Abstract
Genome-wide linkage and association studies of tens of thousands of clinical and molecular traits are currently under way, offering rich data for inferring causality between traits and genetic variation. However, the inference process is based on discovering subtle patterns in the correlation between traits and is therefore challenging and could create a flood of untrustworthy causal inferences. Here we introduce the concerns and show they are valid already in simple scenarios of two traits linked or associated to the same genomic region. We argue that more comprehensive analysis and Bayesian reasoning are needed and can overcome some of these pitfalls, although not in every conceivable case. We conclude that causal inference methods may still be of use in the iterative process of mathematical modeling and biological validation.
Causal inference from genetic data
Understanding how genes, proteins, metabolites and phenotypes connect in networks is a key objective in biology. Genes are transcribed and translated into proteins that can act as enzymes to convert precursor metabolites into product metabolites. These relationships are often depicted informally using graphs with arrows pointing in the assumed direction of causality, for example, from genes to proteins to metabolites to classical phenotypes. These diagrams reflect our assumptions about causality in biological systems and in many cases have been painstakingly validated in controlled experimental settings. Today, more than ever before, we are faced with large-scale “post-genomics” data that have the potential to reveal a multitude of yet unknown but potentially causal relationships.
Methods for causal inference have been introduced as early as the 1920s[1] and have been further developed and applied since then in genetic epidemiology and other fields [2–4]). Causal inference is a formal statistical procedure that aims to establish predictive models. For example, if a reduction in the level of critical metabolite is the cause of a disease, then an intervention that increases the metabolite level should alleviate the disease. By contrast, if the reduced metabolite is a consequence of the disease, then intervention will not have the desired effect. Causal reasoning is thus critical to the process of target discovery in pharmaceutical research.
Recent genome-wide linkage studies (GWLS) on model organisms [5–7] and genome-wide association studies (GWAS) on humans [8] have successfully connected molecular and classical traits into networks with arrows indicating inferred causal relationships [9–17]. Causality cannot be established from data alone. Some assumptions about the causal relationships among the variables being modeled are needed. Once these are established, causal inference can be propagated to additional variables. In GWLS and GWAS settings it is typical to assume that genomic variation (quantitative trait locus; QTL) acts as a causal anchor from which all arrows are directed outward. Although this assumption seems quite natural, caution is warranted when the sample is not random, as in case-control studies.
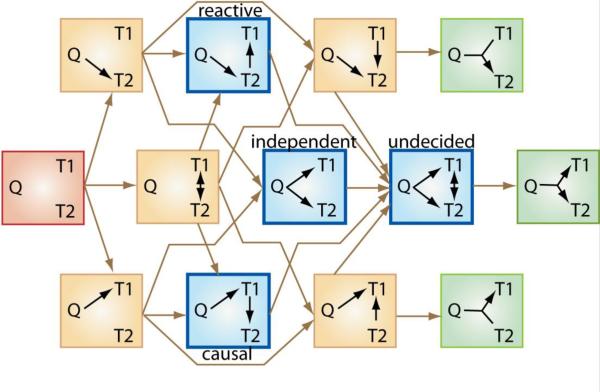
There are many possible causal networks even in a simple system consisting of a genomic locus (QTL) and two traits, T1 and T2 (Figure 1). Causal inference in GWLS and GWAS involves, in its simplest form, the identification of pairs of traits with a common QTL (QTL-trait-trait triads) and determining whether the QTL directly affects each of two traits (independent), or if the QTL affects only one trait which in turn affects the other trait (causal or reactive). If none of these situations apply we assume that the causation is more complex (undecided).
Figure 1. Triad models.
Many different causal relationships are possible among a triad of two traits (T1 and T2) and a QTL (Q). The simplest case (red box) to the left shows no causality, in which case the QTL and the two traits do not influence each other. In the next set of models (yellow), at least one trait is not associated with the QTL. All these models are excluded from consideration based on the assumption that the QTL mapping step has correctly inferred the QTL-trait associations. The models that remain to be discriminated are highlighted in blue and green: the procedure to decide in favor of one of the blue causal topologies is outlined in the text. The three models furthest to the right (green) are extensions of the causal model that include additional interaction terms, e.g. the QTL may modulate the causal effect of T1 on T2. Equivalently, these models may be seen as relaxing the assumption of equal covariance across genotype classes. An extreme scenario is the Simpson's paradox model in which the traits show opposite correlations for different genotypes at the QTL. Such complexities are usually not considered, but may form an important part of actual biological networks. The brown arrows indicate which of the models are nested and can thus be directly compared by statistical testing.
