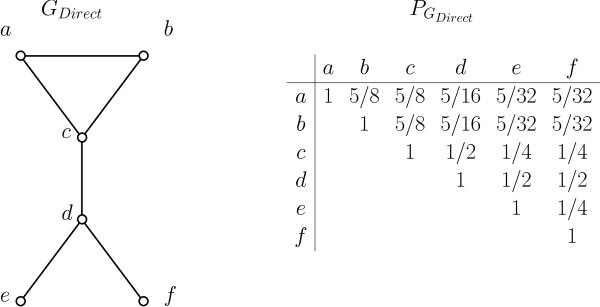Abstract
Background
Affinity purification followed by mass spectrometry identification (AP-MS) is an increasingly popular approach to observe protein-protein interactions (PPI) in vivo. One drawback of AP-MS, however, is that it is prone to detecting indirect interactions mixed with direct physical interactions. Therefore, the ability to distinguish direct interactions from indirect ones is of much interest.
Results
We first propose a simple probabilistic model for the interactions captured by AP-MS experiments, under which the problem of separating direct interactions from indirect ones is formulated. Then, given idealized quantitative AP-MS data, we study the problem of identifying the most likely set of direct interactions that produced the observed data. We address this challenging graph theoretical problem by first characterizing signatures that can identify weakly connected nodes as well as dense regions of the network. The rest of the direct PPI network is then inferred using a genetic algorithm.
Our algorithm shows good performance on both simulated and biological networks with very high sensitivity and specificity. Then the algorithm is used to predict direct interactions from a set of AP-MS PPI data from yeast, and its performance is measured against a high-quality interaction dataset.
Conclusions
As the sensitivity of AP-MS pipeline improves, the fraction of indirect interactions detected will also increase, thereby making the ability to distinguish them even more desirable. Despite the simplicity of our model for indirect interactions, our method provides a good performance on the test networks.
Background
Understanding the organization of protein-protein interactions (PPIs) as a complex network is one of the main pursuits in proteomics today. With the help of high-throughput experimental techniques, a large amount of PPI data has recently become available, providing us with a rough picture of how proteins interact in biological systems. However, the interaction data from these high-throughput experiments suffer from low resolution as compared to data from low-throughput technologies such as protein co-crystallization, and to make matters worse, they are prone to problems including relatively high error rates and protocol-specific biases. Therefore, inferring the direct, physical PPI network from high-throughput data remains a challenge in systems biology.
The leading technologies for identifying PPIs are Yeast 2-Hybrid (Y2H) [1,2] and Affinity Purification followed by Mass Spectrometry (AP-MS) [3-6]. Due to the ability to perform in vivo at biologically reasonable expression levels, as well as the ability to detect protein complexes with fewer false-positives [6], AP-MS approaches have become increasingly popular, although their throughput is lower than Y2 H approaches. In an AP-MS experiment, a protein of interest (the bait) is tagged and expressed in vivo. The bait is then immuno-precipitated (IP), together with all of its interacting partners (the preys), and finally, preys are identified using mass spectrometry. For a more detailed overview of the technique, see [6,7]. Like Y2 H and other high-throughput experimental methods, however, AP-MS suffers from experimental noise. A number of approaches have been proposed to separate true interactions from false-positives. These approaches mostly focus on reducing false-positives due to protein misidentification from MS data [8-10], on detecting contaminants [11], or a combination of both [7,12-16]. These methods often make use of the guilty-by-association principle, and quantify the confidence level of an interaction by considering alternative paths between two protein molecules. In this context, authors say that a true interaction between bait b and prey p is a true positive if, at some point in the set of cells considered, there exists a complex that contains both b and p. We note that as the sensitivity of the AP-MS methods improves and the stability of the complexes that can be detected decreases, the transience of detectable interactions will increase, to a point where, eventually, every protein may be shown to marginally interact with every other protein.
A key property of AP-MS approaches is that a significant number of the co-purified prey proteins are in fact indirect interaction partners of the bait protein, in the sense that they do not interact physically and directly with the bait, but interact with it through a chain of physical interactions involving other proteins in the complex. Therefore, it is critical, when interpreting AP-MS-derived PPI networks, to understand the meaning of the term "interaction". Although not designed to identify physical interactions, AP-MS experiments produce data that may allow separating direct physical interactions from indirect ones. This is the problem we consider in this paper: given the results of a set of AP-MS experiments, filtered for protein misidentifications and contaminants, how can we distinguish direct (physical) interactions from indirect interactions? Note that since the false-positive filtering methods listed above consider indirect interactions as true-positives, they cannot be used to address this problem. Gordân et al. [17] study the related problem of distinguishing direct vs. indirect interactions between transcription factors (TF) and DNA. While the objective of their study is similar to ours, their method makes use of information specific to TF-DNA interactions (e.g. TF binding data, motifs from protein binding microarrays), and thus is not immediately applicable to the problem on general PPI networks. In fact, to our knowledge, no existing approach seems directly applicable.
This paper is organized as follows. We first describe the mathematical modelling of an AP-MS experiment and introduce an algorithmic formulation of the problem. We then describe an overview of our method, which is based on a collection of graph theoretic approaches that succeeds at inferring a large fraction of the network nearly exactly, followed by a genetic algorithm that infers the remainder of the network. The accuracy of the proposed method is assessed using both biological and simulated PPI networks. Finally, we apply our algorithm to the prediction of direct interactions based on a large set of AP-MS PPI data in yeast [18]. Our work opens the way to a number of interesting and challenging problems, and the results obtained indicate that useful inferences can be made despite the simplicity of our modelling.
Results
Because the main contribution of this paper is methodological, we start by giving an overview of the approach developed before detailing the results obtained.
Throughout this work, we make the assumption that appropriate methods have been used to reduce as much as possible protein misidentifications and contaminants, in such a way that all interactions detected are either direct or indirect interactions. Our task is to separate the former from the latter. To avoid confusion, we note that false-positives (resp. false-negatives) henceforth refer to falsely detected (resp. undetected) direct interactions inferred by our algorithm.
Mathematical modelling of AP-MS data
We first describe a simple model of the AP-MS PPI data that shall be used throughout this paper. Although admittedly rather simplistic, our model has the benefit of allowing the formulation of a well-defined computational problem.
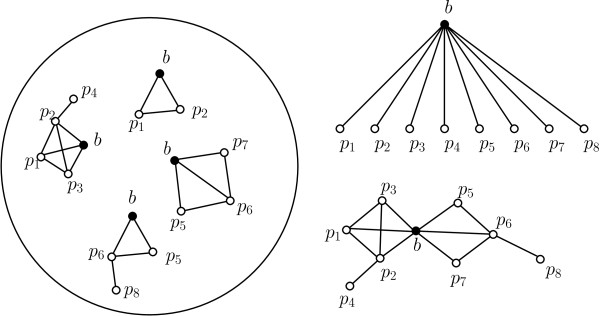
Let Gdirect = (V, Edirect) be an undirected graph whose nodes V represent the set of proteins, and edges Edirect represent direct (physical) interactions between the proteins. Let N(b) = {p ∈ V : (b, p) ∈ Edirect} be the set of direct interaction partners of protein b. We model the physical process through which PPIs are identified in an AP-MS experiment as follows. If a bait protein b is in contact with a direct interaction partner p ∈ N(b), the IP on b will pull p down, which will then be identified through mass spectrometry. In addition, if p interacts with p' ∈ N(p) at the same time as it interacts with b, protein p' may also be pulled down by the IP on b, although the two proteins only indirectly interact. In general, any protein x that is connected to b by a series of simultaneous direct interactions may be pulled down by b. As a result, all interaction partners of b (direct or indirect) will be identified together. Figure 1 depicts an example of this effect. In order to distinguish direct physical interactions from indirect ones, the availability of quantitative AP-MS data is helpful.
