The ability of stem cells to differentiate into specialized lineages within a specific microenvironment is vital for regenerative medicine. For harnessing the full potential of stem cells for regenerative therapies, it is important to investigate and understand the function of three types of micro-environmental cues—soluble signals, cell–cell interactions, and insoluble (physical) signals—that dynamically regulate stem cell differentiation.[1] Neural stem cells (NSCs) are multipotent and differentiate into neurons and glial cells,[2] which can provide essential sources of engraftable neural cells for devastating diseases such as Alzheimer’s disease,[3] Parkinson’s disease[4] and spinal cord injury.[5] One of the major challenges involved in the differentiation of NSCs is to identify and optimize factors which result in an increased proportion of NSCs differentiating into neurons as opposed to glial cells. To this end, soluble cues such as brain-derived neurotrophic factor (BDNF),[6] sonic hedgehog (Shh),[7] retinoic acid (RA),[6c] and neuropathiazol[8] have been shown to significantly increase neuronal differentiation of NSCs in vitro. However, the research toward studying the function of the other two microenvironmental cues (cell–cell interactions and insoluble cues) during the neurodifferentiation of NSCs is limited, mainly due to the lack of availability of methods for the investigation.[9] While various aspects such as cell–cell interactions,[10] combinations of extracellular matrix (ECM) proteins,[1a,11] and physical properties of substrates have been shown to play a vital role in determining the fate of other adult stem cells such as mesenchymal stem cells (MSCs),[12] cardiac stem cells,[13] and hematopoetic stem cells,[14] little is known about the influence of such factors on the neuronal differentiation of NSCs. Therefore, there is a pressing need to develop methods for investigating the role of cell–cell interactions and insoluble signals in selectively inducing the differentiation of NSCs into specific neural cell lineages.
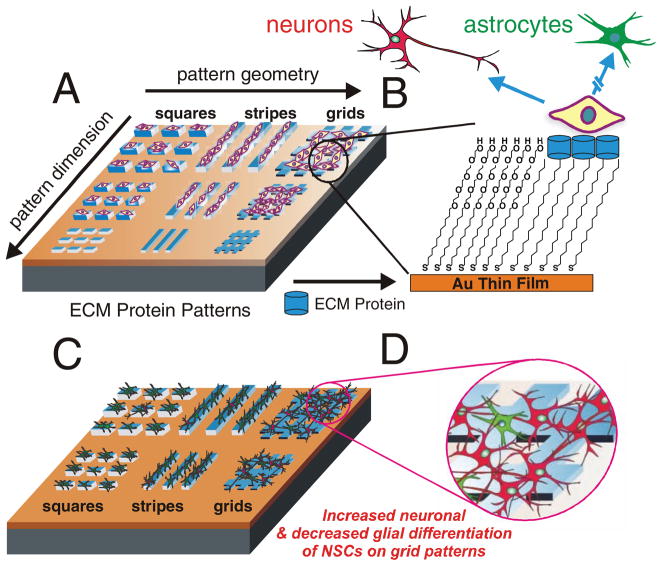
Herein, we demonstrate how ECM protein patterns can be used to investigate the effect of physical cues combined with cell–cell interactions on the differentiation of NSCs. Bio-surface chemistry combined with soft lithography was used to generate combinatorial patterns with varying geometries and dimensions of ECM proteins (e.g., laminin, fibronectin, and collagens) to study the influence of surface features and ECM compositions on the differentiation of NSCs. We hypothesized that the ECM protein patterns with variant geometries and dimensions would provide physical cues (e.g., mechanical or topographical cues), as well as guide cell–cell and cell–ECM interactions in a controlled manner, both of which would ultimately lead to a pattern geometry-dependent and pattern dimension-dependent neuronal and glial differentiation (Figure 1). Our data confirmed that the difference in the extent of neuronal and glial differentiation of NSCs on the ECM protein patterns was entirely due to the pattern geometry and dimension, as all the experiments were carried out in the absence of exogenous factors that promote neurogenesis; this suggests that NSCs can undergo differentiation by purely sensing the difference in ECM pattern geometries and dimensions.
