Abstract
Lymphocyte survival during immune responses is controlled by the relative expression of pro-and anti-apoptotic molecules, regulating the magnitude, quality and duration of the response. Here we investigate the consequences of deleting genes encoding the anti-apoptotic molecules Mcl1 and Bcl2l1 (Bcl-xL) from B cells using an inducible system synchronized with expression of activation induced cytidine deaminase (Aicda) following immunization. This revealed Mcl1 and not Bcl2l1 to be indispensable for formation and persistence of germinal centers (GC). Limiting Mcl1 expression reduced the magnitude of the GC response with an equivalent, but not greater, effect on memory B cell formation and no effect on persistence. Our results identify Mcl1 as the main anti-apoptotic regulator of activated B cell survival and suggest distinct mechanisms controlling survival of GC and memory B cells.
Vertebrate immune responses are characterized by the clonal expansion of antigen-specific lymphocytes, by their differentiation into effector cells and by the production of small, persistent populations of memory cells. An added feature of B cell immunity is the increasing affinity of the antibody response with time, with B cells expressing high affinity antigen receptors (BCR) preferentially recruited into the effector and memory compartments (1). The processes underpinning changes in affinity of immunoglobulin receptors occur within germinal centers (GC), transient structures that arise after T cell dependent immunization (2–4). Thus the factors governing the survival of GC B cells will determine the qualitative and quantitative attributes of the effector cells, which in this case are plasma cells, and memory B cells. Survival of GC B cells is mediated by both the intrinsic and extrinsic apoptotic cell death pathways (5) with roles proposed for FASL/FAS (6, 7), Bcl2l1 (BclxL) (8), Bcl2 (9, 10), and Bim (11). These studies, however, do not identify the pro-survival molecules that are physiologically relevant within the GC
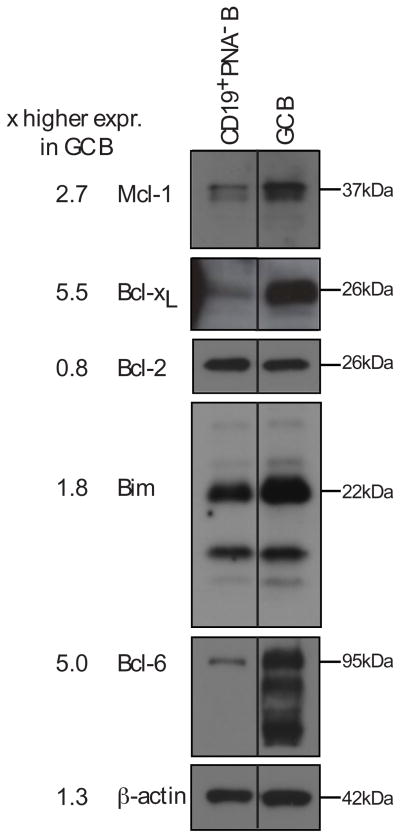
To address the components of the intrinsic apoptotic pathway regulating GC B cell behaviour, we first measured the expression of relevant proteins, comparing GC and follicular B cells (Fig. 1). Besides Bcl2l1 and Bim expression being up-regulated and Bcl-2 down-regulated, as previously reported (6, 12, 13), Mcl1 protein was increased in GC B cells (Fig. 1).
Fig. 1. Expression of anti- and pro-apoptotic proteins in GC B cells.
Immunoblots of lysates from sorted CD19+ PNA− follicular B cells and CD19+ PNA+ germinal center B (GC B) cells. The presence and absence of Bcl6 was used to ascertain the efficiency of cell sorting. Blots were probed with antibodies to Mcl1, Bcl2l1, Bcl2, Bim, Bcl6 and β-actin. Intensities are expressed as fold change relative to follicular B cells. Values from one of two independent experiments are shown. Duplicate lanes were removed from the original image with the solid line indicating the point of rejoining.