Biological variation in the two traits beyond that induced by the common QTL is key to distinguishing between the independent and causal scenarios. If there is a causal link, the biological and QTL variation from T1 will propagate to T2. If the variation propagates in an approximately linear fashion, we can, with simple linear regression (Box 1), subtract the biological and QTL variation in T1 from T2 and are left with the additional or `residual' variation in T2 unrelated to the QTL. If we attempt the reciprocal analysis, the additional variation in T2 may make the linear regression fail to subtract all of the QTL variation from T1. As a result the residual variation in T1 will still relate to the QTL. This reasoning suggests a simple approach to distinguish among the independent and causal models on the basis of the outcome of two reciprocal statistical tests: does the residual variation in T1 still relate to the QTL, and does the residual variation in T2 still relate to the QTL. Traits are declared independent (yes, yes), causal (yes, no), reactive (no, yes), or more complex (no, no) in which case no decision is made (see Box 1 for the statistical details). While the apparent simplicity of this approach is seductive, here we highlight some possible pitfalls illustrated by three simple but realistic scenarios, and discuss avenues to restoring the potential of causal inference.
Concerns about causal inference
It is compelling to explore how this causal inference method for QTL-trait-trait triads performs, particularly in GWAS where the majority of QTL identified explain much less than 5% of the total variance [18]. The method will declare certain triads to be independent and others to be causal, but such inferences are not without error. Of all triads that are truly causal, what proportion can be correctly identified as such? This proportion is referred in statistics as the `sensitivity' of the method. It is good for a method to be sensitive, but not sufficient to make it of practical use. Triads with truly independent traits may also have a chance to be identified, incorrectly, as causal by the method. As a consequence, the potential number of false causal links arising from, say, 80% independent trait-trait pairs can overwhelm the number of true causal links arising from the 20% causal trait-trait pairs. The proportion of true causal links amongst those identified as causal is referred to in statistics as the `positive predictive value'. A good method combines a high positive predictive value, say 90%, with an acceptable sensitivity, say 10% or higher (see Box 1 for the statistical details). A QTL is a genomic region that can contain multiple candidate genes and polymorphisms. Without prior knowledge that two traits sharing a common QTL are biologically or biochemically related, they are more likely to be regulated by different genes or polymorphisms within the QTL region. In which case we would say the traits are independent and that their apparent relationship is explained by linkage disequilibrium and not by a shared biological pathway. Different types of prior knowledge about the (unknown) number of true causal and true independent relationships can be incorporated into the causal inference (Box 2).
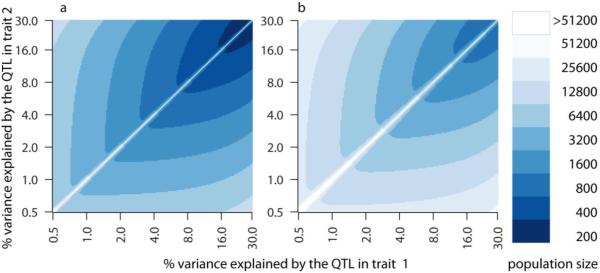
We present three different scenarios to illustrate the properties of the method. In the first scenario T1 is causal for T2, all QTL and biological variation in T1 is propagated to T2 and, on top of this variation, T2 shows additional variation. This additional variation may originate from an independent perturbation such as another QTL affecting T2 but not T1, or an environmental perturbation affecting T2 but not T1. The correlation between T1 and T2 is resulting fully from the causal relationship between the two traits. Exact analytical equations can be used to compute the required population size to attain desired levels of sensitivity and positive predictive value (Box 1). It requires specifying the size of the QTL effect, the frequency in the population of the major QTL allele, and the prior believe that the triad is causal rather than independent. A population size of approximately 200–6,000 (GWLS) to 800–25,000 (GWAS) provides 50% sensitivity and 90% positive predictive value for causal inference with QTL explaining from 30% down to 0.5% of total variance (Figure 2, with parameters as specified in the legend). Lowering the sensitivity to 10% would reduce the required population size, but this effect is visible only in the area close to the diagonal (Figure 2). In this area traits are too tightly correlated and there is little additional variation in T2, making it difficult to infer the correct causal direction, i.e. sensitivity is low.