Figure 1.
A schematic diagram of our approaches. A) The fabrication of ECM protein patterns and their application for NSC differentiation. B) The selective attachment of NSCs on the protein patterns and differentation into two different kinds of neural cells. C) The differentiation of NSCs into either neurons (red) or astrocytes (green) on the protein patterns. D) Increased neuronal differentiation on the grid patterns, as compared to the stripes and squares.
Extracellular matrix protein patterns with variant geometries and dimensions were fabricated by initially patterning octadecanethiol (ODT, 5 mm in ethanol), a hydrophobic alkanethiol, which formed self-assembled monolayers (SAMs) of squares, stripes, and grids on glass substrates coated with a thin film (12 nm) of gold. In order to minimize the nonspecific attachment of laminin, the background of the substrates was passivated by incubating in a solution (5 mm in ethanol) of tetraethylene glycol terminated alkanethiol [EG4-(CH2)11-SH, 12 h] (See Supporting Information for synthesis and characterization). After passivating the background, a solution of ECM protein [e.g., laminin (10 μg mL−1) in phosphate buffered saline (PBS) buffer, pH = 7.4] was added onto the substrates (3 h) and was preferentially adsorbed onto the hydrophobic regions (ODT patterns). The selective adsorption of laminin on hydrophobic regions was consistent with the results of other groups[15] and was also confirmed by immunostaining using anti-laminin IgG (See Supporting Information, Figure S1). Only the patterned regions, coated with ECM proteins, promoted cell adhesion and growth whereas the rest of the substrate remained inert (Figure 1). We similarly patterned several different ECM proteins including fibronectin and collagen, but found that laminin provided the optimum microenviromental cues for NSC adhesion and growth. Hence, all our differentiation studies were carried out using laminin patterns.
To examine the effect of the ECM protein patterns on stem cell differentiation, primary rat hippocampal neural stem cells (Millipore) were first expanded and maintained in an undifferentiated state in a homogeneous monolayer on a polyornithine and laminin-coated Petri dish in a defined serum-free growth medium [DMEM/F12 supplemented with B27 and basic fibroblast growth factor (bFGF, 20 ng mL−1)]. For obtaining reproducible and consistent results, all experiments were carried out using NSCs from passages 2–5 at a constant cell density of 150 000 cells per substrate (1.5 cm × 1.5 cm), which was optimum for cell growth without clustering. Arresting the proliferation of NSCs and initiating their spontaneous differentiation was achieved by withdrawing bFGF from the culture medium (resulting in basal medium), without the additional treatment with exogenous factors (proteins and small molecules). The basal medium (2 mL) containing the NSCs (75 000 cells mL−1) was put in a single well of a 6-well plate containing a substrate with laminin patterns. After the NSCs attached onto the laminin patterns (1 h), the substrates were rinsed with copious amounts of media in order to minimize nonspecific interactions of NSCs with the passivated areas, and then incubated in fresh basal medium. The media was exchanged with fresh media every other day. During our screening approach to investigate the function of physical cues on neuronal differentiation of NSCs, we monitored the differentiation on ECM protein patterns by using two orthogonal assays, namely immunocytochemical and morphological assays. To assess the differentiation of NSCs, the down-regulation of the NSC marker (Nestin) and the geometry-dependent expression of the neuronal marker (β-III Tubulin, TuJ1) and glial marker (glial fibrillary acidic protein, GFAP) were monitored. In addition, the development of branches or spindle-like morphologies, and neurite outgrowths were observed by using an inverted phase contrast microscope (Zeiss Axiovert 200M equipped with AxioCam CCD).