Increases in Bcl2l1 and Mcl1 expression in GC B cells prompted us to examine the contribution of these pro-survival proteins to the production of memory B cells, plasma cells and affinity maturation. We therefore conditionally deleted loxP-flanked alleles of Bcl2l1 or Mcl1 in B cells following antigen activation using a transgene-encoded Cre recombinase expressed concurrently with the activation induced cytidine deaminase (Aicda) locus (14). The loxP-flanked alleles should therefore be deleted in B cells initiating somatic hypermutation (SHM) or class switch recombination (CSR), processes requiring AID (15).
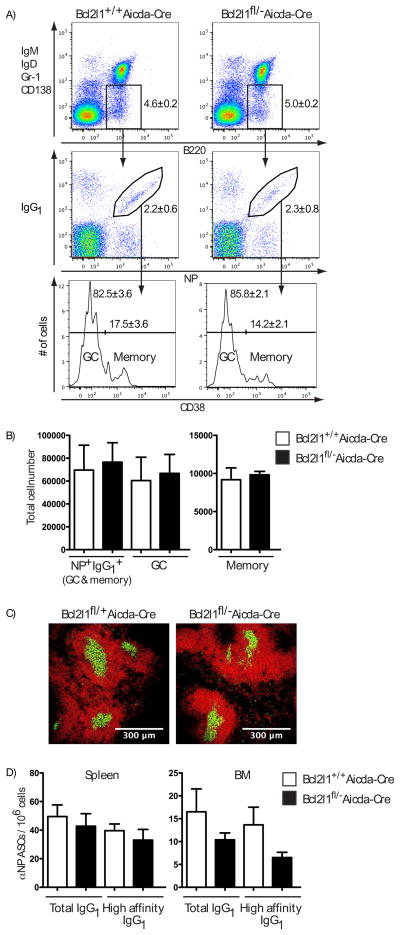
Bcl2l1fl/− Aicda-Cre mice, with one Bcl2l1 allele deleted (16) and one flanked by LoxP (fl) sites (17), were immunized with a T cell dependent antigen comprised of the NP hapten conjugated to Keyhole Limpet Hemocyanin (NP-KLH). Cellular responses were analyzed 21 days later in Bcl2lfl/− Aicda-Cre and control mice, when GC and memory B cells co-exist (6, 18). No significant difference in the frequency or number of antigen-specific (NP+ IgG1+) B cells was observed in the spleens of Bcl2l1fl/− Aicda-Cre mice compared to controls, both in total and after subdivision into GC (CD38−) and memory (CD38+) compartments (Fig. 2, A and B). Similarly, no differences were seen 7 and 14 days after immunization, excluding the possibility of an early deficit being masked by compensation as the response progressed (fig. S1). Immunofluorescent staining of spleen sections confirmed the presence of physical GC (Fig. 2C). Other explanations for a lack of difference, such as incomplete deletion or a failure to express appropriately the Cre recombinase were excluded (fig. S2 and S3). Conditional deletion of Bcl2l1 did, however, affect the bone marrow plasma cell compartment, itself a product of the GC (19, 20). Although the representation of NP-specific IgG1 antibody secreting cells (ASC) in the spleen was normal at day 21, the frequency in bone marrow was reduced (Fig. 2D), a difference that was significant at 14 days post immunization (fig. S4A). Consistent with this, serum titres of anti-NP IgG1 in Bcl2l1fl/− Aicda-Cre mice at day 21 were significantly diminished compared to controls (fig. S4B). Collectively these results demonstrate that Bcl2l1 is dispensable for GC formation, the expansion of antigen-specific B cells and the formation of B cell memory but functions in establishing or maintaining emigrant populations of ASC (21).
Fig. 2. Bcl2l1 is dispensable for GC formation and the subsequent development of NP-specific IgG1 B cells and memory B cells.
