Abstract
Nuclear receptors are important transcriptional factors that share high sequence identity and conserved domains, including a DNA-binding domain (DBD) and a ligand-binding domain (LBD). The LBD plays a crucial role in ligand-mediated nuclear receptor activity. Hundreds of different crystal structures of nuclear receptors have revealed a general mechanism for the molecular basis of ligand binding and ligand-mediated regulation of nuclear receptors. Despite the conserved fold of nuclear receptor LBDs, the ligand-binding pocket is the least conserved region among different nuclear receptor LBDs. Structural comparison and analysis show that several features of the pocket, like the size and also the shape, have contributed to the ligand binding affinity and specificity. In addition, the plastic nature of the ligand-binding pockets in many nuclear receptors provides greater flexibility to further accommodate specific ligands with a variety of conformations. Nuclear receptor coactivators usually contain multiple LXXLL motifs that are used to interact with nuclear receptors. The nuclear receptors respond differently to distinct ligands and readily exchange their ligands in different environments. The conformational flexibility of the AF-2 helix allows the nuclear receptor to sense the presence of the bound ligands, either an agonist or an antagonist, and to recruit the coactivators or corepressors that ultimately determine the transcriptional activation or repression of nuclear receptors.
Keywords: nuclear receptor, crystal structure, ligand binding domain, ligand recognition, cofactor recruitment
1, Introduction on functional domains of nuclear receptors
Nuclear receptors are important transcriptional factors essential for a broad aspect of human physiology, ranging from development and differentiation to metabolic homeostasis [1]. The complete human genome contains 48 nuclear receptors that include classic receptors, adopted orphan receptors and orphan receptors (Table 1). Classic receptors are regulated by endocrine ligands that have been extensively studied, such as steroid hormones, retinoic acids, vitamin D and thyroid hormone. The human nuclear receptors also include a class of orphan receptors for which no ligand was known when the receptor was cloned [2; 3]. Since nuclear receptors are critical in physiology, there has been enormous interest in pursuing the orphan receptors as drug targets. The result of intense research in the past few years is the emergence of a class of so-called “adopted” orphan receptors for which either natural or synthetic ligands have been identified, through the approach of “reverse endocrinology” [4].
Table 1.
Human nuclear receptors and the solved LBD structures
| Classic Receptors | Structure | Adopted Orphan receptors | Structure | Orphan Receptors | Structurea |
|---|---|---|---|---|---|
| AR | [65] | CAR | [19; 77; 78] | COUP-TF (I, II, III)b | [21] |
| ER (α, β) | [66; 67] | ERR (α, β, γ) | [27; 79] | Dax-1b | [10] |
| GR | [44] | FXR | [80; 81] | RORγb | [24] |
| MR | [68; 69; 70] | HNF4 (α, γ) | [54; 55] | GCNF | - |
| PR | [71] | LXR (α, β) | [82; 83] | NGF1-B (α, β, γ) | [56] |
| RAR (α, β, γ) | [15; 73; 74] | PPAR (α, γ, δ) | [30; 52; 84] | PNR | - |
| TR (α, β) | [16; 75] | PXR | [29] | RevErbAb | [36] |
| VDR | [76] | ROR (α, β) | [72; 85] | SHP | - |
| RXR (α, β, γ) | [14; 86] | TLX | - | ||
| LRH | [87] | TR2, TR4 | - | ||
| SF1 | [88; 89] |
A “-” indicates the structure unsolved.
The structures were published in most recent three years (between 2007 and 2010).
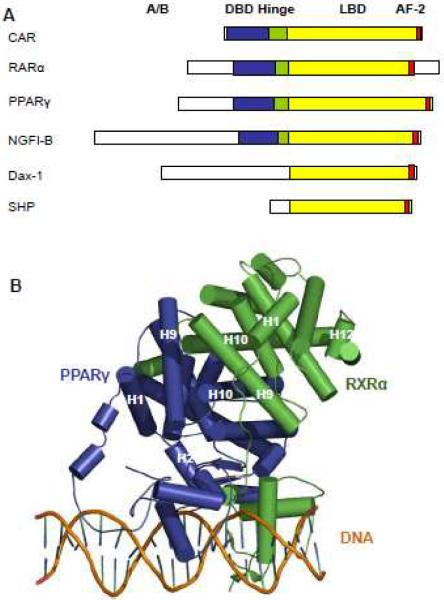
All nuclear receptors share high sequence identity and conserved domains. Hundreds of different crystal structures of nuclear receptors have revealed a general mechanism for domain organization. A typical nuclear receptor usually contains five functional regions: the A/B region that contains an N-terminal activation function-1 domain, the central C region that contains a DNA-binding domain (DBD), the C-terminal E region that contains a ligand-binding domain (LBD), and the D hinge region that links the DBD and the LBD (Figure 1a). Among these domains, DBD and LBD share highest similarity in most nuclear receptors. The DBD is used to recognize promoter and contains structural features allowing the nuclear receptors to bind differentially to target genes [5]. The LBD plays a crucial role in ligand-mediated nuclear receptor activity. In addition to its role in ligand recognition, the LBD also contains an activation function-2 (AF-2) domain, whose conformation is highly dependent on the bound ligand. The hinge D, together with N-terminal A/B region and C-terminal E region, are less conserved and show distinct structural features among different nuclear receptors. First, the length of N-terminal A/B region varies among different nuclear receptors. For example, the sequence of constitutive androstane receptor (CAR) revealed limited A/B region and no AF-1 function [6]. Also the secondary structure of AF-1 mainly consists of unstructured coils instead of helices and beta sheets for LBD and DBD. As such, the folding of AF-1 is very flexible that makes structural determination very difficult. Indeed, no AF-1 structure is available so far despite that hundreds of structures of isolated DBDs and LBDs have already been solved. Accordingly, the molecular basis of AF-1 is least understood.