Figure 2. Population size required for reliable causal inference.
Here we show the required population size in (a) genome-wide linkage studies (GWLS) and (b) genome-wide association studies. Each color represents a different population size; the scale is shown in the right panel. These numbers have been calculated from the equations in Box 1 by using a 10% significance threshold for the t-tests, 90% positive predictive value and 50% sensitivity. We assume that there is only biologically variation and no measurement error. The x (or y) axis indicates the percentage of variance explained by a QTL in trait T1 or T2, respectively on a logarithmic scale ranging from 0.5% to 30%. Allele frequencies of the biallelic QTL are set equal in GWLS, and 10% and 90% in GWAS. Furthermore we use Bayesian reasoning (Box 2): we assume a priori that only 1% (20%) of the QTL-trait-trait connections is truly causal in GWLS (GWAS).
In the second scenario one or more shared hidden factors cause additional correlation between the traits. One can think of undetected QTL with pleiotropic effects on the traits, structural chromosomal variation leading to co-expression of genes in a particular region, physiological variation related to daily circadian rhythms, or environmental variation due to features of the experimental implementation. In a causal model, the effect of the hidden factor acts on T2 in two ways: indirectly through T1, but also directly. For increasing values of hidden factor correlation (while keeping QTL and total variance constant), the linear regression will tend to subtract the effect of the hidden factor and not that of the QTL. As a consequence the causal links will look more like independent (yes, yes); increasing sample size will not help to attain the desired levels of sensitivity and positive predictive value. In an independent model, the effect of the hidden factor acts on T1 and T2 directly, and not indirectly. As with the causal model, for increasing values of hidden factor correlation (while keeping QTL and total variance constant), the linear regression will typically tend to subtract the effect of the hidden factor and not that of the QTL. However, in the special case of equal slopes for hidden factor and QTL, the linear regression will be able to subtract hidden factor and QTL effects. A true independent model then tends to change from correct identification (yes, yes) via either causal (yes, no) or reactive (no, yes) to undecided (no, no). Increasing sample size will help only when slopes are still slightly different, not if they are equal. Note that equal slopes cannot occur in the causal model, because the hidden factor acts directly and indirectly on T2. Sample size shown in Figure 2 is still approximately adequate if the hidden factor variance is small, i.e. equals at most the QTL variance.
In the third scenario, measurement error comes into play, which is realistic for most technologies for scoring molecular and classical traits. Note that the use of surrogate variables, such as RNA expression as a proxy for the causal protein levels, may also introduce a kind of measurement error. Measurement variation is never `biologically' propagated from one trait to another trait, yet it will change (reduce or increase) the correlation between the two traits, and thus the causal inference will be affected. Correlated measurement errors are analogous to the hidden factor scenario described above with one exception. The special case of equal slopes for hidden factor and QTL can now occur also in the causal model: slopes for correlated measurement error and QTL can be equal. In this case, a true causal model can change from correct identification (yes, no) to undecided (no, no). Independent measurement errors will cause the linear regression to fail to subtract the QTL variation in both reciprocal analyses; therefore the causal model will tend to look more like independent (yes, yes) if measurement variance increases. However, an actual causal link from one trait measured with large measurement error to a downstream trait measured with small measurement error can be reported as reactive [13]. Again, increasing sample size will not be helpful to attain the desired levels of sensitivity and positive predictive value.
