Abstract
Actomyosin powers muscle contraction and various cellular activities including cell division, differentiation, intracellular transport and sensory functions. Despite their crucial roles, key aspects of force generation have remained elusive. To perform efficient force generation, the powerstroke must occur while myosin is bound to actin. Paradoxically, this process must be initiated when myosin is in a very low actin-affinity state. Recent results shed light on a kinetic pathway selection mechanism whereby the actin-induced activation of the swing of myosin’s lever enables efficient mechanical functioning. Structural elements and biochemical principles involved in this mechanism are conserved among various NTPase–effector (e.g. kinesin–microtubule, G protein–exchange factor and kinase–scaffold protein) systems that perform chemomechanical or signal transduction.
Allosteric activation in motor and signaling systems
Actomyosin is the best-understood of the ubiquitous mechanochemical systems of eukaryotic organisms [1,2]. The key action of the actomyosin mechanochemical cycle is the powerstroke, a rowing action of the myosin head which results from the swing of its lever (Box 1). To produce effective translocation along actin, the lever swing must occur with the myosin head in an actin-bound state (Fig. 1); the occurrence of a futile lever swing in an actin-detached state leads to an ATP-wasting cycle (Fig. 1). The powerstroke can be viewed as the result of three linked events: binding of the myosin head to actin, structural changes in the head leading to a strong actomyosin interaction, and the lever swing. These events lead to the release of ATP hydrolysis products, Pi (inorganic phosphate) and ADP.
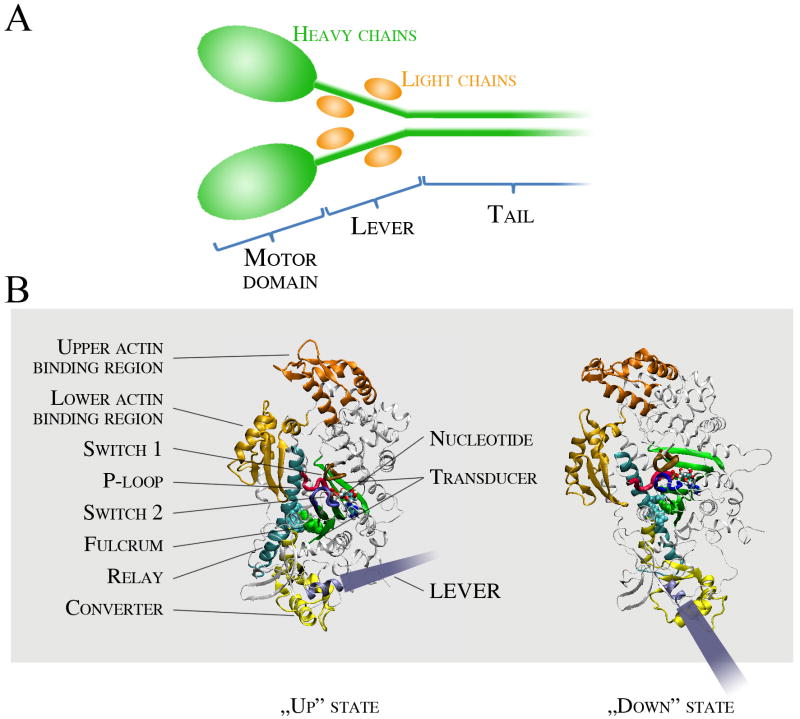
Box 1. Myosin structural elements.
Myosin is a P-loop NTPase, and diverse isoforms of myosin are divided into at least 35 classes [5]. Myosin has three functional parts: the motor domain, the lever and the tail region (Fig. IA). The heavy chain subunit forms the motor domain, the axis of the lever, and the tail, whereas the light chains stabilize the structure of the lever. In some myosin classes, further light chains bind to the tail region. The tail generally confers effector functions and, in some classes, is responsible for heavy chain dimerization. In the myosin II class, which includes the muscle myosins, the tail is a long coiled-coil rod responsible for filament formation. The motor domain confers the mechanoenzymatic function of myosin and moves the lever. It has three main functional parts: the nucleotide pocket, the actin binding region and the relay/converter/transducer region (Fig. IB). The nucleotide binding pocket is conserved throughout P-loop NTPases. The bound nucleotide is complexed by three loops called P-loop (pink), switch 1 (brown) and switch 2 (blue). Switch 1 faces towards the actin binding region, whereas switch 2 makes connections with the relay/converter/transducer element. The actin binding region is divided by a deep, long cleft. The interaction of the “lower” part of the actin binding region (gold) is thought to initiate a weak-binding interaction with actin. Upon cleft closure, the upper part of the actin binding region (orange) forms further interactions with actin, leading to strong actin binding. The relay/converter/transducer elements propagate conformational information from the nucleotide pocket to induce lever swing. During the powerstroke, the lever swings from an up (primed) to a down position. The length of the lever thus determines the stroke size [36]. The relay (cyan) is a long helix which moves as a seesaw around a phenylalanine cluster. The tilt of the relay seesaw swings the converter (yellow), which is directly connected to the lever (grey). The transducer (green) is formed by a central seven-stranded β-sheet, which is another structural element conserved among actin- and microtubule-based motors. The transducer is distorted in certain enzymatic states, and the relaxation of this strain might contribute to force generation during the powerstroke.