Fig. 1. Domain organization of nuclear receptors.
A, A schematic representation showing functional domains of different nuclear receptors. The DBD is labeled in blue, the hinge is in green and the LBD is in yellow. The presence of AF-2 is indicated in red. B, Multi-domain structure of the PPARγ/RXR/DNA complex in ribbon representation. The crystal structures of PPARγ(blue) and RXRα(green) heterodimer (top) on PPAR response DNA sequence (Bottom).
Interestingly, the nuclear receptors Dax-1 and SHP only have LBDs. Although they lack DBDs, Dax-1 and SHP are able to interact with other transcriptional factors and function as corepressors in regulating their target genes [7; 8]. SHP has been shown to bind to LRH-1 using its second LXXLL motif in a way similar to nuclear coactivators [9]. In contrast, the crystal structure of the Dax-1/LRH-1 complex reveals a distinct molecular mechanism for the function of Dax-1 from SHP [10]. Instead of a dimer, Dax-1 and LRH-1 form a trimeric structure, with the existence of a Dax-1 homodimer. Both Dax-1 molecules use the same new conserved PCFXXLP repressor motif to interact with different sites on LRH-1, which does not resemble any dimerization modes between nuclear receptors or interaction of nuclear receptor with coregulators.
The flexibility of the domain arrangement has made the crystallization of nuclear receptor very challenging. Despite that many structures have been determined for isolated nuclear receptor DBD and LBD, the only two-domain crystal structure available is PPARγ/RXRα dimer [11]. The crystal structure described includes DBD, LBD and hinge region of PPARγ/RXRα bound with DNA, thus providing important molecular basis for domain organization and target gene recognition (Figure 1b). In the structure, PPARγ and RXRα form a non-symmetric complex, with multiple interfaces that link these two receptors. In addition to DBD-DBD and LBD-LBD contacts, an unexpected feature revealed is the existence of the interface between PPARγ LBD and RXRα DBD. This observed DBD-LBD interaction shows importance to DNA recognition and transcriptional activation of PPARγ/RXRα dimer as indicated by mutation analysis. Also the 5' extension of the conserved PPAR response DR1 sequence (direct repeats of AGGTCA with one base-pair spacing) on the target gene is also critical for the binding specificity of PPARγ/RXRα complex. Interestingly, the hinge region of PPARγ is also involved in the positioning of the transcription factor dimer on the target gene. However, the structures of the A/B segments are not visible partly due to their highly dynamic nature. Although this PPARγ/RXRα two-domain crystal structure has provided important structural insights into the functional complex of nuclear receptor heterodimer, many structural aspects on domain organization are still elusive. For example, what are the arrangements of domains within the homodimers and monomers? Also the role of ligand binding in regulating DNA recognition is still not clear. As such, further work on multi-domain structures remains a top priority of nuclear receptor structural biology.
2, Structure and function of nuclear receptor LBDs
The nuclear receptor LBD interacts with ligands and mediates transcriptional activation in a ligand-dependent fashion. Specifically, the binding of ligands to the LBD determines the recruiting of transcriptional coregulators which triggers induction or repression of target genes. The coregulators include coactivators like the p160 factors also referred to as the steroid receptor coactivators (SRC) family, and corepressors such as SMART (silencing mediator for retinoid and thyroid hormone receptors) and N-CoR (nuclear corepressor) [12; 13]. In addition to ligand binding, the PPARγ LBD has also been suggested to affect DNA binding through interacting with the DBD of its partner RXRα [11]. Given the critical roles of the LBDs in nuclear receptor signaling, the LBD structures that can reveal important clues to the binding of ligands and cofactors has been the focus of nuclear receptor structural study.
In contrast to limited structural information on multi-domain structures, hundreds of structures are available on isolated nuclear receptor LBDs. The first set of LBD structures of the apo-RXR and ligand-bound RAR and TR were published in 1995 by the Moras and Fletterick groups [14; 15; 16]. During past 15 years, LBD structures have been determined for most nuclear receptors (Table 1), which include representative structures from all classic receptors and adopted orphan receptors. These structures are obtained with various LBDs in complex with agonists or antagonists, some with fragments of coactivators or corepressors, and in the form of monomers, dimers, trimers or even tetramers. Despite the importance of orphan receptors and many attempts on structural determination, there are still a few nuclear receptors whose structures remain unsolved, partly due to a lack of ligand information (Table 1). Nevertheless, the rich information provided by the available structures has made it possible to understand the molecular basis of ligand binding and ligand-mediated regulation of nuclear receptors.
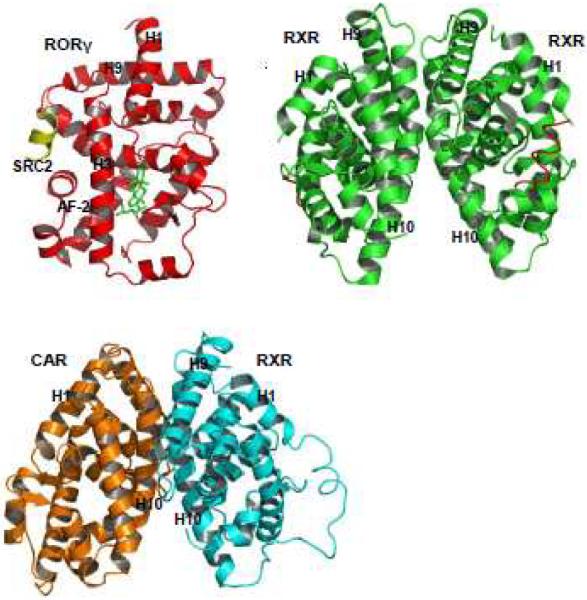
Crystallographic studies have revealed that all nuclear receptors exhibit similar structural features, suggesting similar mechanisms for ligand binding and coregulator recruitment. Table 1 lists some representatives of these three-dimensional structures of nuclear receptor LBDs. The functional units of nuclear receptors can be monomers, homodimers, or heterodimers, depending on specific nuclear receptors (Figure 2). For example, retinoic acid receptor-related orphan receptor γ (RORγ) regulates gene transcription by binding to DNA as a monomer [17]. A typical homodimeric form has been defined with nuclear receptor RXR [18]. In addition, RXR also serves as the common heterodimeric partner for many nuclear receptors, like CAR [19]. Interestingly, the interfaces of both homodimer and heterodimer are mediated through the same helix (H10) from both receptor partners.