Restoring the potential of causal inference
We have explored causal inference in the simple context of QTL-trait-trait triads using a statistical decision procedure (Box 1) to possibly reject the undecided model in favor of one of the nested causal, reactive and independent models. This procedure is similar to other implementations of triad analysis [5, 7, 9] which, although not identical, lead to comparable results [11]. Other computational methods for causal inference such as structural equation modeling [19, 20] or Bayesian network analysis [21] can operate on larger numbers of traits and QTL. These methods also rely on the correlation structure in the data and will therefore suffer from some of the same problems as triad analysis: they require large population size, and can be confounded by hidden factors or measurement noise. This calls for several recommendations to restore the potential of causal inference.
Our first recommendation is to use Bayesian reasoning in the causal inference procedure. Prior belief or knowledge about the number of true causal and true independent links that might be expected in a typical QTL, depending on the study design, should be considered to safeguard against high false positive rates (low positive predictive values). In studies that involve mapping gene expression (eQTL), protein (pQTL) or metabolite (mQTL) traits, information about co-localization of QTL and genes that are functionally linked to the trait provides information about the likelihood of causal links. Lastly, biological annotations such as Gene Ontology [22] or Kyoto Encyclopedia of Genes and Genomes (KEGG) [23] pathways should also be considered when weighing evidence for causal links. The use of more informative priors (Box 2) provides better prioritizing and filtering of the large numbers of possible triads, and may reduce the required population size for reliable causal inference to more realistic numbers.
Our second recommendation is to identify and eliminate or account for experimental factors that can induce spurious correlation. It is not usually possible to measure all relevant factors, yet even some of the most obvious factors such as age or sex of study subjects are often not taken into account. Any variation in diet, time since last feeding or time of sample collection, the size of plant seeds or the size of litter, temperature and light cycles, location in the greenhouse or field, can have profound effects. Such factors can be easily included in the model, but only when they are recorded [24, 25]. While it may not be necessary in inbred line cross studies, it is critical to consider the impact of population structure in almost every other setting where genetic variation is present. Methods are available to estimate kinship and the corresponding structure of the correlation. Combining these methods with causal inference can minimize the effects of spurious genetic correlation [26]. The effects of hidden factors affecting larger numbers of traits can be detected and corrected for by dimension reduction methods ([26–30]). Causal inference can then be applied to the residual data. However, these multivariate analysis methods also have the potential to remove signals relevant for causal inference from data and their application should be considered carefully.
Our third and final recommendation is to consider a richer set of possible models than the four blue models in Figure 1. For example, fitting a model like the top right yellow model in Figure 1 could provide a powerful case for the causal signal in the data [17, 19, 20]. The green models in Figure 1 with more complex correlation structure can also be informative and have been explored [17]. If two traits have multiple QTL in common, then this may be taken as additional evidence that the two traits are connected in the network [31]. This allows for the possibility to generalize the triad analysis to a multiple QTL-trait-trait analysis. A test of the effects of all QTL that propagate from one trait to another can be obtained by modifying step 3 in the decision procedure (Box 1) to assess the combined effect [32].
Concluding remarks
Many in the scientific community share a healthy skepticism of causal inference and for good reasons as we have shown. Nevertheless we conclude that causal inference in linkage or association analysis may soon become a feasible strategy given the rapidly growing prior knowledge of biological networks, the increasing population sizes, the advent of cheaper and more accurate measurement techniques, and the possibility of coupling causal inference methods with Bayesian reasoning. Further development of methods that consider the simultaneous effects of multiple traits and multiple QTL is needed, as well development of techniques that address the effects of experimental factors, study design and population structure. Reasonable caution remains warranted and statistical methods of causal inference should be viewed as a necessary step in an era of high throughput data generation and discovery.
Box 1. Causal inference with triads.
(A) Decision procedure
The triad analysis is a statistical decision procedure consisting of the following steps:
Step 1. Establish that two traits are linked to the same locus. This rules out the red and yellow models (Figure 1). We are ignoring the green models. So we are now reduced to the four blue models (independent, causal, reactive, undecided).
Step 2. Regress T2 on T1 and T1 on T2 to obtain residuals of each trait adjusted for the other. Denote residuals by R2 and R1, respectively.
Step 3. Compute a bivariate t-test for association between the residuals (R1 and R2) and the QTL. Note that R2 is 100% adjusted for both QTL effect under the causal model only (zero expected value; Table I). We note that in other implementations of triad analysis one would compute univariate t-tests of R1 against QTL and R2 against QTL. This ignores the correlation between these two tests and we have amended it here.