Figure 1.
Indirect interactions in AP-MS PPI data. In a cell, multiple copies of a bait protein b are expressed, and interact (directly or indirectly) with other proteins p1, ..., p8 (left); After the pull-down on the bait b, MS detects all prey proteins, including indirect interaction partners (top right); The direct interaction network should, however, contain only the edges between direct interaction partners (bottom right).
Although quantitative AP-MS remains at its infancy, prey abundance can be estimated fairly accurately using approaches such as the peptide count [19], spectral count [20], sequence coverage [21], and protein abundance index [22]. Combined with increasing accuracy and sensitivity of mass spectrometers, these methods are becoming more reliable. Throughout the discussions in this paper, we assume that this quantitative data is available to us.
The strength of a physical interaction can be measured by the energy required to break it. Let A(b, x) denote the abundance of a prey protein x obtained by IP on bait b, and let χ(p1, p2) denote the number of pairs of molecules p1 and p2 that interact directly in the cells considered. When there are more than two interaction partners, we let χ(p1, p2, ..., pk) denote the number of copies of complexes simultaneously containing p1, p2, ..., pk. Since protein interactions may be disrupted by the purification process, we expect A(b, x) to be correlated with the strength of the interaction between b and x. Thus, we assume that a direct interaction between a pair of individual proteins b and x survives the purification process with probability , and breaks with probability . Then the amount of protein x obtained from the pull-down on b would be
Consider now a system of three proteins b, x, x', where (b, x) and (x, x') form direct interactions but b and x' do not interact directly. Then, . In general, the amount of protein x that will be obtained upon pull-down of b will be proportional to the probability that b and x remain connected after each edge (u, v) ∈ Edirect is broken with probability (u, v). Our goal is then to infer Gdirect from the set of observed abundances A(x, y). In this paper, we make the following simplifying assumptions:
1. All direct interactions (u, v) ∈ Edirect survive with the uniform probability , and fail independently with probability 1 - .
2. All possible direct interactions take place at the same time, irrespective of the presence of other interactions, and with the same frequency.
Although these assumptions are clearly unrealistic, they provide a useful starting point for separating direct interactions from indirect ones (see Discussion for possible relaxation of these assumptions). Despite its simplicity, our mathematical modelling of AP-MS does fit existing biological data reasonably well (see Model validation). We note that Asthana et al. [23] have proposed a probabilistic graph model that is similar to ours. However, their model measures the likelihood of a protein's membership in a protein complex, and thus is not applicable to our problem.
Problem formulation
We are now ready to formulate the algorithmic problem addressed in this paper. We henceforth consider the (unknown) direct interaction network Gdirect as a probabilistic graph, where each edge in Gdirect survives the AP-MS process with probability , and fails otherwise. Let be a random graph obtained from Gdirect by removing edges in Edirect independently with probability 1 - . Then, define to be the probability that vertices u and v remain connected (directly or indirectly) in :
We call the connectivity matrix of Gdirect. See Figure 2 for an example of a direct interaction network and its connectivity matrix. Although can be estimated from Gdirect by straight-forward Monte Carlo sampling, its exact computation (known as two-terminal network reliability problem [24,25]) is #P-Complete [24], and so is its approximation within a relative error of ϵ [26].
Figure 2.
Example of a direct interaction network and its connectivity matrix. An example of a direct interaction network GDirect (left) with its connectivity matrix (right) calculated with . Assuming each edge of GDirect survives with probability , the probability of connectivity between each pair of protein can be estimated via sampling of the probabilistic network.
A set of AP-MS experiments where all proteins have been tagged and used as baits yields an approximation of A(x, y) for all pairs of proteins (x, y), which can be transformed into an estimate M(x, y) of through appropriate normalization. We are thus interested in inferring Gdirect from M:
EXACT DIRECT INTERACTION GRAPH FROM CONNECTIVITY MATRIX (E-DIGCOM)
Given: A connectivity matrix Mn × n
Find: A graph G = (V, E) such that PG(u, v) = M(u, v) for each u, v ∈ V.
In a more realistic setting, the connectivity matrix M would not be observed precisely, and the E-DIGCOM problem may not admit a solution. We are thus interested in an approximate, optimization version of the problem:
APPROXIMATE DIRECT INTERACTION GRAPH FROM CONNECTIVITY MATRIX (A-DIGCOM)
Given: A connectivity matrix Mn× n and a tolerance level 0 ≤ δ ≤ 1
Find: A graph G = (V, E) such that the number of pairs (u, v) ∈ V × V such that |PG(u, v) - M(u, v)| ≤ δ is maximized.
Note that although the computational complexity of the DIGCOM problems is currently unknown, the fact that simply verifying a candidate solution is #P-Complete suggests that the problem may be hard and may not belong to - candidate solutions to problems in NP can, by definition, be verified in polynomial time, whereas #P is widely believed to require super-polynomial time computations. Related problems include network design problems that have been studied extensively in the computer networking community. For example, one related, but different, problem is to choose a minimal set of edges over a set of nodes so that the resulting network has at least the prescribed all-pairs terminal reliability; various algorithms including branch-and-bound heuristics [27] and genetic algorithms [28,29] have been proposed.
Algorithm overview
Our algorithm for the A-DIGCOM problem has three main phases outlined here and detailed in Methods.
Phase I. We start by identifying, based on the connectivity matrix M, vertices from Gdirect with low degree, together with edges incident to them. As most PPI networks exhibit the properties of scale-free networks [30], this resolves the edges incident to a significant portion of the vertices (~75% in our networks; see below).
Phase II. At the other end of the spectrum, Gdirect contains densely clustered regions (cliques or quasi-cliques), possibly corresponding to protein complexes. We use a heuristic to detect these dense regions from the connectivity matrix M.
Phase III. To infer the remainder of the network, we use a genetic algorithm. This highly customized genetic algorithm makes use of the findings from the previous two steps in order to dramatically reduce the dimension of the problem space, and to guide the mating process between parent candidates to create good offspring solutions.
In what follows, we highlight the main theoretical results on which these three phases rely. Details are given in Methods and proofs in Appendix.
I-a. Finding cut edges
A cut edge in a graph G is an edge (u, v) whose removal would result in u and v belonging to two distinct connected components (e.g. edge (3,4) in Figure 2). The following theorem allows the identification of all cut edges based on the connectivity matrix PG.
Theorem 1. A pair of vertices u and v from V forms a cut edge in G if and only if the following two conditions hold.
(i)
(ii) V can be partitioned into V = Vu ∪ Vv , where Vu = {x ∈ V : PG(x, u) ≥ PG(x, v)} and Vv = {x ∈ V : PG(x, u) <PG(x, v)} , such that ∀s ∈ Vu and ∀t ∈ Vv, .
The above theorem immediately provides an efficient algorithm, requiring time O(|V|2), to test whether a pair of vertices forms a cut edge. Observe that removing a cut edge (u, v) from a connected graph allows us to decompose the graph into two connected components (subgraphs induced by Vu and Vv, respectively), and the probability of connectivity between every pair of vertices in Vu (Vv, resp.) remains the same after removing (u, v). Therefore, the submatrices that correspond to Vu and Vv can be treated as independent subproblems, and one can recursively detect cut edges in the remaining subproblems. Note that if the input graph is assumed to be a tree, such a recursive algorithm would identify the entire graph exactly. On the other hand, PPI networks are sparse in general, and contain many cut edges and degree-1 vertices. As a result, this algorithm allows a significant simplification of our problem by identifying all cut edges.
I-b. Finding degree-2 vertices
We now consider the problem of identifying degree-2 vertices from the connectivity matrix M. After degree-1 vertices, which are identified in the previous step, they constitute the next most frequent vertices in the biological networks we studied. While we do not have a full characterization of these vertices, the following theorem gives a set of necessary conditions.