(A) Flow cytometric analysis of splenocytes day 21 after intraperitoneal immunization with NP-KLH in alum. Isotype switched B cells (IgM− IgD− Gr-1− CD138− B220+) cells were analyzed for NP+ IgG1+ status with NP+ IgG1+ cells being sub-divided into GC (CD38−) and memory (CD38+) B cells. Numbers in plots and histograms represents mean percentage ± SEM of all mice examined. (B) Absolute cell numbers of the populations identified in (A) are graphed as mean ± SEM of between four and six mice in each group and summarizes two experiments. (C) Frozen spleen sections from Bcl2l1fl/+ Aicda-Cre and Bcl2l1fl/− Aicda-Cre mice, 14 days after immunization, stained with antibodies to B220 to identify follicles (red) and GL7 for germinal centers (yellow). Original magnification × 20 with scale indicated. (D) Frequencies of total and high affinity NP-specific IgG1-secreting ASCs in spleen and bone marrow. Data are the mean ± SEM of replicate wells with between four and six mice in each group and summarize two experiments.
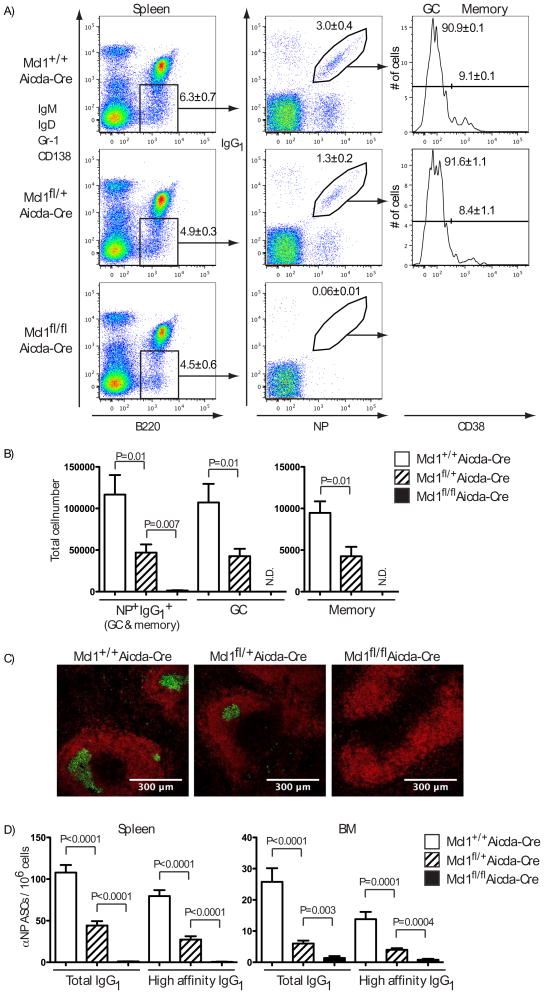
Mcl1 was another candidate for mediating B cell survival within GC (Fig. 1). Although Mcl1 and Bcl2l1 function in the same apoptotic pathway (22), they differ substantially in their ability to bind members of the pro-apoptotic BH3-only and Bax/Bak-like proteins (23) and could thus have distinct roles. We therefore generated mice with Mcl1 alleles flanked by LoxP sites (24) and carrying the Aicda-Cre transgene (14). Analysis of spleens of Mcl1fl/fl Aicda-Cre mice 14 days after immunization revealed the absence of NP+ IgG1+ B cells when both alleles of Mcl1 were conditionally deleted, whereas deleting one Mcl1 allele produced a partial reduction in antigen-specific B cells, with the GC and memory compartments effected equally (Fig. 3, A and B). Thus, Mcl1 is essential for the generation of antigen-specific IgG1 B cells and appears to be a limiting determinant in the magnitude of the GC response. Moreover, GC could not be observed by histology in the spleens of immunized Mcl1fl/fl Aicda-Cre mice and those seen in Mcl1fl/+ Aicda-Cre mice were reduced in size and frequency (Fig. 3C). The frequency of NP-specific IgG1 ASC in both spleen and bone marrow was severely diminished in homozygous Mcl1 deleted mice with a corresponding reduction in heterozygous Mcl1 deleted mice (Fig. 3D). Serum titres of anti-NP IgG1, but not IgM, were correspondingly and significantly reduced (fig. S5). To determine when Mcl1 became critical for the survival of activated B cells we examined Mcl1fl/fl Aicda-Cre mice on day 5 (fig. S6) and day 7 (fig. S7) after immunization with NP-KLH in alum. Although an immune response was detected at both times, no antigen-specific, isotype switched or GC phenotype B cells were detected in the Mcl1fl/fl Aicda-Cre mice, indicating the dependence on Mcl1 precedes GC formation.
