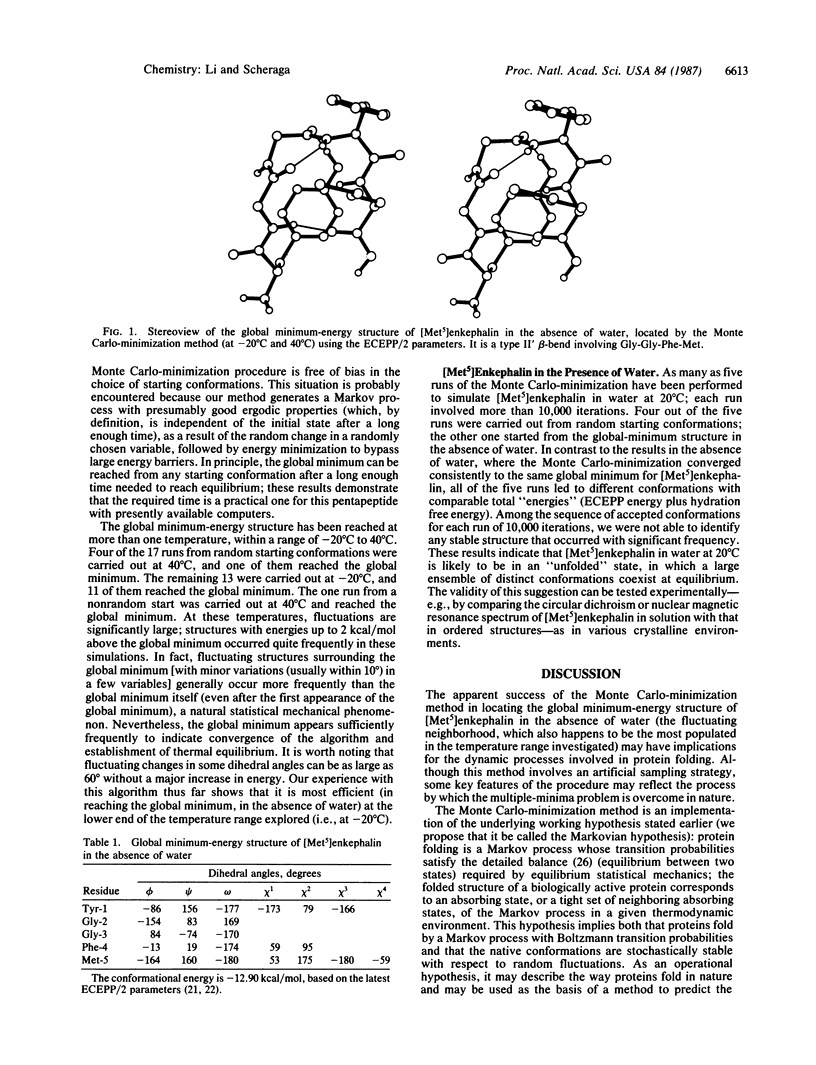
Abstract
A Monte Carlo-minimization method has been developed to overcome the multiple-minima problem. The Metropolis Monte Carlo sampling, assisted by energy minimization, surmounts intervening barriers in moving through successive discrete local minima in the multidimensional energy surface. The method has located the lowest-energy minimum thus far reported for the brain pentapeptide [Met5]enkephalin in the absence of water. Presumably it is the global minimum-energy structure. This supports the concept that protein folding may be a Markov process. In the presence of water, the molecules appear to exist as an ensemble of different conformations.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Protein states and proteinquakes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elber R., Karplus M. Multiple conformational states of proteins: a molecular dynamics analysis of myoglobin. Science. 1987 Jan 16;235(4786):318–321. doi: 10.1126/science.3798113. [DOI] [PubMed] [Google Scholar]
- Go N. Theoretical studies of protein folding. Annu Rev Biophys Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- Gurd F. R., Rothgeb T. M. Motions in proteins. Adv Protein Chem. 1979;33:73–165. doi: 10.1016/s0065-3233(08)60459-3. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. Optimization by simulated annealing. Science. 1983 May 13;220(4598):671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975 Feb 27;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- Paine G. H., Scheraga H. A. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. I. Backbone structure of enkephalin. Biopolymers. 1985 Aug;24(8):1391–1436. doi: 10.1002/bip.360240802. [DOI] [PubMed] [Google Scholar]
- Paine G. H., Scheraga H. A. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. II. Average backbone structure of enkephalin. Biopolymers. 1986 Aug;25(8):1547–1563. doi: 10.1002/bip.360250812. [DOI] [PubMed] [Google Scholar]
- Paine G. H., Scheraga H. A. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. III. Probable and average conformations of enkephalin. Biopolymers. 1987 Jul;26(7):1125–1162. doi: 10.1002/bip.360260711. [DOI] [PubMed] [Google Scholar]
- Piela L., Scheraga H. A. On the multiple-minima problem in the conformational analysis of polypeptides. I. Backbone degrees of freedom for a perturbed alpha-helix. Biopolymers. 1987;26 (Suppl):S33–S58. doi: 10.1002/bip.360260008. [DOI] [PubMed] [Google Scholar]
- Purisima E. O., Scheraga H. A. An approach to the multiple-minima problem by relaxing dimensionality. Proc Natl Acad Sci U S A. 1986 May;83(9):2782–2786. doi: 10.1073/pnas.83.9.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purisima E. O., Scheraga H. A. An approach to the multiple-minima problem in protein folding by relaxing dimensionality. Tests on enkephalin. J Mol Biol. 1987 Aug 5;196(3):697–709. doi: 10.1016/0022-2836(87)90041-6. [DOI] [PubMed] [Google Scholar]
- Robson B., Osguthorpe D. J. Refined models for computer simulation of protein folding. Applications to the study of conserved secondary structure and flexible hinge points during the folding of pancreatic trypsin inhibitor. J Mol Biol. 1979 Jul 25;132(1):19–51. doi: 10.1016/0022-2836(79)90494-7. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A., Paine G. H. Conformational energy calculations on polypeptides and proteins: use of a statistical mechanical procedure for evaluating structure and properties. Ann N Y Acad Sci. 1986;482:60–68. doi: 10.1111/j.1749-6632.1986.tb20937.x. [DOI] [PubMed] [Google Scholar]
- Vásquez M., Scheraga H. A. Use of buildup and energy-minimization procedures to compute low-energy structures of the backbone of enkephalin. Biopolymers. 1985 Aug;24(8):1437–1447. doi: 10.1002/bip.360240803. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B., Ristow S. Acquisition of three-dimensional structure of proteins. Annu Rev Biochem. 1973;42:135–158. doi: 10.1146/annurev.bi.42.070173.001031. [DOI] [PubMed] [Google Scholar]