Theorem 2. Let s be a degree-2 vertex in G such that N(s) = {u, v}. Then, the following three conditions must hold.
(i) Low connectivity: for each t ∈ V, .
(ii) Neighborhood: for each t ∈ V - {s, u, v}, PG (s, t) <PG (s, u) = PG (s, v).
(iii) "Triangle" inequality: for each t ∈ V - {s, u, v}, PG (s, t) <max{PG (u, t), PG (v, t)}.
These necessary conditions allow us to rule out vertices that cannot be of degree 2, and give rise to a O(|V|2) heuristic for predicting degree-2 vertices (see Algorithm 1 in Methods). In practice, our studies have shown that vertices satisfying these conditions while having degree higher than two are extremely rare (see below).
II. Detecting densely connected regions
We now turn to the problem of finding densely connected regions in the network. These regions may correspond to protein complexes, where tagging any one of the members of the complex results in the identification of all other members of the complex with high probability. While correctly predicting the physical interactions within each complex is a difficult task, separating these dense regions from the remainder of the network is essential to improving the accuracy of the genetic algorithm (part III).
Based on the connectivity matrix M, our algorithm identifies (possibly overlapping) clusters of proteins of size at least k such that, for every pair u, v in each cluster, M(u, v) ≥ tk for some threshold tk. For appropriately chosen values of k and tk (see Methods), the set of clusters found corresponds to cliques in Gdirect with high accuracy (see below).
The dense regions discovered at this phase provide us (1) the set of edges within each dense region; and (2) sparse cuts between disjoint dense regions. The edge set within each cluster will be used in the initial candidates for the genetic algorithm, whereas the cuts defined by the clusters will be used as crossover points during the crossover operation in the genetic algorithm.
III. Cut-based genetic algorithm
To predict the remaining section of the network, we use a customized genetic algorithm that aims at finding an optimal solution to the A-DIGCOM problem. We first devise a solution to a generalization of the A-DIGCOM problem, and then show how the results of parts I and II of the algorithm are used to improve performance.
Genetic algorithms have been shown to be an effective family of heuristics for a wide variety of optimization problems [31], including network design under connectivity constraints [28,29]. A genetic algorithm models a set of candidate solutions as individuals of a population. From this population, pairs of promising candidate solutions are mated, and their off spring solutions inherit properties of the parents with some random mutations. Over generations, this process of natural selection improves the fitness of the population.
The A-DIGCOM problem is a hard optimization problem, because (i) the size of the search space is huge - for a graph of size n, and (ii) there is no known polynomial-time algorithm to evaluate a proposed candidate solution (i.e. compute PG from G). For these reasons, a straight-forward genetic algorithm implementation failed to produce satisfactory results (data not shown). Instead, we use a more sophisticated approach by making use of the results obtained in previous sections in order to reduce the search space and to guide the mating operations for more effective search. Details are given in Methods.
Model validation
In order to test our approach, we first sought to validate our model of AP-MS indirect interactions. To this end, we used one of the most comprehensive AP-MS-based networks published to date on yeast, obtained by Krogan et al. [18]. The dataset reports the Mascot score [32] and the number of peptides detected for each bait-prey pair (peptide count). The complete set of interactions reported contains 2186 proteins and 5496 interactions (Krogan et al. Table S six); we call the resulting network GKroganFull. The authors identified a subset of these interactions as high-confidence, based on their Mascot scores (Krogan et al. Table S five). We call this set of high-confidence interactions GKroganHigh; this network consists of 1210 proteins and 2357 interactions. We expect that GKroganHigh is relatively rich in direct interactions, whereas the complete set of interactions GKroganFull consists in part of indirect interactions.
Considering GKroganHigh as a direct interaction network, we simulated Monte Carlo sampling to estimate , using and 50,000 samples, which yields a 95% confidence interval of size at most 0.007 on each entry. Next, we normalized the peptide counts of the interactions in GKroganFull using protein lengths (See Methods). We then compared to the normalized peptide counts of the interactions in GKroganFull. We expect that a significant fraction of low-confidence interactions in GKroganFull - GKroganHigh are likely to be indirect interactions. If our model is correct, their peptide counts should then be correlated with the corresponding entries in . Indeed, the positive linear correlation between the predicted connectivity and the observed normalized peptide counts is very significant (regression p-value of 8.17 × 10-11, Student t-test; see Additional file 1). Furthermore, this correlation is strongest when , as compared to or 0.7, justifying the use of this value in our subsequent analyses.
Accuracy of the prediction algorithm
The ideal validation of the accuracy of our algorithm would involve (i) constructing a connectivity matrix M using actual quantitative AP-MS data; (ii) predict direct interactions based on M using our algorithm; and then (iii) comparing our predictions to experimentally generated direct interaction data. Yeast 2-Hybrid (Y2H) experiments are less prone to detect indirect interactions than are AP-MS methods, and several large-scale efforts have been reported [2,33,34]. Unfortunately, for a number of technical reasons, the overlap between AP-MS PPI networks and Y2H networks remains very small [35]. As a consequence, Y2 H data cannot be used directly to validate predictions made on AP-MS data. Instead, we had to rely on partially-synthetic data set, where an actual network of high-quality Y2 H interactions is assumed to form the direct interaction graph, and a connectivity matrix is generated from it using Monte Carlo sampling, under our model. Two sets of Y2 H interactions were used: (i) GY u is the network constructed from the gold standard dataset of Yu et al. [35]. This network consists of 1090 proteins and 1318 interactions with high confidence of direct interactions; (ii) GDIP is the core, high-quality, physical interaction network of yeast, available at DIP database, version 20090126CR [36], consisting of 1406 proteins and 1967 interactions. These biological networks were complemented with two artificial 1000-vertices networks. The first was generated using the preferential attachment model (PAM) [30]. For the second, we used the duplication model (DM) [37], which, in contrast to the PAM, generates graphs containing several dense clusters. The resulting artificial "direct" interactions graphs are called GPAM and GDM and contain 1500 ~ 2000 interactions each. We then used the Monte Carlo sampling approach described above to estimate the connectivity matrices , and . These will form the input to our inference algorithm, whose output will then be compared to the corresponding direct interaction graph. It is important to note that these input matrices are not perfectly accurate and may contain sampling errors. However, it is easy to bound the size of the errors with high probability and use it as a tolerance level within our algorithm. We also note that the results presented in this section only aim at evaluating the performance of the inference algorithm on input data that was generated exactly according to our probabilistic model. As such, the error rates reported may be considered as lower bounds for those on actual biological data. An assumption-free evaluation in provided later in this section.
Identification of weakly connected vertices
Theorem 1 provides an efficient algorithm that guarantees the identification of all cut edges, provided that the given connectivity matrix is precise. We say that a vertex v is a 1-cut vertex if all edges incident on v are cut edges. By applying Theorem 1 recursively to detect cut edges and decomposing the graph into two connected components, we can detect and remove all 1-cut vertices from the input connectivity matrix.
Table 1 (i) reports the number of 1-cut vertices that are detected by the recursive algorithm from Theorem 1. In both the Yu and the DIP network, 1-cut vertices constitute approximately 50% of the network, and identifying them allows a significant reduction in the problem size. We note that the inaccuracies in the input connectivity matrices could, in principle, have introduced errors in the detection of cut edges. However, this rare event was never observed on any of our networks.
Table 1.