Fig. 3. Conditional deletion of Mcl1 abrogates the GC response and the formation of NP-specific memory B cells.
(A) Flow cytometric analysis of splenocytes 14 days after intraperitoneal immunization with NP-KLH in alum. Isotype switched B cells (IgM− IgD− Gr-1− CD138− B220+) cells were analyzed for NP+ IgG1+ status. NP+ IgG1+ cells were further sub-divided into GC (CD38−) or memory (CD38+) B cells. Numbers in dotplots and histograms represents mean percentage ± SEM of all mice examined. (B) Absolute cell numbers of the populations identified in (A) are shown in the graphs as the mean ± SEM of between five and ten mice in each group, summarizing four experiments. (C) Frozen spleen sections from Mcl1+/+ Aicda-Cre, Mcl1fl/+ Aicda-Cre and Mcl1fl/fl Aicda-Cre mice, 14 days after immunization, stained with antibodies to B220 to show B cell follicles (red) and GL7 to show germinal centres (yellow). Original magnification ×20 with scale indicated. (D) Frequencies of total and high affinity NP-specific IgG1 ASCs in spleen and bone marrow. Data are the mean ± SEM of replicate wells from between five and ten mice in each group and summarize four experiments. Significant differences in (B) and (D), determined by Student’s T-test, are indicated.
The effects of Mcl1 deletion in GC formation extended beyond the NP response since the number of GC-phenotype B cells (CD19+ PNA+ IgDlo) in Mcl1fl/+ Aicda-Cre mice was reduced and in Mcl-1fl/fl Aicda-Cre mice, no hCD2t+ (and therefore Cre expressing) GC B cells were found (fig. S8A and B). Furthermore, analysis of Peyer’s patches in the Mcl1 strains revealed the absence of GC-phenotype B cells in Mcl-1fl/fl Aicda-Cre mice and their reduction in Mcl1fl/+ Aicda-Cre mice compared to controls (fig. S8C), emphasizing the generality of the requirement for Mcl1 in GC formation. The conditional Mcl1 allele was constructed such that after deletion of Mcl1, hCD4 is expressed on the cell surface under the control of the Mcl1 regulatory elements, thereby serving as a reporter of Mcl1 deletion and Mcl1 expression (fig. S9). The persisting GC and memory B cells in Mcl1fl/+ Aicda-Cre mice were all hCD4+ and had thus deleted one allele of Mcl1 (fig. S10).
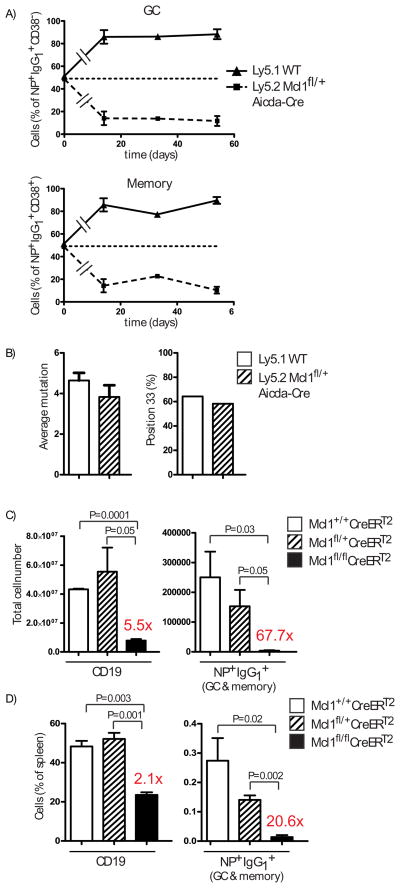
The reduced number of NP+ IgG1+ cells in Mcl1fl/+ Aicda-Cre mice compared to controls revealed a relationship between the Mcl1 gene dosage and the survival of antigen reactive B cells, suggesting that amounts of Mcl1 could underpin B cell competition within the GC or affect persistence in the memory B cell compartment. To determine if this was the case, lethally irradiated Ly5.1 recipients were reconstituted with an equal mixture of bone marrow from Mcl1fl/+ Aicda-Cre (Ly5.2) and WT (Ly5.1) mice, then immunized with NP-KLH in alum and analyzed 14, 33 and 54 days later. The representation of antigen-reactive B cells with one allele of Mcl1 deleted upon activation (Ly5.2) was reduced significantly in day 14 GC compared to parity in the naive B cell populations and remained constant thereafter (Fig. 4A), which suggested a disadvantage in initiation but not persistence. The decreased representation of Mcl1fl/+ B cells within the GC was reflected in an identical fold reduction amongst memory B cells at all time points measured (Fig. 4A and fig. S11), suggesting that entry into and persistence within the GC-derived memory compartment is not influenced by amounts of Mcl1. Other routes of memory B cell formation may have distinct survival requirements (25).