Step 4. Choose a model based on outcomes of the bivariate t-tests using a p-value of, e.g., 10%: independent if (yes, yes), causal if (yes, no), reactive if (no, yes). If none of these apply we default to the “undecided” case.
(B) Properties of procedure
We describe two statistical measures and derive implications for population size:
Sensitivity
The sensitivity of the method is the probability of correctly detecting a true causal relationship. This probability is obtained from the non-central bivariate t-distribution (QTL effect of residuals determine the non-centrality; Table I).
Positive predictive value
The positive predictive value is probability of a declared causal connection being true. We incorporate prior knowledge (Box 2): P1 is the product of the prior probability of a link to be causal times the probability to correctly identify a causal link as such; P2 is the product of the prior probability of a link to be independent times the probability to incorrectly identify an independent link as causal. Then the positive predictive value is P1 / (P1+P2).
Required population size
The above process is repeated for all combinations of QTL variance in the two traits, and for sample size ranging from 200 to 51,200. The minimum sample size to achieve both 50% sensitivity and 90% positive predictive value is plotted (Figure 2).
Box 2. Bayesian Reasoning.
Bayes rule [33] is a probability property that allows one to combine evidence from data with existing knowledge and expertise through the inclusion of priors in an inference process. The definition of the prior in a causal inference on a QTL-trait-trait triad is the result of a partly subjective process that can be guided by the following considerations:
QTL confidence interval size. The larger the confidence intervals of the QTL are, the more likely it is that distinct polymorphisms control the traits. In GWLS, linkage disequilibrium is pervasive leading to large confidence intervals.
SNP density in the QTL region within the population. The more polymorphic the QTL region is, the more likely it is that the traits are actually controlled by distinct polymorphisms. In GWAS, populations are heterogeneous leading to a lot of allelic diversity along the genome.
Gene density within the confidence interval. Polymorphisms that lie within gene coding regions are more likely to propagate variation at phenotypic level than polymorphisms in non-coding regions. The fewer the number of genes within the QTL confidence interval, the more likely that the two traits are affected by the same polymorphism.
Local or distant eQTL. If a gene expression trait is locally regulated by an eQTL and the other trait is distantly regulated by the eQTL, then the gene with the local eQTL is more likely to be causal for the other trait than the other way around[14].
Additional shared QTL. The sharing of multiple additional QTL between the two traits may be taken as additional evidence that they are connected in the network[31]. It is more likely that these QTL affect the traits through the same polymorphisms than it is that locations of multiple distinct polymorphisms coincide by chance.
QTL hotspot. Regions of the genome, known as QTL hotspots, have been reported that harbor QTL for large numbers of traits. These could be the result of a single major polymorphism or of many polymorphisms in linkage disequilibrium and each affecting different traits independently. Further investigation and experience in understanding this phenomenon is needed to determine which is more likely.
Independent biological knowledge. Biological knowledge about the two traits (for example if the two genes belong to a same KEGG pathway) can be used as a priori evidence that the traits are related.
Box 1 Table I.
Equations for regression parameters in the basic independent and causal model (first scenario in the main text)a,b
| Independent model | Causal model | ||
|---|---|---|---|
| T1 = QTL + e1 | T1 = QTL + e1 | ||
| T2 = QTL + e2 | T2 = T1 + e2 | ||
| Regress T1 on T2 | Slope | 1 − v2/vt2 | 1 − v2/vt2 |
| Regress residual R1 on QTL | QTL effect | 2v2/vt2 | 2v2/vt2 |
| Variancec | v1 + v2(v2/vt2−1)2 | v2(v2/vt2−1)2 + v1(v2/vt2)2 | |
| Regress T2 on T1 | Slope | 1 − v1/vt1 | 1 |
| Regress residual R2 on QTL | QTL effect | 2v1/vt1 | 0 |
| Variancec | v2 + v1(v1/vt1−1)2 | v2 | |
| Covariation of QTL effects | Covariancec | v1 (v1/vt1 − 1) + v2(v2/vt2 − 1) | v2(v2/vt2 − 1) |
T1 and T2 have mean zero and equal QTL effect; this can always be achieved by subtracting the means and re-scaling.