Performance of detecting weakly connected vertices
| (i) 1-cut vertices | (ii) Degree 2 vertices | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Network | Total | real | pred. | FDR(%) | FNR(%) | real | pred. | FDR(%) | FNR(%) | Remaining |
| Yu | 1090 | 552 | 552 | 0 | 0 | 195 | 207 | 7.7 | 2.05 | 331 |
| DIP | 1406 | 656 | 656 | 0 | 0 | 309 | 326 | 5.82 | 0.64 | 424 |
| PAM | 1000 | 457 | 457 | 0 | 0 | 351 | 363 | 3.58 | 0.28 | 180 |
| DM | 1000 | 323 | 323 | 0 | 0 | 117 | 126 | 11.9 | 5.12 | 551 |
Number of vertices detected as (i) 1-cut vertices, and (ii) degree 2 vertices in the real network and the predicted network. False discovery (FDR) and false negative (FNR) ratios are given in percentage. Remaining: the number of vertices remaining after identifying 1-cut vertices and degree 2 vertices.
Algorithm 1 (see Methods) guarantees to efficiently identify all degree-2 vertices (again, provided that the connectivity matrix is known), but may also incorrectly flag some higher-degree vertices. As seen in Table 1 (ii), nearly all degree-2 vertices were identified, with a low false-discovery rate ranging from 6 to 9%. Moreover, the false-positives incorrectly detected as degree-2 vertices indeed had small degrees, and their predicted neighbors were mostly correct (but incomplete) predictions. Flagging degree-2 vertices reduces the problem size further by 15 to 36%.
After repeatedly detecting and removing 1-cut vertices and degree-2 vertices from the problem space, the edges adjacent to approximately 70% of the vertices are detected with very low error rate. The remaining vertices only constitute approximately 30% of the original network. We call this remaining subset the hard core of the connectivity matrix. Because it is more densely connected than the rest of the network, the topology of hard core is more difficult to reconstruct.
Running our algorithm on the PAM simulated data yields similar resolution and error rate as on the Y2 H networks. However, our DM network is found to be less amenable to these strategies, leaving 55% of vertices unresolved and resulting in an error rate approximately twice that seen for other networks. This is simply due to the fact that networks generated by the duplication model do not contain as many 1-cut vertices or degree 2 vertices when compared to other networks, including the biological Y2 H networks.
Identification of dense regions
Our dense region detection algorithm aims at identifying all edges that belong to a k-clique in GDirect, for a given value of k. We report the accuracy of the algorithm in Table 2. As expected, our algorithm achieves extremely high sensitivity for clique edges. However, the false-discovery rate is quite high, especially for smaller values of k (e.g. k = 5). This is due to the fact that distinguishing a 5-clique from, say, a quasi-clique of size 7 is extremely difficult, causing false-positive predictions. We note however that these erroneous predictions are mostly inconsequential, as the intra-cluster topology of each dense region shall only be used in generating the initial candidate solutions for the genetic algorithm.
Table 2.
Performance of quasi-clique predictions
| Network | k = 7 | k = 6 | k = 5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| real | pred. | FD(%) | FN(%) | real | pred. | FD(%) | FN(%) | real | pred. | FD(%) | FN(%) | |
| Yu | 42 | 54 | 22.22 | 0 | 66 | 104 | 36.53 | 0 | 146 | 308 | 55.19 | 5.48 |
| DIP | 0 | 42 | 100 | 0 | 86 | 112 | 26.79 | 4.65 | 184 | 266 | 35.34 | 6.52 |
| PAM | 0 | 31 | 100 | 0 | 0 | 45 | 100 | 0 | 0 | 96 | 100 | 0 |
| DM | 254 | 346 | 28.32 | 2.36 | 194 | 267 | 29.21 | 2.57 | 488 | 718 | 34.96 | 4.30 |
Number of edges that belong to maximal cliques of size k. Real: actual number of edges that belong to maximal cliques of size k; pred: predicted number of maximal k-clique edges; FD(false discovery ratio): percentage of false-positives in the predicted set; FN(false negative ratio): percentage of false-negatives in the real set.
Cut-based genetic algorithm
The various parameters of the genetic algorithm (population size, mate selection probability, mutation rate, etc.) were optimized for the running time and accuracy of the solution based on GYu. Although our genetic algorithm could in principle be used on any connectivity matrix, running it on the full matrix of > 1000 proteins is impossible: the search space is huge, and the amount of time required to evaluate the fitness of a given candidate solution is too large. However, as discussed previously, applying first the 1-cut and degree-2 vertex detection algorithms significantly reduces the problem size and makes it accessible to our genetic algorithm. Table 3 (i) reports the accuracy of the genetic algorithm predictions on the hard core of each connectivity matrix. We note that since the network to be inferred is relatively highly connected, the problem is significantly more difficult than the identification of 1-cuts and degree-2 vertices. Indeed, the false-discovery and false-negative rates range from 35% to 55% for most datasets. For a comparison, an algorithm that would pick edges randomly would achieve 98.75% false-discovery and false-negative rates. Combining the three phases of the algorithm, the overall error rate obtained on each data set ranges from 10 to 20% false-discovery and false-negative rates, except for the DM data set, which fares considerably worse, for the reasons explained earlier.
Table 3.
Performance of genetic algorithm and overall algorithm
| Network | (i) reduced network | (ii) overall network | ||||||
|---|---|---|---|---|---|---|---|---|
| real | pred. | FDR(%) | FNR(%) | real | pred. | FDR(%) | FNR(%) | |
| Yu | 563 | 552 | 43.65 | 44.76 | 1318 | 1390 | 14.96 | 10.31 |
| DIP | 931 | 890 | 35.50 | 38.34 | 1967 | 2041 | 17.34 | 14.23 |
| PAM | 473 | 421 | 49.88 | 55.39 | 1538 | 1462 | 16.14 | 20.28 |
| DM | 1138 | 1295 | 43.39 | 35.58 | 1869 | 1804 | 32.81 | 35.15 |
(i) Performance of the genetic algorithm on the reduced network obtained from removing 1-cut vertices and degree-2 vertices. (ii) Overall performance of the combined prediction pipeline on the complete connectivity matrix. Real and predicted describe the number of edges in the real and predicted networks, respectively.
To the best of our knowledge, there has been no other efforts to solve the DIGCOM problems (neither exact, nor approximate version). We thus compared our approach to a simple hill climbing search algorithm on the Yu et al. data set (see Methods). We let this algorithm run over several days (as opposed to few hours spent using our approach), with multiple restarts, and discovered that it provides very poor sensitivity and specificity (see Table 4 for the best results obtained). This is not surprising since the hill climbing method is highly dependent on the initial solution (in this case, a spanning tree chosen randomly based on the connectivity matrix) and the search space is simply too large to exhaustively search for the a good initial solution. We also tested the hill-climbing approach in the same setting as the genetic algorithm, i.e. combining it with the 1-cut edges and degree-2 vertices detection algorithms. Here, the modified hill-climbing approach showed a better sensitivity and specificity than the pure hill-climbing approach, but still performed much worse than our genetic algorithm (Table 4). Furthermore, the improvement over the pure hill-climbing approach was mostly due to the high sensitivity and specificity of our algorithm for detecting weakly connected vertices.
Table 4.
Comparison of our method to simple hill-climbing approach
| Network | Real | Predicted | FDR(%) | FNR(%) |
|---|---|---|---|---|
| (i) Hill-climbing | 1318 | 2108 | 86.67 | 78.68 |
| (ii) Hill-climbing + weakly conn. nodes | 1318 | 1517 | 43.57 | 35.05 |
| (iii) Our approach (GA) | 1318 | 1390 | 14.96 | 10.31 |
Comparison of our method to simple hill-climbing approach. (i) accuracy of the hill-climbing approach used over the complete network; (ii) accuracy of the hill-climbing approach after fixing the weakly connected nodes using our algorithm; (iii) accuracy of our combined pipeline using the genetic algorithm.