Fig. 2. Stereoviews of monomeric and dimeric nuclear receptor structures.
Crystal structure of monomeric LBD of hRORγ bound to 22(R)-hydroxycholesterol (A), RXR homodimer (B) and heterodimer LBDs of CAR and RXR. The interfaces of both RXR homodimer and CAR/RXR heterodimer are mediated through the same helix (H10) from both receptor partners.
The overall structure of nuclear receptor LBD is composed of about 11–13 α-helices that are arranged into a three-layer antiparallel α-helical sandwich, with the three long helices (helices 3, 7, and 10) forming the two outer layers (Figure 2). The middle layer of helices (helices 4, 5, 8 and 9) is present only in the top half of the domain but is missing from the bottom half, thereby creating a cavity, so called ligand-binding pocket, for ligand binding in most receptors. The bound ligands stabilize the nuclear receptor conformation through direct contacts with multiple structural elements of the receptor, including helices H3, H5, H6, H7, H10, and the loop preceding the AF-2 helix. The C-terminal activation region also forms an α-helix (AF-2), which can adopt multiple conformations depending on the nature of the bound ligand. Helices 3, 4 and 12 enclose a shallow hydrophobic groove which is the site for coregulator binding.
2.1. The plastic nature of ligand-binding pocket
Small molecules known as ligands play important roles in modulating the activity of nuclear receptors, since the binding of ligands can induce the conformational changes that determine the recruitment of coactivators or corepressors. As such, the functions of nuclear receptors are tightly associated with their cognate ligands. Given the critical roles of these ligands in human disease, they have been studied intensively in pharmaceutical development.
The first step of nuclear receptor activation is initiated by ligand binding which occurs at the ligand-binding pocket. Interestingly, these well defined pockets of nuclear receptors also are promising sites for drug discovery research. As such, the ligand-binding pocket is an important structural feature of nuclear receptors. Hundreds of structures of nuclear receptors complexed with ligands have revealed a detailed molecular basis for ligand/receptor interaction. The overall hydrophobic nature of the ligand-binding pocket allows the nuclear receptors to interact with many lipid soluble ligands, which explains the promiscuity of some nuclear receptors ability to bind to ligands and also raises challenging questions on ligand selectivity among 48 nuclear receptor members. For example, all-trans retinoic acid is capable of binding to retinoid receptors as well as the retinoic acid receptor-related orphan receptor β (RORβ), and even the chicken ovalbumin upstream promoter-transcription factors (Coup-TFII), albeit with different affinity and functional activity [20; 21]. Another example is that bile salts can activate multiple nuclear receptors, including farnesoid X receptor (FXR), vitamin D receptor (VDR) and pregnane X receptor (PXR) [22]. Recently, natural hydroxycholesterols have been shown to serve as ligands to both LXRs and RORγ [23; 24]. Indeed, many undesired side effects of drugs targeting nuclear receptors are associated with the cross-reactivity of these ligands with other members in the nuclear receptor family. On the other hand, cross-reactivity may also offer opportunities to improve therapeutic efficacy of the ligands by providing additive or complementary effects through simultaneously regulating several related targets. As such, there is a pressing need to develop detailed structure-function relationships of nuclear receptor and ligand interaction to facilitate the discovery of potent ligands that have improved selectivity and reduced side effects. For example, PPAR pan agonists that activate all three α, γ and δ have been shown to have better therapeutic effects than the PPARγ agonists [25]. Taking advantage of multiple available structures of PPAR subtypes, a scaffold-based drug design was used to search for compounds that modulate the activities of all three PPARs [26]. The resulting lead compound, indeglitazar, revealed strong beneficial effects for treating diabetes but with less potency in promoting adipocyte differentiation.
Despite the conserved fold of nuclear receptor LBDs, the ligand-binding pocket is the least conserved region among different nuclear receptor LBDs. Structural comparison and analysis show that several features of the pocket have contributed to the ligand binding affinity and specificity. First, the size of ligand-binding pocket varies greatly, from 100 Å3 in the estrogen-related receptor α (ERRα) to 1200 Å3 in the pregnane X receptor (PXR) and 1400 Å3 in the subtypes of PPARs. The small pocket seen in the ERRα suggests that only ligands with 4–5 carbon atoms or less can fit [27]. In contrast, the large pocket in PXR allows the binding of antibiotic rifampicin, one of the largest structural ligands for nuclear receptors [28]. Aside from large ligands, PXR also has enough room to accommodate the binding of smaller ligands like cholesterol-lowering drug SR12813 in multiple conformations [29]. The specificity of ligand binding is also determined by the shape of the ligand-binding pocket, a property that varies greatly from receptor subtype to subtype, to accommodate a variety of functions mediated by these receptors. The large pocket seen in PPARs has a distinct three-arm Y-shape, allowing these receptors to bind ligands with multiple branches (such as phospholipids and synthetic fibrates), or to bind singly-branched ligands, such as fatty acids, in multiple conformations [30]. Interestingly, homology modeling suggests that the size and shape of the ligand-binding pocket of FXR varies across species to accommodate different bile salt found in fish and mammals [31]. It is likely that cross-species comparison of nuclear receptors may provide insight into the structural features mediating specificity in the ligand recognition.