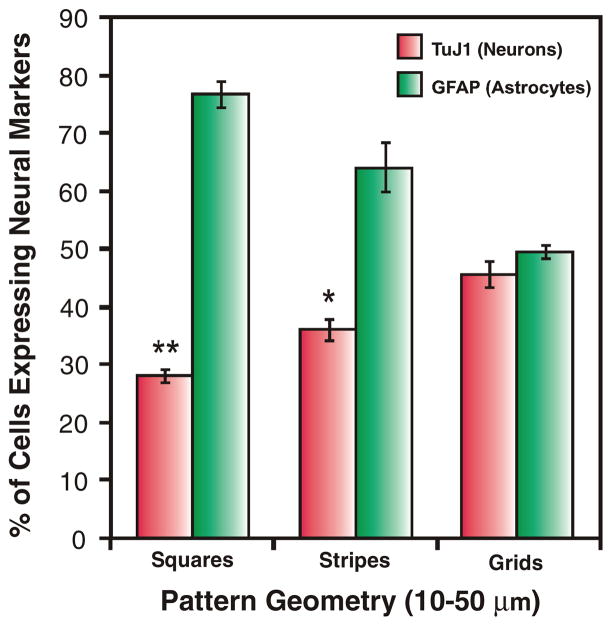
Patterns of ECM proteins with different geometries contributing to adhesion, proliferation, growth and migration of various cells (including stem cells) have been reported.[16] In addition, reports from the literature have shown cell–cell interactions to play a critical role in the differentiation of adult stem cells. For instance, it was recently shown that cell–cell interactions played an important role in the osteogenic (bone) differentiation of MSCs.[10] To study the influences pattern geometries and cell–cell interactions on the differentiation of NSCs, we initially patterned the NSCs on stripes of laminin, which promoted one-way interactions in a controlled manner (Figure 2.A1). We found that after six days, 36% of NSCs on the isolated stripes differentiated into neurons (Figure 2.A2 and Figure 3). At the same time we observed that 64.3% of NSCs on these stripes differentiated into astrocytes (Figure 2.A3 and Figure 3).
Figure 2.
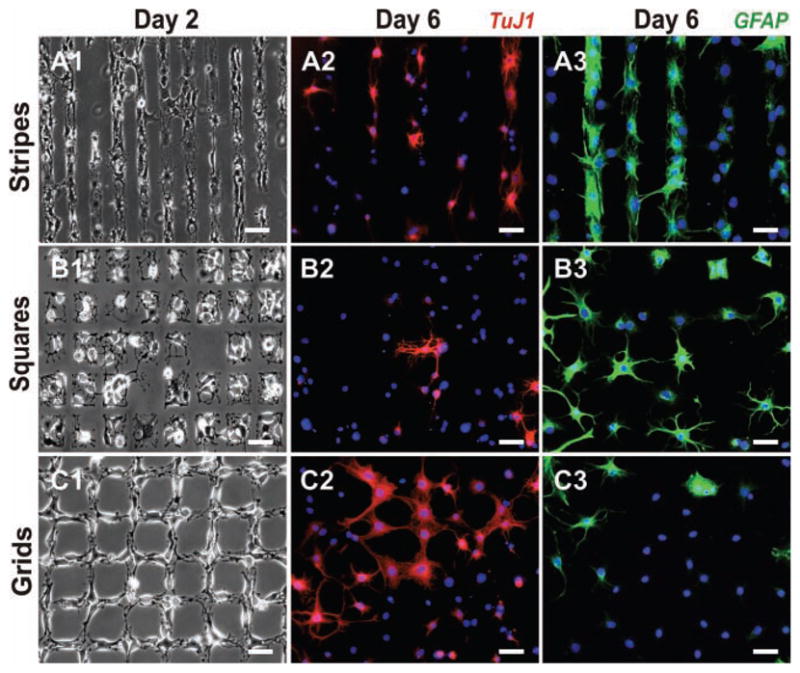
Growth and differentiation of NSCs on the laminin patterns. Phase contrast images show NSC attachment and growth on stripes (A1), squares (B1), and grids (C1) on Day 2 after seeding. Fluorescent images of cells stained for the neuronal marker TuJ1 (red) and nucleus (blue) show the extent of neuronal differentiation of NSCs on stripes (A2), squares (B2), and grids (C2) on Day 6 after seeding. Similarly, cells stained for astrocyte marker GFAP (green) and nucleus (blue) show the extent of glial differentiation on stripes (A3), squares (B3), and grids (C3) on Day 6 after seeding. Scale bars: 50 μm.