To provide an idea of the running times, Table 5 gives the empirical data from our experiments. The first two phases (detecting weaking connected vertices and recognizing dense regions) were run within seconds while the genetic algorithm was run for a fixed amount of time, and the top scoring candidates where chosen as shown in Table 3.
Table 5.
Running times of the algorithm
| Network | (i) 1-cut & degree 2 vertices (secs) | (ii) Quasi-clique predictions | (iii) Genetic algorithm |
|---|---|---|---|
| Yu | 0:00:29 | 0:31:02 | 15:00:00 |
| DIP | 0:00:41 | 0:48:14 | 15:00:00 |
| PAM | 0:00:19 | 0:29:49 | 15:00:00 |
| DM | 0:00:24 | 1:18:03 | 15:00:00 |
Running times of our method on the model networks for each phase of the algorithm: (i) detecting 1-cut vertices and degree two vertices; (ii) predicting quasi-clique clusters; and (iii) running the genetic algorithm. We are reporting the average run time over three runs on each network. The implementation was tested on a Powermac G5 2 Ghz with 4 GB of RAM. Note that the genetic algorithm was run for a fixed amount of time, and the top scoring candidates achieved the quality as shown in Table 3. The times are shown in hh.mm.ss format.
Inferring direct interactions from AP-MS experimental data
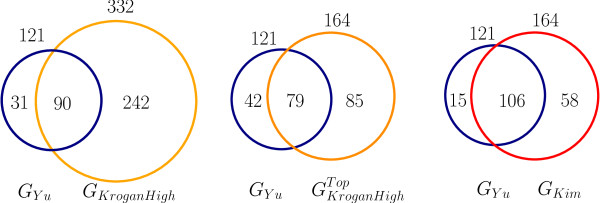
In order to apply our algorithm to biological data from AP-MS experiments, we used the raw data reported by Krogan et al. [18] for the 2186 putative interactions of GKroganFull. We only considered the subnetwork of tagged proteins, and further focussed our efforts on the analysis of 77 proteins that are well separated in the tag-induced subnetwork. Quantitative abundance estimates were derived from the peptide counts reported for each prey, and an experimentally derived connectivity matrix M was obtained after normalization (see Methods). Our full prediction algorithm was then run on the estimated connectivity matrix, resulting in a direct interaction graph prediction we call GKim that consists of 164 interactions (See Additional File 2). The network GKim was compared to GKroganHigh, the set of high-confidence interactions reported by Krogan et al., and to , a subset of GKroganHigh consisting of the 164 (to compare against GKim) most confident interactions they reported.
Both GKroganFull and GKroganHigh overlap GKim quite substantially. These three sets of predictions were then compared against a set of high-quality binary interactions from GYu. In Y2 H experiments, the interaction partners are separately screened using a genetic readout. Therefore, interactions from GY u are believed to be direct, and thus used to test against the predictions from AP-MS data. On the other hand, these interactions may reflect only a subset of all direct interactions among the 77 proteins.
As shown in Figure 3, our results show that the high-confidence AP-MS data GKroganHigh exhibited very little overlap with the direct binary interaction set GYu. 72.6% of interactions in GKroganHigh is disjoint from GY u, and 25% of GYu remains undetected by GKroganHigh. Furthermore, even the top scoring set of interactions showed high discrepancy ratios against GYu. In contrast, GKim produced by our algorithm coincide with GYu with better sensitivity and specificity. Given the crudeness of the method in translating the AP-MS data into a connectivity matrix, our algorithm has thus performed relatively well in predicting direct interactions from real AP-MS data.
Figure 3.
Inferring direct interactions from actual AP-MS dataset. Overlap between the Y2 H interaction network of Yu et al. and various AP-MS-based networks: (a) High-confidence set of interactions from Krogan et al. (b) Set of 164 highest scoring interactions from Krogan et al. (c) Set of 164 interactions predicted as direct interactions by our algorithms, based on the AP-MS data from Krogan et al.
Discussion and conclusion
The approaches for determining bait-prey abundance remain in their infancy, and to date, no large-scale PPI networks have this type of quantitative data. As these approaches gain in accuracy, so will the results of our approach. Furthermore, as the sensitivity of AP-MS pipelines improves, the fraction of indirect interactions detected will also increase, thereby making the ability to distinguish them even more critical. In this paper, we lay the bases of modelling the indirect interactions in AP-MS experiments. We formulate the DIGCOM problem, which aims at distinguishing direct interactions from indirect ones, and provide a set of theoretical and heuristic approaches that are shown to be highly accurate on both biological PPI networks and simulated networks. Despite the unrealistic assumptions that should eventually be relaxed, our results show that the predicted set of interactions fits the experimental data reasonably well. In addition, applying our algorithms to a large-scale AP-MS data set from Krogan et al. results in predictions that overlap Y2H data approximately 35% more often than the equivalent number of top-scoring interactions reported by these authors.
The DIGCOM problems raise a number of challenging, yet fascinating computational and mathematical problems to investigate. Is the solution to the exact DIGCOM problem, if it exists, always unique? We suspect it is. What is the computational complexity of the exact and approximate DIGCOM problems?
We believe they are NP-hard, and possibly not even in NP. Are there types of graph substructures, other than those discussed here, that can be unambiguously inferred from PG? Are there special properties of PPI networks, other than the power-law degree distribution, of which an algorithm can take advantage to make more accurate predictions and/or provide approximation or probabilistic guarantees?
The modelling and algorithm proposed here is only a first step toward an accurate detection of direct interactions from AP-MS data. Several generalizations and improvements are worth investigating. First, the abundance of an interaction is not constant and needs to be modelled more accurately. Second, the strength of all physical interactions is non-uniform, and some interactions may be more prone to disruption by the affinity purification process than others. Given sufficient quantitative AP-MS data, one may study a generalization of the DIGCOM problem that aims at identifying not only the set of direct interactions, but also their individual strengths and abundances. While modelling these aspects is in theory possible, the amount and quality of experimental data required is currently unavailable, and the computational complexity of the resulting problems is likely to be daunting.
Perhaps a more significant limitation of our model is that all direct interactions are assumed to occur simultaneously, though it is clear that certain interactions are either mutually exclusive, or restricted to specific subcellular compartments or conditions. We are currently investigating approaches to decompose the observed network into a family of simultaneously occurring interactions in such a way that the observed interaction abundances are the sum of the direct and indirect interactions over all cell compartments and conditions. However, it is clear that complementary experimental data, such as comprehensive protein localization assays or cell cycle expression data, would be required to reduce the space of possible solutions in a biologically meaningful manner.
An additional assumption that may need to be relaxed is the independence of the edge failures, which may not hold in cases where the loss of an interaction between two proteins causes a significant destabilization of the larger complex they belong to. Unfortunately, in the presence of strong dependencies between edge failures, it becomes almost impossible to distinguish direct from indirect interactions. Nonetheless, it may be possible to at least identify complexes where such dependencies hold, by studying subsets of proteins for which the AP-MS data differs significantly from our model.
In conclusion, this paper opens the door to a number of fascinating modelling and algorithmic questions that will lead to important implications in systems biology. Any improvements in tackling these questions would take us one step further towards this goal.
Methods
In this section, we describe the algorithmic details of our approach to the DIGCOM problem.
Identification of weakly connected vertices
The algorithm to identify 1-cut vertices is trivial given Theorem 1: recursively find edges satisfying conditions in Theorem 1, and decompose the input matrix into two independent subproblems. On the other hand, the conditions in Theorem 2 yields Algorithm 1, which predicts the set of degree 2 vertices as well as the edges adjacent to them. It is easy to see that the algorithm to identify 1-cut vertices runs in time O(|V|4) and Algorithm 1 runs in time O(|V|2).