Here, e1 and e2 represent variance in the biological process, not measurement errors; v1 and v2 denote the variances of e1 and e2; and vt1 and vt2 denote the total variance which is sum of the QTL and the biological variances. The ratio v1/vt1 is the proportion of total variance that is not explained by the QTL.
Multiply by 1/nA+1/nB in case of two genotypes where nA (nB) is the number of samples with genotype A (B); multiply by 4n/(n(nA + nB) − (nA − nB)2) in case of three genotypes where n = nA + nH + nB is the total number of samples. Note that 4n/(n(nA + nB) − (nA − nB)2) = 1/nA + 1/nB if nH=0.
Acknowledgements
This work was funded by EU 7th Framework Programme under the Research Project PANACEA, Contract No. 222936 to YL, and by the BioRange programme from the Netherlands Bioinformatics Centre (NBIC), which is supported by a BSIK grant through the Netherlands Genomics Initiative (NGI) to BMT.
Glossary
- Allele Frequencies
At a given polymorphic locus, the different alleles may have different predominance within the studied population. In GWLS using a cross originating from two inbred founders, the QTL has two alleles in equal frequencies in the population under study. By contrast, in GWAS due to a combination of random segregation, drift and selection, allele frequencies can be markedly different from equal. Imbalanced allele frequencies are less optimal for QTL detection
- Causal anchor
Causal anchors are causal relationships that are provided by knowledge external to the data. Because meiotic recombination is a random process that predates the establishment of phenotypes, correlation between DNA variation (QTL) and a trait implies causation of the DNA variation on the trait variation in experimental populations: QTL can therefore be used as causal anchors. The assumption should be carefully evaluated in natural populations, which may have hidden structure, or in case-control studies where sampling may indirectly alter allelic associations.
- Causal inference
A process of determining whether variation observed in a trait is a cause or a consequence of variation observed in another trait. Here we adopt the definition used in [3] that causality is defined by the effects of intervention in a system. If X is a cause of Y, then we can predict that an intervention that alters the level of X will result in a change in Y.
- Correlation
Correlation is a statistical measure of how much two variables change together. Correlation best captures linear relationships between variables (on original scale or after a transformation).
- Distant eQTL
A distant (or trans) eQTL is an eQTL which is located far from the gene it controls (for example on a different chromosome).
- eQTL
An expression Quantitative Trait Locus is a region in the genome at which allelic variation correlates with the mRNA expression level variation of a certain gene.
- Genome-wide association studies (GWAS)
A genome wide association study is an experiment in which the genomes of unrelated individuals is screened for genetic markers (typically millions of single nucleotide polymorphisms) at which allelic variation correlates with variation in studied traits.
- Genome-wide linkage studies (GWLS)
A genome wide association study is an experiment in which the genomes of related individuals is screened for genetic markers (typically a few hundreds or thousands of single nucleotide polymorphisms) at which allelic variation correlates with variation in studied traits. Examples of GWLS include experimental crosses such as recombinant inbred panels, intercrosses and backcrosses.
- Local eQTL
A local (or cis) eQTL is an eQTL which is located nearby the gene it controls in the genome. Often a local eQTL will be caused by allelic variation in the regulatory region of the gene or within the gene itself.
- mQTL
A metabolite Quantitative Trait Locus is a region in the genome at which allelic variation correlates with the abundance variation of a certain metabolite.
- pQTL
A protein Quantitative Trait Locus is a region in the genome at which allelic variation correlates with the abundance variation of a certain protein. Just like eQTL, pQTL can be local or distant according to the genomic position of the gene encoding for the protein relative to the QTL.
- Prior
A prior (or prior probability) reflects the initial belief in a given proposition (such as “Trait T1 is causal for trait T2”) before observing the data. The application of Bayes' rule combines the evidence provided by observed data with the prior to provide a measure of evidence of the proposition that accounts for previous experience or external knowledge.