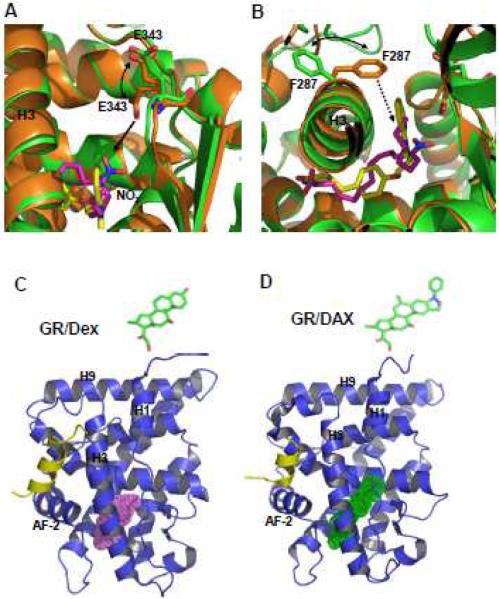
Other than the size and the shape of the pocket, the most pronounced feature of many nuclear receptors is their plastic nature of the ligand-binding pockets which provides the flexibility for nuclear receptors to accommodate specific ligands with a variety of conformations. As such, many nuclear receptors show differential binding modes to different ligands and it's difficult to define a conserved ligand binding pattern for nuclear receptors, adding another layer to the complexity and uncertainty of ligand-mediated nuclear receptor activity. For example, in response to the binding of natural ligand nitrated linoleic acid (LNO2), PPARγ displays a great conformational flexibility to accommodate the bound ligand [32]. Conformational changes in two pocket residues (E343 and F287) are evidenced when the LNO2/PPARγ complex is overlaid on the rosiglitazone/PPARγ structure (Fig 3A and 3B). In response to LNO2 binding, the charged side chain of E343 adopts a second conformation, allowing the receptor to form hydrogen bond with the NO2 group (Fig. 3A). The hydrophobic side chain of F287 also shifts from its rosiglitazone-bound conformation toward the hydrophobic tail (C18) of LNO2, thus stabilizing LNO2 binding by making additional hydrophobic interactions with the LNO2 backbone (Fig. 3B).
Fig. 3. The flexibility of the ligand binding pocket.
(A & B), PPARγ displays conformational flexibility to accommodate natural LNO2 and synthetic compound rosiglitazone. Overlays of the PPARγ/LNO2 structure with the PPARγ/rosiglitazone structure, where LNO2-bound PPARγ is in green and rosiglitazone-bound PPARγ is in gold. The conformational shift of E343 toward NO2 group (A) and the shift of F287 toward the LNO2 backbone (B) are indicated. The hydrophobic interaction between F287 and the LNO2 backbone is shown with a dashed line. (C & D) The pocket size of glucocorticoid receptor shows dramatic changes upon the binding of different ligands. The LBD structures and ligand binding pockets of glucocorticoid receptor are illustrated by pink (for Dex) and green (for DAC) surfaces, respectively. The chemical structures of DAC and Dex are also shown on top of the crystal structures.
In addition to the conformational changes of individual side chains, nuclear receptors can also shift backbones to change the size of ligand binding pocket. Deacylcortivazol (DAC) is a potent glucocorticoid that has been an effective therapy for childhood acute leukemia shown resistant to treatment with other glucocorticoids. The chemical structure of DAC contains a phenyl-pyrazole moiety fused to the 2–3 position of the traditional glucocorticoids like dexamethasone (DEX) (Fig. 3C and 3D). The structure reveals that the DAC-binding pocket in glucocorticoid receptor (GR) LBD is expanded to a volume of 1,070 Å3, which is almost twice the size of the DEX-binding pocket (540 Å3), to fit the binding to the larger ligand DAC [33]. The conformational differences in both the protein backbone and side chains contribute to the expansion of the ligand-binding pocket. A similar example is the pocket in estrogen receptor alpha (ERα) [34]. An estradiol derivative with a prosthetic group, ortho- trifluoromethlyphenylvinyl, binds in a novel extended pocket in the ERα ligand-binding domain and acts as a potent agonist. Such structural plasticity has also been observed for the ecdysone receptor [35]. Structural comparisons indicate that steroidal and non-steroidal ligands dock into different and only partially overlapping ligand-binding pockets.
The structural comparison of the apo-REV-ERBβ and heme-REV-ERBβ complex reveals an even more dramatic change of the REV-ERBβ ligand binding pocket [36; 37]. In contrast to the absence of a pocket observed in apo-REV-ERBβ structure, the binding of heme opens up a pocket of approximately 500 A3 that allows the entry and the docking of heme ligand. The creation of a new REV-ERBβ pocket described here provides an extreme example of the fact that nuclear receptors may have an even greater degree of conformational capacity to adopt a wide range of ligands including various low-affinity metabolic molecules.
Taken together, nuclear receptors have evolved remarkably down to the single residue level to recognize specific ligands by changing the size, shape, and the polar/nonpolar nature of their ligand-binding pockets. More importantly, the plastic nature of the nuclear receptor pocket suggests that these transcriptional factors may be regulated by a variety of small molecules in an unpredicted manner in vivo. From the drug discovery point of view, nuclear receptors may possess even greater potential since the flexible ligand binding pocket allow them to interact with a wider array of pharmacophores. The further characterization of this plastic nature of the ligand-binding pocket may also provide important clues to adopt the remaining orphan receptors whose ligands remain elusive. However, the flexible pocket in nuclear receptors also has made it very challenging to predict the precise mode of ligand binding using virtual ligand screening methodologies. Indeed, it has been shown that the binding pose for more than 50% ligands was not correctly predicted if the information of receptor flexibility was not incorporated [38].