Note that the identification algorithm for 1-cut vertices allows us to remove these vertices, i.e., the corresponding rows and columns in the input matrix. This is possible due to the fact that removing a cut edge does not change the connectivity between any two nodes on the same side of the cut. On the other hand, we cannot simply remove degree-2 vertices without affecting the remaining entries in the matrix. Therefore, as shown in Algorithm 1, degree-2 vertices and their incident edges will be marked as such in the solution, but are not removed from the input matrix.
Predict degree-2 vertices
Input: Probability matrix M and vertex set V
Output: Degree 2 vertices V 2 and their incident edges E2
foreach vertex s ∈ V do
if s satisfies conditions in Theorem 2 then
V 2 ← V 2 ∪ {s};
u, v ← the two vertices with Ms, u = Ms, v = max{Ms, t : ∀t ∈ V - {s}};
E2 ← E2 ∪ {(s, u), (s, v)};
end
end
Return V 2 and E2
Algorithm 1: Prediction of degree 2 vertices
Detect dense clusters in the network
Input: Probability matrix M, and minimum cluster size k
Output: Set of possible k-cliques in Gdirect.
t ← CliqueConn(k) (the connectivity between two vertices in an m-clique; see Appendix)
Gt ← (V, Et), where Et = {(u, v): u, v ∈ V and M(u, v) ≥ t}
foreach connected component S ⊆ V do
Find cliques of size k in S
end
Return cliques discovered.
Algorithm 2: Detecting dense clusters in the network
Identification of densely connected regions
Densely connected regions are identified using a clique-cover algorithm (see Algorithm 2). We note that the algorithm guarantees to identify all cliques of size k' ≥ k contained within Gdirect. However, sets of vertices that do not form a k-clique may also be reported, provided that they are sufficiently connected among themselves, possibly via vertices outside the set. However, for sufficiently large values of k, we found this to be a very rare occurrence. While finding cliques in a graph is a computationally intensive task in general, the construction of Gt for large values of k creates few small connected components and leaves the remaining vertices isolated. Therefore, in practice, Algorithm 2 can be implemented to run in a reasonable amount of time.
Cut-based genetic algorithm
The genetic algorithm aims at solving a generalization of the A-DIGCOM. First, we allow each edge (u, v) in the network to survive with a non-uniform probability , instead of one probability over all edges. Secondly, we assume that we are given two sets of edges EYES and ENO that indicate the set of edges that are guaranteed to be in the solution, and guaranteed not to be in the solution, respectively. This will later allow us to factor in the outcome of the previous sections. Therefore, the edges whose presence remains to be determined are .
Encoding of candidate solutions
To represent a candidate solution, we first create a hash table that maps each putative edge in EMAYBE to an integer. Each candidate is then encoded as a list of integers (edges). Edges in EYES, which are part of all solutions, are not explicitly listed, in order to save space. Since the networks we consider are sparse (|E| = O(|V|)), such an encoding technique significantly reduces the space requirements.
Initial population
The initial population of candidates is generated using a preferential attachment model [30] using the following observations: (i) The average connectivity of vertex u, is strongly positively correlated with the degree of u in Gdirect; (ii) the age of a vertex, measured by when the vertex was introduced to the graph, is positively correlated with the degree of the vertex. Therefore, during the generation of each candidate, we choose the next vertex to be added with probability proportional to its average connectivity. This results in a candidate solution where the degree of most vertices is likely to be close to the their true degree in Gdirect. Furthermore, in order to create candidates that are clustered similarly to the true direct interaction graph, we include the set of edges predicted by Algorithm 2 to each initial candidate.
Fitness function
The fitness of a candidate solution G, fitness(G) is obtained by first estimating the probability matrix PG using 500 Monte Carlo samples, and then counting the number of vertex pairs (u, v) whose estimated connectivity PG(u, v) is within the tolerance level δ, i.e., M(u, v) ± δ (See below for choosing the tolerance level δ ).
Crossover
The crossover operation needs to hybridize two parent candidates to produce off springs preserving the good properties of the parents. This operation will be guided by a randomly chosen balanced cut V = V1 ∪ V2. Let G1 and G2 denote the two parent networks and let E1(Gi) and E2(Gi) denote the edges of Gi such that both endpoints lie in V1 and V2, respectively. Furthermore, let E1,2(Gi) denote the edges of Gi that crosses from V1 and V2. Mating G1 and G2 results in two children G' = (V, E'), and G'' = (V, E'') such that:
While choosing a random cut as the crossover point is a reasonable strategy to construct a new pair of offsprings, our studies have shown that a planned strategy in choosing the crossover points results in better performance and less chance of premature convergence. In particular, if the crossover point is chosen at a dense cut in the parent networks, then the connectivity among vertices within each partition would be deteriorated significantly. This results in offsprings with much poorer fitness than their parents. On the other hand, if the parents are hybridized at a sparse cut, the connectivity among vertices within each partition are more localized. Therefore, crossover operations are best done by selecting sparse balanced cuts (|V1| ≈ |V2|). Finding sparse balanced cuts is a well-studied problem in combinatorial optimization, for which various approximation algorithms exist [38,39]. However, these algorithms assume that the graph itself, not the connectivity matrix M, is given as input. We therefore use a simple heuristic that avoids cutting through the dense regions of the network. To generate these sparse cuts, we contract each dense region identified in Algorithm 2 to a single vertex, and then generate weighted (by the number of vertices in each dense region) balanced partitioning of the vertices at random.
Mutation
In order to introduce variability to the population of candidates, a small number of edges (5 ~ 10%) are randomly inserted or deleted. Moreover, observe that the child network constructed as above may not remain connected. Aside from the random mutation, therefore, we employ a simple local search that greedily adds edges to keep the network connected.
Genetic algorithm parameter selection
The various parameters of the genetic algorithm were selected based on the resulting performance on the Yu et al. data set. Two main parameters that affect the performance significantly are the population size and the selection criteria. For selection criteria, we tested several different selection criteria by setting the probability of choosing a candidate as a parent. The best compromise between running time and accuracy was obtained using a population size of 500, and selection probability for a parent proportional to fitness(ci) - minFit, where minFit is the fitness of the worst candidate in the population (data not shown).
Restricting the solution space
While our genetic algorithm offers a plausible method for the A-DIGCOM problem, one can reduce the size of the solution space, which typically results in faster convergence to better solutions, using the results in Theorem 1 and 2. First, recall that finding all cut edges decomposes the problem into independent subproblems on 2-edge-connected components. Second, the identification of degree-2 vertices defines two sets of edges EYES and ENO that constitute all putative edges incident to the identified degree-2 vertices. In other words, EMAY BE forms the subgraph of G induced by the set V 3+ of vertices with degree ≥ 3. Furthermore, observe that the edges in EY ES form parallel paths between vertices in V 3+. A classical result in network reliability (see Fact 2 in Appendix) suggests that these parallel paths can be merged into a single meta-edge whose reliability can be efficiently computed. To be more formal, let (u, v) = {P1(u, v), P2(u, v), ..., Pk(u, v)} be the set of paths between u and v in EY ES. These paths can then be replaced by a single edge (u, v) with its survival probability . By merging every set of parallel paths, we obtain a compact network over V3+ that efficiently encodes the edges in EY ES. Since our genetic algorithm handles the case where the edge survival probability is non-uniform, this compact encoding results in substantial gains in running time for estimating the fitness of the candidates, as well as in time and space requirements for handling large population sizes. In our applications, this allows us to remove approximately 70 ~ 75% of the original set of vertices.