Fig. 4. Mcl1 determines the magnitude and the persistence of the GC response.
(A) The percentage of Ly5.1 (WT) versus Ly5.2 (Mcl1fl/+ Aicda-Cre) GC (NP+ IgG1+ CD38−) and memory (NP+ IgG1+ CD38+) B cells in Mcl1fl/+ Aicda-Cre/Ly5.1 chimeras at 14, 33 and 54 days after immunization. Data are the mean ± SEM of between 2 and 3 mice for each time point. (B) Analysis of somatic hypermutation assessed by comparing the VH186.2 genes of single NP+ IgG1+ Mcl1fl/+ Aicda-Cre B cells with those in Ly5.1 control B cells, sorted 14 days after immunization of the chimeric mice described in (A). Average Mutation per VH186.2 gene segment compares the region encoding amino acids 10 to 96 with the germline sequence from 12 Mcl1fl/− NP+IgG1+ B cells and 14 control cells. Position 33 (%) measures the frequency of the Trp to Leu exchange within CDR1 of VH186.2 that alone confers a 10-fold increase in affinity of binding NP (34). (C) Absolute number of B cells and NP+ IgG1+ B cells following Tamoxifen treatment of chimeric mice in which Mcl1 is amenable to deletion only in B cells. Mice, immunized with NP-KLH in alum intraperitoneally, were treated with Tamoxifen on days 7 and 8, and analyzed on day 9 for B cells (CD19+) and NP+ IgG1+ cells amongst isotype switched B cells (IgM− IgD− Gr-1− CD138− B220+) cells. Status of the Mcl1 alleles in B cells is indicated. (D) Proportion of B cells (CD19+) and NP+ IgG1+ B cells in the spleens of the mice described in (C). Fold changes between the indicated B cell compartments in Tamoxifen treated Mcl1+/+ and Mcl1fl/fl mice for are shown. Significant differences in (C) and (D), determined by Student’s T-test, are indicated.
It was possible that GC B cells lacking either Bcl2l1 or Mcl1 had compensated for a survival disadvantage by acquiring higher than normal affinity, thereby permitting their persistence in the plasma cell and memory B cell compartments. Affinity amongst antigen-specific ASC, measured using the ratio of high affinity:total binding (20, 26), was unaffected in both cases (fig. S12A and B), indicating no change in selection into this compartment. Affinity amongst NP memory B cells was determined by sequence analysis of the immunoglobulin VH186.2 genes (27, 28). The distribution, frequency and nature of somatic mutations in the VH186.2 genes (27, 28) of sorted, single NP+ IgG1+ CD38hi memory B cells 21 days after immunization of Bcl2l1fl/ Aicda-Cre mice was similar to controls (fig. S12C). The same analysis of NP+ IgG1+ B cells from Mcl1fl/+ Aicda-Cre (Ly5.2) and WT (Ly5.1) chimeric mice, 14 days after immunization, also revealed no differences in distribution or frequency of somatic mutations (Fig. 4B), indicating there was no compensation for reduced Mcl1 protein in GC B cells through enhanced selection.