2.2. Structural features that determine the differential recruiting of coactivators and corepressors
The physiological and pharmacological actions of nuclear receptor ligands are carried out through recruiting nuclear receptor coregulators which in turn regulate the expression of the downstream target genes. It has been well established that agonist activates target genes by recruiting coactivator while antagonists repress transcription by inducing the binding of corepressors, referred to as ligand dependent function of nuclear receptors [39]. Moreover, a group of selective nuclear receptor modulators, such as the selective estrogen receptor modulators (SERMs), can selectively regulate the target genes through more complex mechanisms, including the recruitment of specific cofactors in a tissue specific manner [40].
Nuclear receptor coactivators such as SRC-1 contain multiple LXXLL motifs that are used to interact with nuclear receptors. X-ray structures of various nuclear receptor LBDs have revealed a conserved mode of coregulator binding and the critical roles of the AF-2 helix in coregulator binding selectivity. The LXXLL coactivator motif adopts a two-turn α-helix with its three-leucine side chains fitting into the hydrophobic pocket of the LBD. This coactivator docking is further stabilized by two charge clamp residues, which are formed by a highly conserved glutamate residue from the AF-2 helix and a lysine residue from H3. On the other hand, the antagonist-bound receptor is in an inactive state and recruits corepressors [41; 42]. The corepressors bind to LBDs via a conserved LXXXIXXXL/I motif, which is similar to the LXXLL coactivator motif but contains an N-terminal extension. In contrast to coactivators, the longer corepressor motif adopts a three-turn α helix instead of two turns for the coactivator motif, and binds to the same overlapped site as for the LXXLL helix. As such, the AF-2 helix undergoes major conformation change upon ligand binding and must shift to an inactive form to accommodate the larger corepressor helix. It seems clear that the nature of the ligand determines the actual packing position of AF-2. The conformational flexibility of this helix allows the nuclear receptor to sense the presence of the bound ligand, either an agonist or an antagonist, and to recruit the coactivators or corepressors that ultimately determine the transcriptional activation or repression of nuclear receptors.
2.3. Structural features that determine coactivator binding specificity
Upon the binding of an agonist, nuclear receptors use a charge clamp pocket, in part composed of the C-terminal AF-2 helix, to form a hydrophobic groove for binding of the LXXLL motif of the coactivators. However, there are numerous coactivators with distinct functions, each containing multiple LXXLL motifs. Currently there are approximately 300 nuclear receptor coregulators, including the steroid receptor coactivators (SRC1, 2 and 3) and nuclear corepressor N-CoR and SMRT [43]. The functional profile of each nuclear receptor in response to ligand binding is largely determined by the selective usage of transcriptional coregulators since ligand-specific recruitment of coregulators ultimately controls transcriptional output of target genes. Thus, the structural basis for the interaction of a nuclear receptor with a given coactivator will help to elucidate the mechanism of ligand-regulated nuclear receptor activity.
In addition to LXXLL motifs, several structural features that determine cofactor binding have also been revealed by crystal structures. First, nuclear receptors can achieve specific recognition of coactivators by interacting with the variable residues within or flanking the LXXLL motifs. In the case of GR, the specific recognition of the coactivator SRC2 third LXXLL motif is mediated by two additional charged residues of GR that form a second charge clamp to interact with the charged residues specific to that SRC2 motif [44]. In the case of PPARs, the studies reveal that the strong interaction of coactivator PGC-1α with PPARγ is mediated through both hydrophobic and specific polar interactions between PGC-1α and PPARγ [45]. PGC-1α contains several unique features that define its high affinity binding to PPARγ within the structure. The first one is that K145 in the core region of PGC-1α ID1 forms a direct hydrogen bond with N312 in PPARγ. This H-bond further stabilizes the binding of PPARγ and PGC-1α in addition to the hydrophobic interaction between these two molecules. The second feature is the remarkable stability of the PGC-1α ID1 helix by its intramolecular interaction. In the structure, residue S142 forms a direct hydrogen bond that caps the backbone amide of E140 of the LXXLL helix. These intramolecular interactions are likely to stabilize the overall helical structure of the PGC-1α ID1 motif, thus facilitating the hydrophobic docking of this helix into the PPARγ. Together, both of these unique intermolecular and intramolecular contacts serve as a basis for the high affinity and specific binding of PPARγ toward PGC-1α.
The second structural feature that determines the coactivator binding specificity is the presence of atypical motifs other than LXXLL for some coactivators. It turns out that the conserved LXXLL motifs are not always preferred by some nuclear receptors when they recruit coactivators. The coactivators can interact with different receptors using alternative interaction sites. Mapping of PGC-1α defined one nuclear receptor interaction domain with consensus LXXLL motif (residues 144–148, ID1) that is critical for binding nuclear receptors including PPARγ [46]. Subsequent studies identified atypical leucine rich sites that also play roles in recognizing some nuclear receptors. For example, both biochemical data and crystal structure of the ERRα LBD bound to the PGC-1α LLKYL motif (residues 210–214, ID2) reveal the specific binding of this inverted leucine-rich motif to ERRα [27; 47]. While ID2 is not required for PGC-1α to interact with PPARγ, this motif is shown to bind to the nuclear receptor ERRα [27; 47]. Interestingly the ID2 contains an atypical LLXYL motif, which is an inverted LXXLL sequence. Instead of three hydrophobic leucine side chains, ID2 uses two leucine side chains to dock into the groove of the ERRα coactivator binding site. The interaction is further strengthened by the favorable van der Waals contacts between the tyrosine in the PGC-1α ID2 core and ERRα residues L333, I336 and L509 [27]. Another example is steroid hormone receptor androgen receptor (AR). Although the basic mechanism of hormone-dependent activation of AR through LBD resembles those for other nuclear receptors, one key difference is that AR does not interact well with typical LXXLL motifs. Also the coactivators containing such motifs (such as the SRC-1/p160 family) do not potentiate AR activation well. Instead, AR prefers to interact with the FXXLF motif presented in the N-terminal domains of many AR coactivators [48; 49]. The structural studies revealed that FXXLF causes conformational changes in AR side chains contacting the peptide through an induced-fit mechanism [50].