Randomized hill-climbing algorithm
For performance comparisons, we tested our algorithm against a simple randomized hill-climbing approach. In this approach, we start with a randomly chosen spanning tree G1 of the vertex set V. At the ith iteration, we first sample the connectivity probability of Gi, using the Monte Carlo simulation. Then, we randomly pick a vertex pair u, v with probability proportional to
where . If the selected pair u, v are connected by an edge in Gi, but M(u, v) > (u, v), then we remove (u, v) from Gi. On the other hand, if u and v are not connected by an edge, but M(u, v) < (u, v), then we add (u, v) to Gi. We repeat this local optimization heuristic while making sure the candidate solution remains connected.
Choosing a tolerance level δ and handling numerical errors
In order to deal with numerical errors from Monte Carlo sampling, we use a well-defined tolerance level δ as additive errors. Note that the sampling process for estimating the probability matrix PG is a binomial process, which, by the central limit theorem, is closely approximated by a normal distribution. The confidence interval is largest when the estimated probability is equal to 0.5, in which case we obtain a confidence interval of
where denotes the fraction of samples where the two vertices are connected after n samples, and z1 - α/2 is the z-value for desired level of confidence. Using this formula, we can conclude:
1. When n = 20000 (computation of our input matrix M from test networks), we obtain a 95% confidence interval of size at most 2·δ = 2·0.007 = 0.014.
2. When n = 500 (computation of the connectivity matrix for each candidate solution in our genetic algorithm), the 95% confidence interval is of size at most 2·δ = 2·0.04 = 0.08.
With the chosen tolerance level δ, we modify our algorithm as appropriate each time we compare two connectivity probabilities. For example, in Theorem 1, the first condition PG(u, v) = is modified to; and in Theorem 2, we modify the first condition to .
Generation of scale-free networks
In order to generate artificial scale-free networks, we used two generation models: the preferential attachment model, and the duplication model. In the preferential attachment model, we evaluated the degree distribution of the two biological networks (GY u and GDIP ) and used the Barabási-Albert algorithm to construct a scale-free network with attachment factor 1.5 (each iteration adds a new vertex with 1 ~ 2 edges attached to existing vertices). In the duplication model, at each iteration, we randomly pick a vertex to duplicate with probability proportional to its degree and randomly drop the duplicated edges with probability at 0.5 in order to t the degree distributions and sparsity of biological networks.
Calculation of connectivity matrix from peptide counts
The peptide count of a prey protein in an AP-MS experiment is the number of different peptides that have been observed by MS for that protein. We note that the peptide counts are biased towards preys with longer protein sequences, and to rectify this propensity, we normalized the abundance data by the protein sequence lengths to obtain the abundance ratios R(i, j). In order to turn the normalized abundance ratios into the connectivity matrix for our probabilistic graph model, we used a simple logistic function
where the parameters α, β are chosen so that the computed distribution of fits the simulated connectivity distribution of GY u, using a χ2 test (α = 2.8921, β = -0.6318). In the cases where R(i, j) differ from R(j, i), we choose the average of the two entries to symmetrize the matrix.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors jointly contributed to the design and analysis of the algorithms. EK and MB jointly contributed to the formulation of the problem, interpretation of the experiments, and preparation of the manuscript. Implementation and testing were done by EK. All authors have read and approved the final manuscript.
Appendix
We start with two basic results that will prove useful when proving more complex theorems. Let G1 = (V1, E1) and G2 = (V2, E2) be two graphs. Then the following are true.
Fact 1. (Series composition) Suppose V1 ∩ V2 = {c}, and a new graph G is constructed by joining G1 and G2 at c. Then, for any s ∈ V1 - {c} and for any .
Fact 2. (Parallel composition) Suppose V1 ∩ V2 = {s, t}, and a new graph G is constructed by joining G1 and G2 at s and t (possibly leading to parallel edges between s and t). Then, .
Proof of Theorem 1
Proof. Necessity is trivial. For sufficiency, suppose the conditions (i) and (ii) hold, and (u, v) is not a cut edge. Then, to keep the graph connected, there must be an edge (s, t) ≠ (u, v) joining Vu and Vv. Since (s, t) is an edge, . However, by assumption, we have , which is a contradiction. □
Proofs of Theorem 2
Proof. We prove each condition separately.
Condition (i) Low connectivity
Since s has degree 2, it becomes disconnected from the rest of the graph with probability . Thus, . The equality can only hold if PG(s, u) = PG(s, v) = 1, which is impossible.
Condition (ii) Neighborhood
We first show PG(s, u) = PG(s, v). Let PE -{(s, u)}(s, u) denote the connectivity of s and u when edge (s, u) is removed from E. Then, we have:
Next, we show that for any t ∈ V - {s, u, v}, PG(s, t) <PG(s, u). For a subgraph H ⊆ G, let p(H) denote the probability of observing H from a probabilistic graph G. We can write this probability as
Thus, for any two subgraphs Hi, Hj ⊆ G, such that |E(Hi)| = |E(Hj)|, p(Hi) = p(Hj). Let ℋ(s, t) be the set of subgraphs of G where s and t are connected. We can then write the probabilities PG(s, t) and PG(s, u) as the sum of the probabilities of these subgraphs.
Observe that these two sets of subgraphs may overlap:
where ℋ(s, t, u) is the set of subgraphs such that s, t, and u are all connected, while is the set of subgraphs where s, t belong to the same connected component that doesn't contain u. Thus, we now focus on and . The subgraphs in can be partitioned as follows, depending on which component v belongs to:
(i) {s, u, v}, {t}: s, u, v belong to the same component, and t belongs to another component.
(ii) {s, u}, {v}, {t}: s, u belong to the same component, and each of v and t belongs to distinct components.
(iii) {s, u}, {v, t}: s, u belong to the same component, and v, t belong to another component.
It is easy to see that the cases (i) and (ii) are nonempty, since (s, u), (s, v) ∈ E(G).
Finally, consider the set of subgraphs in . Let H be a subgraph in this set. Since s and u belong to distinct components, (s, u) ∉ E(H), and thus (s, v) ∈ E(H) in order to have s and t connected. We then make the following operation on H to construct H': (1) remove (s, v) from H, and (2) insert (s, u). Then, H' now has s and u in the same component, and v and t in another component - and therefore, H' belongs the case (iii) of . Furthermore, note that H and H' have the same number of edges. Therefore, there is a mapping from subgraphs in to subgraphs in case (iii) of with equal number of edges. Since the cases (i) and (ii) are nonempty, it follows that PG(s, t) <PG(s, u).
Condition (iii) "Triangle" inequality
Given the probabilistic graph G, we partition the subgraphs of G into four cases depending on the existence of the two edges (s, u) and (s, v).
• (s, u), (s, v) ∈ E: occurs with probability .
• (s, u) ∈ E, (s, v) ∉ E: occurs with probability .
• (s, u) ∉ E, (s, v) ∈ E: occurs with probability.
• (s, u), (s, v) ∉ E: occurs with probability.
We can thus rewrite the probability PG(s, t) as follows.
| (1) |
where PG - {s}(u or v, t) denotes the probability that u or v is connected to t in the graph G - {s}. We write PG(u, t) and PG(v, t) similarly:
| (2) |
| (3) |
Subtracting (1) from (2), and (1) from (3) gives:
If , it follows that PG(u, t) > PG(s, t). Otherwise, implies , and it follows that PG(v, t) >PG(s, t). □
Connectivity in an n-clique
Here we construct a formula to compute the connectivity of an n-clique Kn, defined as CliqueConn(n) in Algorithm 2.
First, we recall a classical result in graph enumeration.
Lemma 1
[40,41]Let γn denote the number of connected simple graphs over n labeled vertices. We can count this number by the recurrence.
Now we show the formula for the case when , which is the value we used for our analyses. The formula for other values of can be formulated using a similar proof.