Finally, we examined if survival of GC B cells after formation also required Mcl1. We created groups of mice through bone marrow reconstitution in which B cells were Mcl1fl/fl, Mcl1fl/+ or Mcl1+/+, in each case with Rosa26-CreERT2 (fig. S13). Deletion of Mcl1 in these mice was thus Tamoxifen (Tam) inducible and restricted to B cells. These mice were immunized with NP-KLH in alum, treated with Tam on days 7 and 8, and analyzed on day 9 for the presence of NP+ IgG1+ B cells. Relative to Rosa26-CreERT2 controls, NP+ IgG1+ cells were deleted from the Mcl1fl/fl Rosa26-CreERT2 B cell group and reduced in the Mcl1fl/+ Rosa26-CreERT2 group (Fig. 4C and D). While Tam treatment of Mcl1fl/fl Cre-ErT2 mice affected the survival of naïve B cells in spleen (fig. S13), reducing their number more than 5-fold, the loss of NP+ IgG1+ B cells was considerably greater at 68-fold (Fig. 4C and D). The 2-fold reduction in frequency of NP+ IgG1+ B cells in the spleens of Tam treated Mcl1fl/+ Cre-ErT2 mice in the context of no change in overall B cell representation (Fig. 4D) emphasizes the unique sensitivity of GC B cells to changes in amounts of Mcl1. Immunohistology of the spleens of the Tam treated immunized mice revealed IgM+ blasts as clusters in control spleens but only as rare, single cells in spleens from Mcl1fl/fl Rosa26-CreERT2 mice (fig. S14), indicating the survival of activated, unswitched B cells also depends on Mcl1. Thus, in addition to a role in naïve B cells (29), Mcl1 is required for the survival of activated B cells, for the formation of GC and for the persistence of GC once established.
Differential cell survival is central to regulating the quality and quantity of lymphocyte responses to antigen (30, 31). Previous experiments modulating expression of Bcl-2 family member proteins in B cells (6, 9–11) led to models of GC function in which affinity for antigen enhances B cell survival and therefore representation through modulation of those Bcl-2 family member proteins (30, 31). The data presented here show unequivocally that Mcl-1 has a unique role in maintaining antigen-reactive B cells after their initial exposure and then within the GC. This dependency of GC B cells on Mcl1 is distinguished from that of naïve B cells by two features. First, the sensitivity of GC B cells to reduced amounts of Mcl1 is greater than that of naïve B cells. Second, compounds that specifically antagonize Bcl2 and Bcl2l1 (32) diminish naïve B cells in spleen but leave GC B cells unaffected (21), a result complementary to the data presented here. These results, showing that particular pro-survival proteins can become limiting at particular stages of immune responses, are further exemplified by the differential sensitivity of post-GC plasma cells to altered Bcl2 and Bcl2l1 (21); these cells are sensitive en route, but resistant once established in their survival niches in the bone marrow(19, 21). Collectively, our results challenge the current view of what regulates GC survival, offer significant insights into the formation and maintenance of B cell memory and also suggest Mcl1 as a potential target in GC-derived lymphomas that do not over-express Bcl2l1 (33).
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Health and Medical Research Council (NHMRC) Australia (356202 to DMT and SLN; 461221 to PB and AS), the Leukemia and Lymphoma Society (SCOR grant 7413) and the NIH (CA43540, CA80188). DMT and AS are supported by fellowships from the NHMRC; SLN by a Pfizer Australia Research Fellowship; IV by a fellowship from the Olle Engkvist Byggmastare foundation; KL by a fellowship from the German Academic Exchange Service; PB by the Charles and Sylvia Viertel Charitable Foundation and MB by Boehringer Ingelheim. DNA sequence data have been submitted to GenBank (Submission ID 1370215). We are indebted to the facilities of our respective institutes, particularly those responsible for animal husbandry and flow cytometry. We also acknowledge the assistance of L. O’Reilly for Western blot antibodies and the many helpful discussions with members of the WEHI B Cell Program. Author contributions are as follows: IV and DMT designed the research; IV, KL, SG, VP, SC and PJ performed experiments and contributed to interpretation and discussion. MB, PB, AS and SLN contributed to the design of experiments, interpretation of results and drafting the manuscript. IV, KL and VP analyzed data and prepared figures. IV and DMT wrote the manuscript.
