;4:1247–64. doi: 10.1002/pmic.200300713. [DOI] [PubMed] [Google Scholar]
- 19.Bosch G, Skovran E, Xia Q, Wang T, Taub F, et al. Comprehensive proteomics of Methylobacterium extorquens AM1 metabolism under single carbon and nonmethylotrophic conditions. Proteomics. 2008;8:3494–505. doi: 10.1002/pmic.200800152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx CJ, Van Dien SJ, Lidstrom ME. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 2005;3:e16. doi: 10.1371/journal.pbio.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowther GJ, Kosály G, Lidstrom ME. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J Bacteriol. 2008;190:5057–62. doi: 10.1128/JB.00228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krömer JO, Sorgenfrei O, Klopprogge K, Heinzle E, Wittmann C. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol. 2004;186:1769–84. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, et al. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36:486–91. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 24.Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–7. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 25.Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J boil chem. 2004;279:6613–9. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 26.Chistoserdova L, Laukel M, Portais JC, Vorholt JA, Lidstrom ME. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J Bacteriol. 2004;186:22–8. doi: 10.1128/JB.186.1.22-28.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais JC, et al. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J Bacteriol. 2007;189:9076–81. doi: 10.1128/JB.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JA. Doctoral thesis. Seattle, WA: University of Washington; March 2009. Formaldehyde stress response in Methylobacterium extorquens AM1. [Google Scholar]
- 29.Vorholt JA, Marx CJ, Lidstrom ME, Thauer RK. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol. 2000;182:6645–50. doi: 10.1128/jb.182.23.6645-6650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx CJ, Lidstrom ME. Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology. 2004;150:9–19. doi: 10.1099/mic.0.26587-0. [DOI] [PubMed] [Google Scholar]
- 31.Harder W, Quayle JR. Aspects of glycine and serine biosynthesis during growth of Pseudomonas putida I. Synthesis of enzymes by the wild type. Biochem J. 1971;121:763–9. doi: 10.1042/bj1210763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hepinstall J, Quayle JR. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970;117:563–72. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNerney T, O'Connor ML. Regulation of enzymes associated with C-1 metabolism in three facultative methylotrophs. Appl Environ Microbiol. 1980;40:370–5. doi: 10.1128/aem.40.2.370-375.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korotkova N, Chistoserdova L, Kuska B, Lidstrom ME. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J Bacterial. 2002;184:1750–8. doi: 10.1128/JB.184.6.1750-1758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korotkova N, Lidstrom ME. A connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J Bacteriol. 2001;183:1038–46. doi: 10.1128/JB.183.3.1038-1046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korotkova N, Chistoserdova L, Lidstrom ME. Poly-β-hydroxybutyrate biosynthesis in the facultative methylotroph Methylobacterium extorquens AM1: identification and mutation of gap11, gap20, and phaR. J Bacteriol. 2002;184:6174–81. doi: 10.1128/JB.184.22.6174-6181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee IY, Kim MK, Park YH, Lee SY. Regulatory effects of cellular nicotinamide nucleotides and enzyme activities on poly(3-hydroxybutyrate) synthesis in recombinant Escherichia coli. Biotechnol Bioeng. 1996;52:707–12. doi: 10.1002/(SICI)1097-0290(19961220)52:6<707::AID-BIT8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 38.Lee YH, Kim TW, Park JS, Huh TL. Effect of the supplementation of metabolites on cell growth and poly-β-hydroxybutyrate biosynthesis of Alcaligenes eutrophus. J Microbiol Biotechnol. 1996;6:120–7. [Google Scholar]
- 39.Mangos TJ, Haas MJ. A spectrophotometric assay for the enzymatic demethoxylation of pectins and the determination of pectinesterase activity. Anal Biochem. 1997;244:357–66. doi: 10.1006/abio.1996.9908. [DOI] [PubMed] [Google Scholar]
- 40.Nash T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J. 1953;55:416–21. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50–69. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 42.Vorholt JA, Chistoserdova L, Lidstrom ME, Thauer The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol. 1998;180:5351–6. doi: 10.1128/jb.180.20.5351-5356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruer MJ, Guest JR. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–41. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 44.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, et al. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: The ethylmalonyl-CoA pathway, Proc Natl Acad Sci U S A. 2007;104:10631–6. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spencer ME, Guest JR. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973;114:563–70. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skovran E, Downs DM. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium, implications for thiamine synthesis. J Bacteriol. 2002;182:3896–903. doi: 10.1128/jb.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergmeyer HU, Gawehn K, Grassl M. New York, NY: Academic Press; 1974. Methods of enzymatic analysis, 2 edn. pp. 501–3. [Google Scholar]
- 48.Yang S, Sadilek M, Synovec RE, Lidstrom ME. Liquid chromatography-tandem quadrupole mass spectrometry and two-dimensional gas chromatography-time-of-flight mass spectrometry measurement of targeted metabolites of Methylobacterium extorquens AM1 grown on two different carbon sources. J Chromatogr A. 2009;1216:3280–9. doi: 10.1016/j.chroma.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldberg I, Rock JS, Ben-Bassat A, Mateles RI. Bacterial yields on methanol, methylamine, formaldehyde, and formate. Biotechnol Bioeng. 1976;18:1657–68. doi: 10.1002/bit.260181202. [DOI] [PubMed] [Google Scholar]
- 50.Marx CJ, Chistoserdova L, Lidstrom ME. Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J Bacteriol. 2003;185:7160–8. doi: 10.1128/JB.185.23.7160-7168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Large PJ, Quayle JR. Microbial growth on C(1) compounds. 5. Enzyme activities in extracts of Pseudomonas AM1. Biochem J. 1963;87:386–96. doi: 10.1042/bj0870386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pathway schematics depicting changes that occurred in measured metabolites, gene expression, and enzymatic activities for central metabolism after cells were transitioned from succinate to methanol growth. A boxed gene/protein name indicates the activity of this enzyme was measured. Red lettering for the protein designation indicates an increase in activity; green, decrease; black, no change. Metabolites appearing more than once are connected by gray lines. Diagrams are shown for the following time periods: (A) pre-methanol addition, time = 0 min; (B) initial response, time = 10–30 min post methanol addition; (C) just prior to/at the start of cell growth, time = 1–2 h post methanol addition; and (D) exponential cell growth, time = 3–6 h post methanol addition with (E) serving as a legend. Gene expression intensities are represented by arrow thickness while gene expression fold changes are depicted by arrowhead number. Abbreviations and reaction descriptions are included in Supplementary Table II along with gene expression intensities, LogRatios, fold changes and p-values. Mesaconyl-CoA, ethylmalonyl-CoA, methylsuccinyl-CoA were measured as free acids. **Color at T = 0 represents the concentration before methanol was added for all metabolites except for methanol which was calculated as 50 mM for the initial T = 0 value.
(0.21 MB PDF)
Gene expression data for cellular processes that significantly changed during the transition from succinate to methanol growth.
(0.96 MB XLS)
Gene expression data for central metabolic pathways during the transition from succinate to methanol growth.
(0.40 MB XLS)