Lemma 2
Proof. We write the probability as follows.
where σi denotes the number of subgraphs of Kn of size i where two given vertices u and v are connected. To compute σi, there are ways to pick i - 2 vertices (in addition to u and v) for the connected components, and γi ways to keep them all connected, and finally, ways to assign edges among the remaining vertices. So we have
and finally, we have
□
Supplementary Material
ModelValidation. Correlation plot of the connectivity from and normalized peptide counts from Krogan et al. Each data point corresponds to an interaction in GKroganFull - GKrogranHigh, many of which are expected to be indirect. x-axis: Connectivity entry in , using . y-axis: Peptide count, normalized by protein lengths. The resulting linear regression showed a strong correlation (regression p-value 8.17 × 10-11, Student t-test), suggesting that many interactions in GKroganFull - GKrogranHigh are indirect and are the result of direct interactions in GKroganHigh.
PredictedInteractions. Set of direct interactions among 77 yeast proteins, predicted by our algorithm. The connectivity matrix is generated from normalized peptide counts, and then our algorithm is run to predict the direct interactions.
Contributor Information
Ethan DH Kim, Email: ethan@cs.mcgill.ca.
Ashish Sabharwal, Email: sabhar@cs.cornell.edu.
Adrian R Vetta, Email: vetta@math.mcgill.ca.
Mathieu Blanchette, Email: blanchem@mcb.mcgill.ca.
Acknowledgements
We thank Mathieu Lavallée-Adam, Benoit Coulombe and Javad Sadri for useful discussions, and three anonymous reviewers for their useful suggestions. This work was funded by NSERC grants to MB and AV, and by an NSERC PGS-D to EK. AS was supported by IISI, Cornell University (AFOSR grant FA9550-04-1-0151) and by NSF (Expeditions grant 0832782 and IIS grant 0514429).
References
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(3):1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotech. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Gavin A, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A, Cruciat C, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier M, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sørensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Coulombe B, Blanchette M, Jeronimo C. Steps towards a repertoire of comprehensive maps of human protein interaction networks: the Human Proteotheque Initiative (HuPI) Biochemistry and Cell Biology. 2008;86(2):149–156. doi: 10.1139/O08-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Al-Khoury R, Lavallée-Adam M, Faubert D, Jiang H, Poitras C, Bouchard A, Forget D, Blanchette M, Coulombe B. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods (San Diego, Calif.) 2009;48(4):381–386. doi: 10.1016/j.ymeth.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Park B, Karpinets T, Samatova N. From pull-down data to protein interaction networks and complexes with biological relevance. Bioinformatics. 2008;24(7):979–86. doi: 10.1093/bioinformatics/btn036. [DOI] [PubMed] [Google Scholar]
- Gilmore J, Auberry D, Sharp J, White A, Anderson K, Daly D. A Bayesian estimator of protein-protein association probabilities. Bioinformatics. 2008;24(13):1554–5. doi: 10.1093/bioinformatics/btn238. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol Biol. 2007;367:87–119. doi: 10.1385/1-59745-275-0:87. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom J, Boucher L, Neduva V, Larsen B, Breitkreutz B, Stark C, Liu G, Ahn J, Dewar-Darch D, Qin Z, Pawson T, Gingras A, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Suzuki H, Hayashizaki Y. Interaction generality, a measurement to assess the reliability of a protein-protein interaction. Nucl Acids Res. 2002;30(5):1163–1168. doi: 10.1093/nar/30.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Suzuki H, Hayashizaki Y. Construction of reliable protein-protein interaction networks with a new interaction generality measure. Bioinformatics. 2003;19(6):756–763. doi: 10.1093/bioinformatics/btg070. [DOI] [PubMed] [Google Scholar]
- Pei P, Zhang A. A topological measurement for weighted protein interaction network. Computational Systems Bioinformatics Conference, 2005. Proceedings. 2005 IEEE. 2005. pp. 268–278. [DOI] [PubMed]
- Chen J, Hsu W, Lee ML, Ng S. Increasing confidence of protein interactomes using network topological metrics. Bioinformatics. 2006;22(16):1998–2004. doi: 10.1093/bioinformatics/btl335. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao X, Greenblatt JF, Spencer F, Holstege FCP, Weissman JS, Krogan NJ. Toward a Comprehensive Atlas of the Physical Interactome of Saccharomyces cerevisiae. Molecular & Cellular Proteomics. 2007;6(3):439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Gordân R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Research. 2009;19(11):2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, Onge PS, Ghanny S, Lam MHY, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Gao J, Opiteck G, Friedrichs M, Dongre A, Hefta S. Changes in the protein expression of yeast as a function of carbon source. J Proteome Res. 2003;2(6):643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov R, Yates Jr. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn M, Raine J, Anthony R, Grainger M, Haynes J, Moch J, Muster N, Sacci J, Tabb D. et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Asthana S. Predicting Protein Complex Membership Using Probabilistic Network Reliability. Genome Research. 2004;14(6):1170–1175. doi: 10.1101/gr.2203804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiant LG. The Complexity of Enumeration and Reliability Problems. SIAM Journal on Computing. 1979;8(3):410–421. doi: 10.1137/0208032. [DOI] [Google Scholar]
- Colbourn CJ. The Combinatorics of Network Reliability. Oxford University Press, Inc; 1987. [Google Scholar]
- Provan JS, Ball MO. The Complexity of Counting Cuts and of Computing the Probability that a Graph is Connected. SIAM Journal on Computing. 1983;12(4):777–788. doi: 10.1137/0212053. [DOI] [Google Scholar]
- Jan R, Hwang F, Chen S. Topological optimization of a communication network subject to a reliability constraint. Reliability, IEEE Transactions on. 1993;42:63–70. doi: 10.1109/24.210272. [DOI] [Google Scholar]
- Dengiz B, Altiparmak F, Smith A. Efficient optimization of all-terminal reliable networks, using an evolutionary approach. Reliability, IEEE Transactions on. 1997;46:18–26. doi: 10.1109/24.589921. [DOI] [Google Scholar]
- Dengiz B, Altiparmak F, Smith A. Local search genetic algorithm for optimal design of reliable networks. Evolutionary Computation, IEEE Transactions on. 1997;1(3):179–188. doi: 10.1109/4235.661548. [DOI] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Goldberg DE. Genetic Algorithms in Search, Optimization, and Machine Learning. Addison-Wesley Professional; 1989. [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain J, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16(3):277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual J, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet A, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi A, Tavernier J, Hill DE, Vidal M. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwínski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Galas D, Dewey T. A duplication growth model of gene expression networks. Bioinformatics. 2002;18:1486–1493. doi: 10.1093/bioinformatics/18.11.1486. [DOI] [PubMed] [Google Scholar]
- Leighton T, Rao S. Multicommodity max-flow min-cut theorems and their use in designing approximation algorithms. Journal of the ACM. 1999;48:787–832. doi: 10.1145/331524.331526. [DOI] [Google Scholar]
- Vazirani V. Approximation algorithms. Springer; 2004. [Google Scholar]
- Gilbert E. Enumeration of labeled graphs. Canad J Math. 1956;8:405–411. [Google Scholar]
- Gross J, Yellen J. Handbook of Graph Theory (Discrete Mathematics and Its Applications) CRC; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ModelValidation. Correlation plot of the connectivity from and normalized peptide counts from Krogan et al. Each data point corresponds to an interaction in GKroganFull - GKrogranHigh, many of which are expected to be indirect. x-axis: Connectivity entry in , using . y-axis: Peptide count, normalized by protein lengths. The resulting linear regression showed a strong correlation (regression p-value 8.17 × 10-11, Student t-test), suggesting that many interactions in GKroganFull - GKrogranHigh are indirect and are the result of direct interactions in GKroganHigh.
PredictedInteractions. Set of direct interactions among 77 yeast proteins, predicted by our algorithm. The connectivity matrix is generated from normalized peptide counts, and then our algorithm is run to predict the direct interactions.