In addition to the small coactivator binding surface on the nuclear receptors, several other regions have also been suggested to contribute to the selective binding of coactivators [51]. For example, the residues flanking the PGC-1α LLXYL motif formed contacts with several other sites of the ERRα LBD including helix 4, the helix 8–9 loop, and the C terminus. More importantly, these interactions are functionally required for ERRα to specifically recruit PGC-1α.
Although many structures of nuclear receptor bound to short peptides of the LXXLL motifs have been solved, little structural information is available for the longer coactivators that help determine the binding specificities, partly due to the large size and also the folding flexibility of most coactivators. For example, all three SRC coactivators are 160 kDa proteins. Without a large piece coactivator structure, it's very difficult to understand how coactivator helps organizing and assembling the nuclear monomer, homodimer and heterodimer using multiple LXXLL motifs. The precise mechanism for recruitment of specific coactivators by nuclear receptors remains to be further explored.
3, Multiple mechanisms for regulating nuclear receptor activity
Ligand binding and ligand-induced cofactor recruitment directly regulates the transcriptional output of nuclear receptors. Generally, nuclear receptors respond differently to distinct ligands and readily exchange their ligands in different environments. As such, ligands play a pivotal role in modulating nuclear receptor activity. Ligands can initiate direct interaction with the AF-2 helix, thereby stabilizing the AF-2 helix in the active conformation as observed in the structures of LBD/ligand complexes of the glucocorticoid receptor and PPARs [44; 52]. A conserved mechanism for ligand-induced activation of nuclear receptors involves positioning the C-terminal AF-2 helix to form a charge clamp pocket, which permits the receptor to interact efficiently with coactivator proteins [53]. However, it is also possible that the bound ligands serve as structural components for nuclear receptors in cells that provide structural integrity without regulatory functions. One known such example is the binding of fatty acids to the HNF4 family of nuclear receptors where fatty acids are used as a structural cofactor and can not be exchanged [54; 55].
The understanding of nuclear receptor activation is further complicated by the ligand-independent effects that exist in nuclear receptors like CAR, Coup and Nurr1. Crystal structures of nuclear receptors have provided us rich insights into the high basal activity that is independent of ligand binding. The followings are several possible molecular mechanisms regulating ligand-independent activity of nuclear receptors. 1) The first suggested structural basis is the rigid small hydrophobic pocket that can mimic the roles of ligands to stabilize the position of AF-2. One example of this structural mechanism is seen in Nurr1 structure [56]. As an orphan nuclear receptor, Nurr1 contains a cavity filled with side chains from several hydrophobic residues instead of ligands. The AF-2 helix is predisposed in the active conformation that is stabilized by intra-molecular interactions. 2) A second mechanism is the expanded dimer interface that stabilizes the transcriptional complex and also the contribution from the RXR partner as seen from CAR/RXR. Indeed, the RXR dimerization has been shown to facilitate the transcriptional complex to bind DNA, recruit coactivators and modulate the target gene expression. 3) Regulation of nuclear receptor AF-1 provides another mechanism for regulation of ligand-independent nuclear receptor activity. Many coregulators have been shown to regulate nuclear receptor activity through binding to AF-1. For example, the chromatin-modifying protein Brahma-related gene-1 (BRG1) is able to bind to AF-1 and regulates nuclear receptor transcriptional activity through targeting chromatin remodeling [57]. 4) Post-transcriptional modifications, like phosphorylaton or methylation, may also be responsible for some aspects of ligand-independent action. One such example is the phosphorylation of serine residue 112 of PPARγ which is located at the hinge region. This genetic modulation of PPARγ reduces its transcriptional activity and also regulates insulin sensitivity in vivo [58; 59], suggesting the importance of post-transcriptional modification in regulating nuclear receptor activity. 5) Possible unidentified endogenous ligands that are difficult to detect due to their level may be tissue specific and transient. Given the plastic nature of the ligand binding pocket, the binding mode for ligand shows ligand-specificity and it's difficult to define a conserved pattern for all ligands. Structure of Coup-TFII in apo form purified from E. coli clearly shows repressive conformation since the AF-2 occupies the coactivator binding site [21]. However, Coup-TFII is able to interact with coactivators and activate reporter gene expression in cell based assays, suggesting that some unknown ligands in vivo can induce conformational changes of Coup-TFII to an activated state.
Indeed, the constitutive activity of many orphan receptors that was strongly believed ligand-independent. However, the constitutive activity has turned out to be ligand-dependent owing to the identification of ligands later on. Crystallography and biochemical approaches revealed the phospholipids as ligands for LRH and SF1 and demonstrated that these ligands indeed are able to regulate coactivators binding by these nuclear receptors [60; 61]. Recent studies also suggest that heme serves as ligand for orphan nuclear receptor Rev-erbs [62; 63]. The heme ligand plays functional roles in Rev-erbs dependent corepressor recruitment and gene regulation. For Nurr1, although its apo-structure shows no pocket for the ligand binding, the potential ligand may be able induce conformational changes that open up the pocket to accommodate ligand docking. Indeed, it has been shown that a small-molecule ligand, cytosporone B, activates Nur77, which is structurally similar to Nurr1 [64]. Overall, the constitutive activities of these nuclear receptors are not truly ligand-independent and the recent discoveries of their ligands have led to unexpected insights into signaling mechanisms.