References and Notes
- 1.Rajewsky K. Nature. 1996;381:751. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Haberman AM, Shlomchik MJ. Nat Rev Immunol. 2003;3:757. doi: 10.1038/nri1178. [DOI] [PubMed] [Google Scholar]
- 3.Lanzavecchia A, Sallusto F. Nat Rev Immunol. 2002;2:982. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 4.MacLennan IC. Annu Rev Immunol. 1994;12:117. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 5.Strasser A, Jost PJ, Nagata S. Immunity. 2009;30:180. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi Y, Ohta H, Takemori T. Immunity. 2001;14:181. doi: 10.1016/s1074-7613(01)00100-5. [DOI] [PubMed] [Google Scholar]
- 7.Hao Z, et al. Immunity. 2008;29:615. doi: 10.1016/j.immuni.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y, et al. J Exp Med. 1999;190:399. doi: 10.1084/jem.190.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KG, Weiss U, Rajewsky K, Nossal GJ, Tarlinton DM. Immunity. 1994;1:803. doi: 10.1016/s1074-7613(94)80022-7. [DOI] [PubMed] [Google Scholar]
- 10.Smith KG, et al. J Exp Med. 2000;191:475. doi: 10.1084/jem.191.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer SF, et al. Blood. 2007;110:3978. doi: 10.1182/blood-2007-05-091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein U, et al. Proc Natl Acad Sci U S A. 2003;100:2639. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama T, et al. Immunol Lett. 2002;81:107. doi: 10.1016/s0165-2478(02)00003-2. [DOI] [PubMed] [Google Scholar]
- 14.Kwon K, et al. Immunity. 2008;28:751. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu M, et al. Cell. 2000;102:553. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 16.Motoyama N, et al. Science. 1995;267:1506. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 17.Wagner KU, et al. Development. 2000;127:4949. doi: 10.1242/dev.127.22.4949. [DOI] [PubMed] [Google Scholar]
- 18.Ridderstad A, Tarlinton DM. J Immunol. 1998;160:4688. [PubMed] [Google Scholar]
- 19.Radbruch A, et al. Nat Rev Immunol. 2006;6:741. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 20.Smith KG, Light A, Nossal GJ, Tarlinton DM. Embo J. 1997;16:2996. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrington EM, et al. Proc Natl Acad Sci U S A. 2010;107:10967. doi: 10.1073/pnas.1005256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams JM, Cory S. Oncogene. 2007;26:1324. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, et al. Mol Cell. 2005;17:393. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Willis SN, et al. Science. 2007;315:856. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 25.Toyama H, et al. Immunity. 2002;17:329. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 26.Roes J, Rajewsky K. J Exp Med. 1993;177:45. doi: 10.1084/jem.177.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen D, Simon T, Sablitzky F, Rajewsky K, Cumano A. Embo J. 1988;7:1995. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cumano A, Rajewsky K. Embo J. 1986;5:2459. doi: 10.1002/j.1460-2075.1986.tb04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opferman JT, et al. Nature. 2003;426:671. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 30.Allen CD, Okada T, Cyster JG. Immunity. 2007;27:190. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarlinton D. Nat Rev Immunol. 2006;6:785. doi: 10.1038/nri1938. [DOI] [PubMed] [Google Scholar]
- 32.Oltersdorf T, et al. Nature. 2005;435:677. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 33.Cho-Vega JH, et al. Hum Pathol. 2004;35:1095. doi: 10.1016/j.humpath.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Allen D, et al. Immunol Rev. 1987;96:5. doi: 10.1111/j.1600-065x.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.