- QTL confidence interval
QTL mapping identifies regions of the genome in which allelic variation is linked or associated with a certain trait. The sample size, the density of available genotyped markers and the extent of recombination in the QTL region within the studied population are among the factors that influence the size of the confidence interval. Confidence intervals can extend from only a few hundred kilo base pairs to several mega base pairs complicating the identification of the actual polymorphism behind the QTL.
- QTL mapping
A genomic region is said to be a Quantitative Trait Locus for a trait if allelic variation in this region correlates with trait variation. QTL can be mapped through GWAS or GWLS.
- QTL-trait-trait triads
A set constituted by a QTL and two traits mapping to that QTL. Since a QTL can affect directly a trait, or indirectly through another intermediary trait, multiple causal scenarios can explain this triad as illustrated in particular by the blue models in Figure 1. This article discusses our ability to discriminate between those different scenarios.
- Regression
Regression is a statistical procedure which evaluates the dependence between a variable (e.g. a trait) and one or multiple other variables (e.g. another trait, or QTL genotypes).
- Residuals
In a regression, residuals are the differences between the observed values and the values fitted by the regression.
- Variance
Variance is a statistical parameter that quantifies the spread in the distribution of a variable. For phenotypic traits variance originates from both genetic and non-genetic sources and we can estimate the proportion of trait variance that is contributed by a given QTL
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright S. Correlation and causation. J. Agric. Res. 1921;20:557–585. [Google Scholar]
- 2.Duffy DL, Martin NG. Inferring the direction of causation in cross-sectional twin data: theoretical and empirical considerations. Genet Epidemiol. 1994;11:483–502. doi: 10.1002/gepi.1370110606. [DOI] [PubMed] [Google Scholar]
- 3.Pearl J. Causality: models, reasoning, and inference. Cambridge University Press; 2000. [Google Scholar]
- 4.Spirtes P, et al. Causation, Prediction, and Search. Springer-Verlag; 1993. [Google Scholar]
- 5.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 9.Chen LS, et al. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome Biol. 2007;8:R219. doi: 10.1186/gb-2007-8-10-r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aten JE, et al. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst Biol. 2008;2:34. doi: 10.1186/1752-0509-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millstein J, et al. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaibub Neto E, et al. Inferring causal phenotype networks from segregating populations. Genetics. 2008;179:1089–1100. doi: 10.1534/genetics.107.085167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockman MV. Reverse engineering the genotype-phenotype map with natural genetic variation. Nature. 2008;456:738–744. doi: 10.1038/nature07633. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- 15.Bing N, Hoeschele I. Genetical genomics analysis of a yeast segregant population for transcription network inference. Genetics. 2005;170:533–542. doi: 10.1534/genetics.105.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, et al. Inferring gene transcriptional modulatory relations: a genetical genomics approach. Hum Mol Genet. 2005;14:1119–1125. doi: 10.1093/hmg/ddi124. [DOI] [PubMed] [Google Scholar]
- 17.Kulp DC, Jagalur M. Causal inference of regulator-target pairs by gene mapping of expression phenotypes. BMC Genomics. 2006;7:125. doi: 10.1186/1471-2164-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visscher PM, et al. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 19.Li R, et al. Structural model analysis of multiple quantitative traits. PLoS Genet. 2006;2:e114. doi: 10.1371/journal.pgen.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, et al. Gene network inference via structural equation modeling in genetical genomics experiments. Genetics. 2008;178:1763–1776. doi: 10.1534/genetics.107.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. Generalizing genetical genomics: getting added value from environmental perturbation. Trends Genet. 2008;24:518–524. doi: 10.1016/j.tig.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Akey JM, et al. On the design and analysis of gene expression studies in human populations. Nat Genet. 2007;39:807–808. doi: 10.1038/ng0707-807. author reply 808–809. [DOI] [PubMed] [Google Scholar]
- 26.Kang HM, et al. Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics. 2008;180:1909–1925. doi: 10.1534/genetics.108.094201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehrmann RS, et al. A new perspective on transcriptional system regulation (TSR): towards TSR profiling. PLoS One. 2008;3:e1656. doi: 10.1371/journal.pone.0001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegle O, et al. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 32.Sargon JD. The estimation of economic relationships using instrumental variables. Econometrica. 1958;26:393–415. [Google Scholar]
- 33.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]