Figure 3.
Quantitative comparison of the percentage of cells expressing the neuronal marker TuJ1 and astrocyte marker GFAP on laminin patterns of squares, stripes and grids. Six days after seeding, the differentiated cells were counted and plotted as a ratio of TuJ1-positive cells or GFAP-positive cells to the total number of cells (n = 3). Student’s unpaired t-test was used for evaluating the statistical significance for cells stained for TuJ1 on stripes and squares, compared to those on grids. (* = P < 0.01, ** = P < 0.001).
To further confirm the influence of such interactions on the differentiation of NSCs, we used square patterns of laminin to isolate NSCs and restrict their growth within the square patterns (Figure 2.B1). We hypothesized that the differentiation behaviour of NSCs can be considerably influenced by limiting cell–cell interactions. We observed that NSCs patterned on squares, having the same dimensions and spaces as the stripes, differentiated into neurons to a considerably lesser extent (28.1%, Figure 2.B2 and 3) as compared to the NSCs involved in one-way interactions on the striped laminin patterns. At the same time, the number of NSCs that differentiated into astrocytes increased considerably on squares –76.9% on squares as compared to 64.3% on stripes (Figure 2.B3 and 3). Thus, the reduced cell–cell interactions with the NSCs on the surrounding patterns may have led to reduced neuronal differentiation and increased glial differentiation of the NSCs. Based on the observed differentiation of NSCs on stripes and squares, we further hypothesized that using specific pattern geometries promoting cell–cell interactions could lead to higher neuronal differentiation. For this purpose, we used grid patterns of laminin, having the same dimensions as the stripe and square patterns, for NSC growth and differentiation. The grid patterns were specifically designed to increase cell–cell interactions in a controlled manner (by promoting two-way interactions, Figure 2.C1). After six days in basal medium, as compared to the NSCs patterned on stripes and squares of laminin, we observed a remarkable increase in the number of NSCs that underwent neuronal differentiation (45.6%, Figure 2.C2 and 3) and a decrease in the number of cells that underwent glial differentiation on grid patterns of laminin (49.6%, Figure 2.C3 and 3). All the experiments were repeated several times under the same conditions. To maintain consistency and minimize the effects from other variables, we fabricated and used PDMS stamps to generate ECM protein patterns of all the three geometries (having the same dimensions and spacing) on the same substrate. Using this method, we could reproduce and confirm our results with relative ease. Neuronal and glial differentiation of NSCs was also monitored on control substrates which included substrates coated with laminin (unpatterned) and substrates without laminin. The NSCs on substrates without laminin did not attach and failed to survive, whereas 32.5% of the NSCs on the unpatterned substrates coated with laminin differentiated into neurons and 71.2% of the NSCs differentiated into astrocytes six days after seeding.
In addition to investigating the effect of pattern-geometry, we also studied the effect of dimensions on NSC differentiation. To this end, we generated ten different dimensions for each of the geometries, ranging from sizes as small as 10 μm and as large as 250 μm (Figure 4B). Interestingly, for the three different geometries above 50 μm, we observed little difference in the percentage of NSCs undergoing neuronal and glial differentiation. The result observed for pattern dimensions above 50 μm was similar to that observed with unpatterned substrates. We believe the cells may not be able to sense the difference in pattern geometries above 50 μm and thus show similar behaviour to the cells on unpatterned substrates. Since the NSCs showed remarkable difference in differentiation on patterns ranging from 10–50 μm, all of our statistical analysis and investigation was done using pattern features within this range.