4, Concluding remarks
The biological functions of nuclear receptors have been studied extensively, and their medical importance has been highlighted by the therapeutic uses of their ligands on human diseases. However, clinical use of many ligands is limited by a number of side effects. It is thus critical to develop detailed structure-function relationships and to discover potent nuclear receptor ligands that retain the beneficial activities without the undesired side effects. Crystal structures have revealed critical insights into the mechanisms of ligand-mediated nuclear signaling, including ligand binding affinity and specificity, and differential recruitment of coregulators. The structure-based design of agonists and antagonists that can either induce or block the activities of nuclear receptors will provide promising therapeutic strategies.
As a DNA-binding and ligand-regulated transcription factor, the function of nuclear receptor requires an integrated structure of the DBD followed by an intact hinge region and the LBD. Functional analysis of integrated DBD-LBD structures will provide a comprehensive understanding on how the signaling information flow from nuclear receptor ligands to their target genes. However, the limited amount of information on multi-domain structures is a serious deficiency considering the roles and functions of these receptors in biology. As much as the surprise we learned from LBD structures, the excitement for the integrated multi-domain structures of nuclear receptors is yet to come.
Acknowledgements
The authors thank the grants support from the National Institutes of Health Grant DK081757, the American Heart Association, and the Science Planning Program of Fujian Province (2009J1010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Willson TM, Moore JT. Genomics versus orphan nuclear receptors--a half-time report. Mol Endocrinol. 2002;16:1135–44. doi: 10.1210/mend.16.6.0849. [DOI] [PubMed] [Google Scholar]
- [3].Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- [4].Kliewer SA, Lehmann JM, Willson TM. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284:757–60. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- [5].Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–90. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- [6].Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–52. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–26. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- [8].Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–54. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- [9].Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci U S A. 2005;102:9505–10. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci U S A. 2008;105:18390–5. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–6. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- [13].Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu Rev Physiol. 2005;67:309–33. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- [14].Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–82. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- [15].Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–9. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- [16].Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–7. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- [17].Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, Redinbo MR. Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. 2007;21:1028–38. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- [18].Egea PF, Mitschler A, Moras D. Molecular recognition of agonist ligands by RXRs. Mol Endocrinol. 2002;16:987–97. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- [19].Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The Nuclear Xenobiotic Receptor CAR; Structural Determinants of Constitutive Activation and Heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- [20].Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol. 2003;10:820–5. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- [21].Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, Xu HE. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krasowski MD, Ni A, Hagey LR, Ekins S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol Cell Endocrinol. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–31. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- [24].Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 24:923–9. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Evans JL, Lin JJ, Goldfine ID. Novel approach to treat insulin resistance, type 2 diabetes, and the metabolic syndrome: simultaneous activation of PPARalpha, PPARgamma, and PPARdelta. Curr Diabetes Rev. 2005;1:299–307. doi: 10.2174/157339905774574365. [DOI] [PubMed] [Google Scholar]
- [26].Artis DR, Lin JJ, Zhang C, Wang W, Mehra U, Perreault M, Erbe D, Krupka HI, England BP, Arnold J, Plotnikov AN, Marimuthu A, Nguyen H, Will S, Signaevsky M, Kral J, Cantwell J, Settachatgull C, Yan DS, Fong D, Oh A, Shi S, Womack P, Powell B, Habets G, West BL, Zhang KY, Milburn MV, Vlasuk GP, Hirth KP, Nolop K, Bollag G, Ibrahim PN, Tobin JF. Scaffold-based discovery of indeglitazar, a PPAR pan-active anti-diabetic agent. Proc Natl Acad Sci U S A. 2009;106:262–7. doi: 10.1073/pnas.0811325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kallen J, Schlaeppi JM, Bitsch F, Filipuzzi I, Schilb A, Riou V, Graham A, Strauss A, Geiser M, Fournier B. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem. 2004;279:49330–7. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- [28].Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19:1125–34. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- [29].Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–33. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- [30].Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator- activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- [31].Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008;49:1577–87. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li Y, Zhang J, Schopfer FJ, Martynowski D, Garcia-Barrio MT, Kovach A, Suino-Powell K, Baker PR, Freeman BA, Chen YE, Xu HE. Molecular recognition of nitrated fatty acids by PPAR gamma. Nat Struct Mol Biol. 2008;15:865–7. doi: 10.1038/nsmb.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Suino-Powell K, Xu Y, Zhang C, Tao YG, Tolbert WD, Simons SS, Jr., Xu HE. Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol. Mol Cell Biol. 2008;28:1915–23. doi: 10.1128/MCB.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nettles KW, Bruning JB, Gil G, O'Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, Joachimiak A, Greene GL. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–8. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Billas IM, Iwema T, Garnier JM, Mitschler A, Rochel N, Moras D. Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature. 2003;426:91–6. doi: 10.1038/nature02112. [DOI] [PubMed] [Google Scholar]
- [36].Woo EJ, Jeong DG, Lim MY, Jun Kim S, Kim KJ, Yoon SM, Park BC, Eon Ryu S. Structural insight into the constitutive repression function of the nuclear receptor Rev-erbbeta. J Mol Biol. 2007;373:735–44. doi: 10.1016/j.jmb.2007.08.037. [DOI] [PubMed] [Google Scholar]