Figure 4.
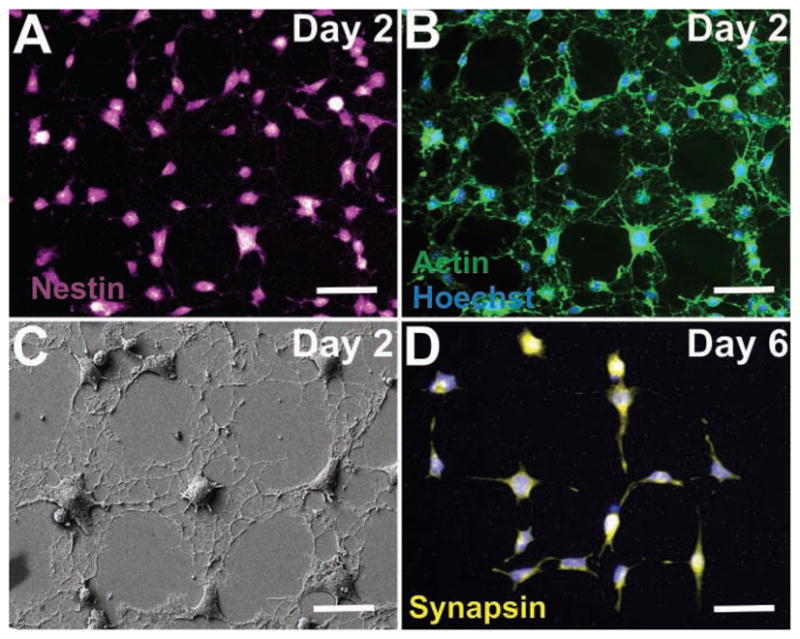
NSC alignment and differentiation on combinatorial ECM patterns. A) NSCs on grids of laminin express the neural stem cell marker, nestin (purple) on Day 2 after seeding, thus confirming that the NSCs are undifferentiated. B) NSCs stained for actin (green) show extensive spreading and cell–cell interactions on grid patterns of laminin on Day 2 after seeding, confirming that the NSCs, while still in the undifferentiated state, extensively interact with each other. C) SEM image of NSCs on Day 2 after seeding, showing the early alignment and extension of processes on grid patterns of laminin. D) NSCs previously shown to extend and grow on the grid patterns of laminin undergo neuronal differentiation and express the neuronal marker synapsin (pseudocolored yellow) on Day 6 after seeding. Scale bars: 20 μm.
We observed that the laminin patterns of all three geometries enabled the NSCs to attach and grow within a day or two day after seeding. By staining for actin using phalloidin and using field emission scanning electron microscopy (FESEM, Zeiss Gemini), we further observed that the cytoskeleton of the NSCs aligned well within the laminin patterns, guiding cellular morphology and interactions (Figure 4B,C). To confirm that the laminin patterns influenced morphological changes before differentiation (as opposed to an early differentiation of NSCs which might have caused a change in alignment and morphology), the NSCs were immunostained for the neural stem cell marker nestin two days after seeding in basal medium. We observed that most of the NSCs that aligned along the patterns, stained positive for nestin (Figure 4A), confirming that cells were in an undifferentiated (multipotent) state when they aligned along the patterns (See Supporting information, Figure S2 for NSCs on squares and stripes stained for actin and nestin). We further confirmed neuronal differentiation of NSCs on the laminin patterns using synapsin as another neuronal marker in addition to TuJ1. After six days in basal medium, a remarkably high number of the NSCs growing along the grid patterns of laminin expressed synapsin (Figure 4D). In addition, colocalization of TuJ1 and synapsin was observed within the NSCs differentiated on the grid patterns, confirming that the neurons expressed both neuronal markers (Supporting information, Figure S3).