- [37].Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, Botchkarev A, Krause HM, Edwards A. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta. PLoS Biol. 2009;7:e43. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr Opin Struct Biol. 2008;18:178–84. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–87. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- [40].Wu YL, Yang X, Ren Z, McDonnell DP, Norris JD, Willson TM, Greene GL. Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol Cell. 2005;18:413–24. doi: 10.1016/j.molcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- [41].Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–7. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- [42].Phelan CA, Gampe RT, Jr., Lambert MH, Parks DJ, Montana V, Bynum J, Broderick TM, Hu X, Williams SP, Nolte RT, Lazar MA. Structure of Rev-erbalpha bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 17:808–14. doi: 10.1038/nsmb.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lonard DM, O'Malley B W. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- [44].Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- [45].Li Y, Kovach A, Suino-Powell K, Martynowski D, Xu HE. Structural and biochemical basis for the binding selectivity of peroxisome proliferator-activated receptor gamma to PGC-1alpha. J Biol Chem. 2008;283:19132–9. doi: 10.1074/jbc.M802040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu Y, Chin WW, Wang Y, Burris TP. Ligand and coactivator identity determines the requirement of the charge clamp for coactivation of the peroxisome proliferator-activated receptor gamma. J Biol Chem. 2003;278:8637–44. doi: 10.1074/jbc.M210910200. [DOI] [PubMed] [Google Scholar]
- [47].Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–74. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- [48].He B, Minges JT, Lee LW, Wilson EM. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J Biol Chem. 2002;277:10226–35. doi: 10.1074/jbc.M111975200. [DOI] [PubMed] [Google Scholar]
- [49].Hsu CL, Chen YL, Yeh S, Ting HJ, Hu YC, Lin H, Wang X, Chang C. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J Biol Chem. 2003;278:23691–8. doi: 10.1074/jbc.M211908200. [DOI] [PubMed] [Google Scholar]
- [50].He B, Gampe RT, Jr., Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell. 2004;16:425–38. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- [51].Greschik H, Althage M, Flaig R, Sato Y, Chavant V, Peluso-Iltis C, Choulier L, Cronet P, Rochel N, Schule R, Stromstedt PE, Moras D. Communication between the ERRalpha homodimer interface and the PGC-1alpha binding surface via the helix 8–9 loop. J Biol Chem. 2008;283:20220–30. doi: 10.1074/jbc.M801920200. [DOI] [PubMed] [Google Scholar]
- [52].Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, Jr., McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–24. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure (Camb) 2003;11:741–6. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- [54].Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;21:21. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- [55].Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr., Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Willson TM, Williams SP. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure (Camb) 2002;10:1225–34. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- [56].Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–60. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- [57].Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 1998;396:377–80. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- [59].Rangwala SM, Rhoades B, Shapiro JS, Rich AS, Kim JK, Shulman GI, Kaestner KH, Lazar MA. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell. 2003;5:657–63. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- [60].Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15:708–15. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- [61].Forman BM. Are those phospholipids in your pocket? Cell Metab. 2005;1:153–5. doi: 10.1016/j.cmet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- [62].Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–13. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–9. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- [64].Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, Li G, Xu Q, Zhang M, Weimer BC, Chen D, Cheng Z, Zhang L, Li Q, Li S, Zheng Z, Song S, Huang Y, Ye Z, Su W, Lin SC, Shen Y, Wu Q. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4:548–56. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- [65].Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek SR, Jr., Weinmann R, Einspahr HM. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc Natl Acad Sci U S A. 2001;98:4904–9. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–8. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- [67].Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. Embo J. 1999;18:4608–18. doi: 10.1093/emboj/18.17.4608. In Process Citation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li Y, Suino K, Daugherty J, Xu HE. Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol Cell. 2005;19:367–80. doi: 10.1016/j.molcel.2005.06.026. [DOI] [PubMed] [Google Scholar]
- [69].Fagart J, Huyet J, Pinon GM, Rochel M, Mayer C, Rafestin-Oblin ME. Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension. Nat Struct Mol Biol. 2005;12:554–5. doi: 10.1038/nsmb939. [DOI] [PubMed] [Google Scholar]
- [70].Bledsoe RK, Madauss KP, Holt JA, Apolito CJ, Lambert MH, Pearce KH, Stanley TB, Stewart EL, Trump RP, Willson TM, Williams SP. A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J Biol Chem. 2005;280:31283–93. doi: 10.1074/jbc.M504098200. [DOI] [PubMed] [Google Scholar]
- [71].Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–6. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- [72].Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. Embo J. 2001;20:5822–31. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5:289–98. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- [74].Germain P, Kammerer S, Perez E, Peluso-Iltis C, Tortolani D, Zusi FC, Starrett J, Lapointe P, Daris JP, Marinier A, de Lera AR, Rochel N, Gronemeyer H. Rational design of RAR-selective ligands revealed by RARbeta crystal stucture. EMBO Rep. 2004;5:877–82. doi: 10.1038/sj.embor.7400235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5:173–9. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- [77].Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, Sherman MA, Forman BM, Fernandez EJ. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–17. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16:919–28. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- [79].Greschik H, Wurtz JM, Sanglier S, Bourguet W, van Dorsselaer A, Moras D, Renaud JP. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol Cell. 2002;9:303–13. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- [80].Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, Rosenfeld JM, Alvarez JG, Noel JP, Nicolaou KC, Evans RM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11:1079–92. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mi LZ, Devarakonda S, Harp JM, Han Q, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol Cell. 2003;11:1093–100. doi: 10.1016/s1097-2765(03)00112-6. [DOI] [PubMed] [Google Scholar]
- [82].Williams S, Bledsoe RK, Collins JL, Boggs S, Lambert MH, Miller AB, Moore J, McKee DD, Moore L, Nichols J, Parks D, Watson M, Wisely B, Willson TM. X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by a histidine-tryptophan switch. J Biol Chem. 2003;278:27138–43. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]
- [83].Svensson S, Ostberg T, Jacobsson M, Norstrom C, Stefansson K, Hallen D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. Embo J. 2003;22:4625–33. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator- activated receptor-gamma. Nature. 1998;395:137–43. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- [85].Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- [86].Love JD, Gooch JT, Benko S, Li C, Nagy L, Chatterjee VK, Evans RM, Schwabe JW. The structural basis for the specificity of retinoid-X receptor-selective agonists: new insights into the role of helix H12. J Biol Chem. 2002;277:11385–91. doi: 10.1074/jbc.M110869200. [DOI] [PubMed] [Google Scholar]
- [87].Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol Cell. 2003;11:1575–85. doi: 10.1016/s1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- [88].Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–55. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- [89].Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]