In summary, we fabricated and utilized patterns of ECM proteins for modulating the extent of neuronal and glial differentiation of NSCs in the absence of soluble cues such as small molecules and exogenous proteins. Potentially, our approach and methodology can be helpful for deconvoluting physical cues and cell–cell interactions from complex micro-environmental cues. More detailed mechanistic studies on how physical cues modulate the signaling cascades and the signaling pathways that are primarily involved in stem cell differentiation induced by such factors are currently under investigation. The implications of our results could also potentially be significant for tissue engineering for brain and spinal cord injuries, where NSCs or NSC-based differentiated cells can be transplanted into the damaged regions with scaffolds. For example, scaffolds having patterns promoting cell–cell interactions in a controlled manner could potentially lead to increased neuronal differentiation in vivo. Even though we have explored only proof-of-concept experiments focusing on differentiation of NSCs, a similar strategy could be extended to study and control the fate of other stem cells, such as MSCs and embryonic stem cells (work in progress). Our results substantiate the importance of pattern dimensions, pattern geometries, and cell–cell interactions in controlling stem cell fate.
Supplementary Material
Acknowledgments
A. S. and S. S. have contributed equally to the authorship of this paper. We acknowledge John Kim and Neal Patel for their help in the experimental procedures. This work was supported by the NIH Director’s Innovator Award [(1DP20D006462–01), K.-B. L.] and the N.J. Commission on Spinal Cord grant [(09–3085-SCR-E-0), K.-B. L.]. S. H. acknowledges the support from the NRF grant (2009–0079103) and the System 2010 program. K.-B. L. acknowledges partial support from Bioforce Nanosciences.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Aniruddh Solanki, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Shreyas Shah, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Kevin A. Memoli, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Sung Young Park, Department of Physics, Department of Biophysics and Chemical Biology, Interdisciplinary Program in Nano-Science and Technology, Seoul National University, Seoul, South Korea.
Prof. Seunghun Hong, Interdisciplinary Program in Nano-Science and Technology, Seoul National University, Seoul, South Korea
Prof. Ki-Bum Lee, Email: kblee@rutgers.edu, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA, http://rutchem.rutgers.edu/~kbleeweb/
References
- 1.a) Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Flaim CJ, Chien S, Bhatia SN. Nat Methods. 2005;2:119. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]; c) Flaim CJ, Teng D, Chien S, Bhatia SN. Stem Cells and Dev. 2008;17:29. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 2.a) Gage FH, Ray J, Fisher LJ. Annu Rev Neurosci. 1995;18:159. doi: 10.1146/annurev.ne.18.030195.001111. [DOI] [PubMed] [Google Scholar]; b) Gage FH. Science. 2000;287:1433. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 3.Roberson ED, Mucke L. Science. 2006;314:781. doi: 10.1126/science.1132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Bjorklund A, Lindvall O. Nat Neurosci. 2000;3:537. doi: 10.1038/75705. [DOI] [PubMed] [Google Scholar]; b) Kim JH, Auerbach JM, Rodriguez-Gomez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sanchez-Pernaute R, Bankiewicz K, McKay R. Nature. 2002;418:50. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]; c) Lindvall O, Kokaia Z. Nature. 2006;441:1094. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 5.a) Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisen J. Cell. 1999;96:25. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]; b) Martino G, Pluchino S. Nat Rev Neurosci. 2006;7:395. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]; c) Barnabe-Heider F, Frisen J. Cell Stem Cell. 2008;3:16. doi: 10.1016/j.stem.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 6.a) Bath KG, Lee FS. Dev Neurobiol. 2010;70:339. doi: 10.1002/dneu.20781. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Matsuda N, Lu H, Fukata Y, Noritake J, Gao HF, Mukherjee S, Nemoto T, Fukata M, Poo MM. J Neurosci. 2009;29:14185. doi: 10.1523/JNEUROSCI.1863-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Song HJ, Stevens CF, Gage FH. Nat Neurosci. 2002;5:438. doi: 10.1038/nn844. [DOI] [PubMed] [Google Scholar]
- 7.a) Li XJ, Zhang XQ, Johnson MA, Wang ZB, LaVaute T, Zhang SC. Development. 2009;136:4055. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Szabo NE, Zhao TY, Zhou XL, Alvarez-Bolado G. J Neurosci. 2009;29:2453. doi: 10.1523/JNEUROSCI.4524-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warashina M, Min KH, Kuwabara T, Huynh A, Gage FH, Schultz PG, Ding S. Angew Chem, Int Ed. 2006;45:591. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]
- 9.a) Banerjee A, Arha M, Choudhary S, Ashton RS, Bhatia SR, Schaffer DV, Kane RS. Biomaterials. 2009;30:4695. doi: 10.1016/j.biomaterials.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Leipzig ND, Shoichet MS. Biomaterials. 2009;30:6867. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]; c) Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophys J. 2008;95:4426. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Magnusson AK, Linderholm P, Vieider C, Ulfendahl M, Erlandsson A. J Neurosci Res. 2008;86:2363. doi: 10.1002/jnr.21688. [DOI] [PubMed] [Google Scholar]
- 10.Tang J, Peng R, Ding JD. Biomaterials. 2010;31:2470. doi: 10.1016/j.biomaterials.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Harper MM, Ye EA, Blong CC, Jacobson ML, Sakaguchi DS. J Mol Neurosci. 2010;40:269. doi: 10.1007/s12031-009-9211-x. [DOI] [PubMed] [Google Scholar]
- 12.a) Park SY, Namgung S, Kim B, Im J, Kim Y, Sun K, Lee KB, Nam JM, Park Y, Hong S. Adv Mater. 2007;19:2530. [Google Scholar]; b) Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Proc Natl Acad Sci USA. 2005;102:11594. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ruiz SA, Chen CS. Stem Cells. 2008;26:2921. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Forte G, Carotenuro F, Pagliari F, Pagliari S, Cossa P, Fiaccavento R, Ahiuwalia A, Vozzi G, Vinci B, Serafino A, Rinaldi A, Tizaversa E, Carosella L, Minieri M, Di Nardo P. Stem Cells. 2008;26:2093. doi: 10.1634/stemcells.2008-0061. [DOI] [PubMed] [Google Scholar]; b) Kraehenbuehl TP, Zammaretti P, Van der Vlies AJ, Schoenmakers RG, Lutolf MP, Jaconi ME, Hubbell JA. Biomaterials. 2008;29:2757. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]; c) Kraehenbuehl TP, Ferreira LS, Zammaretti P, Hubbell JA, Langer R. Biomaterials. 2009;30:4318. doi: 10.1016/j.biomaterials.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Taichman RS. Blood. 2005;105:2631. doi: 10.1182/blood-2004-06-2480. [DOI] [PubMed] [Google Scholar]; b) Kiel MJ, Morrison SJ. Nat Rev Immunol. 2008;8:290. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]; c) Adams GB, Scadden DT. Nat Immunol. 2006;7:333. doi: 10.1038/ni1331. [DOI] [PubMed] [Google Scholar]
- 15.Agheli H, Malmstrom J, Larsson EM, Textor M, Sutherland DS. Nano Lett. 2006;6:1165. doi: 10.1021/nl060403i. [DOI] [PubMed] [Google Scholar]
- 16.a) Nakajima M, Ishimuro T, Kato K, Ko IK, Hirata I, Arima Y, Iwata H. Biomaterials. 2007;28:1048. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]; b) Ruiz A, Buzanska L, Gilliland D, Rauscher H, Sirghi L, Sobanski T, Zychowicz M, Ceriotti L, Bretagnol F, Coecke S, Colpo P, Ross F. Biomaterials. 2008;29:4766. doi: 10.1016/j.biomaterials.2008.08.017. [DOI] [PubMed] [Google Scholar]; c) Knoll B, Weinl C, Nordheim A, Bonhoeffer F. Nat Protoc. 2007;2:1216. doi: 10.1038/nprot.2007.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.