Box 1, Figure I. Structure of the myosin holoenzyme and the motor domain.
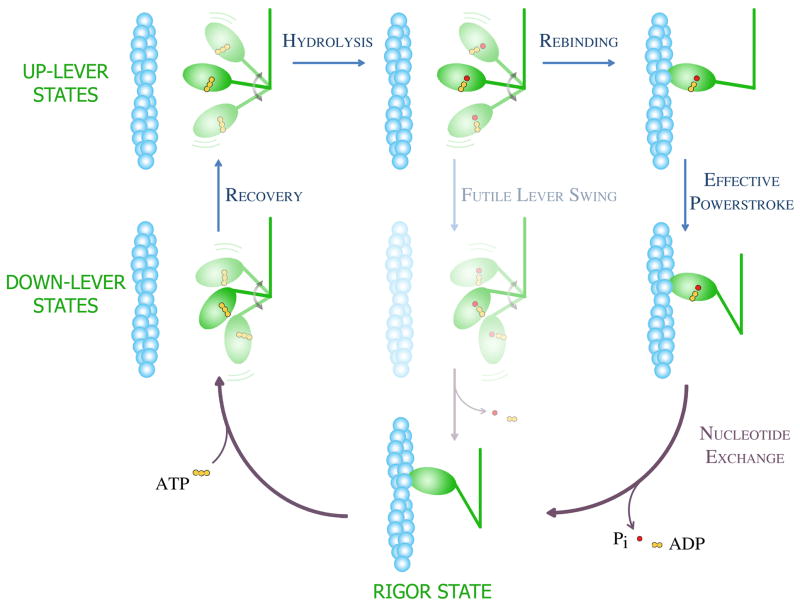
Figure 1. Mechanochemical cycle of the actomyosin motor.
The mechanism shown incorporates the Lymn-Taylor model in conjunction with major conceptual advances gained from recent structural and kinetic investigations. The myosin head and the actin filament are shown in green and blue, respectively. During the working cycle, ATP binding to the myosin head dissociates the strongly-bound actomyosin “rigor” complex (lowest panel). During the recovery step (left two panels), which occurs in ATP-bound myosin, the myosin lever moves from a “down” to an “up” position. Note the change in lever orientation relative to the motor domain (see also Box 1, Fig. IB). In the actin-detached states (upper left four panels), the myosin head (motor domain plus lever) is shown in several different orientations to indicate its free rotation about a flexible joint that connects it to the distal part of the molecule (and the thick filament in muscle). The hydrolysis of ATP to ADP and Pi (inorganic phosphate) occurs only in the up-lever state (upper left panel). The post-hydrolytic up-lever complex (upper middle panel) can continue the cycle in two pathways. If the lever swings back to a down position when the head is detached from actin (futile lever swing, middle panels), the ATP hydrolysis cycle is completed without work production. In order to undergo an effective powerstroke leading to force generation, the head must rebind to actin (upper right panel) before the lever swing (right two panels). When the lever swing is completed, the hydrolysis products are exchanged to a new ATP molecule. Note that this scheme does not indicate changes in actin affinity and motor domain structure.
Despite several decades of intense study, important problems related to ATP hydrolysis-linked enzymatic force generation remain unresolved [1,3,4]. The complex process of the powerstroke is not readily accessible owing to the low abundance and short lifetime of reaction intermediates [2]. The most intriguing question is how the powerstroke is initiated and why post-hydrolytic myosin heads bind to actin prior to lever arm swing when they are still in a low-actin-affinity state. Such a mechanistic question cannot be adequately addressed by models merely consisting of a succession of structural states. More detailed dynamic approaches are needed, such as that of Lymn and Taylor who were the first to account for the mechanistic role of actin-activated product release (Box 2). Although they are fundamentally important processes, the dissection of the lever swing and product release steps have proven especially problematic. Here we provide a synthesis of recent results to show that enzymatic activation of the myosin lever swing is a key factor in channeling the system into effective force generating pathways.
Box 2. The road to our current understanding of actomyosin.
In the classical Studies from the Institute of Medical Chemistry Albert Szent-Györgyi and colleagues described that muscle contraction is based on the cyclic interaction of two proteins, coupled to the hydrolysis of ATP [37]. They were the first to isolate actin and myosin. A few years later Andrew Szent-Györgyi and Endre Bíró described that actin markedly accelerates the ATPase activity of myosin, a phenomenon now termed “actin activation” [38].
A. F. Huxley and Niedergerke, in parallel with H. E. Huxley and Hanson, established the sliding filament model, which describes contractile movement as the sliding of intercalating thin (actin) and thick (myosin) filaments past each other [39,40]. They proposed that sliding was driven by the swing of crossbridges (later identified as myosin heads) while bound to the thin filament. Lymn and Taylor associated the biochemical cycle of actomyosin with its mechanical cycle (Fig. 1) [41], whereas Bagshaw and Trentham provided a detailed characterization of the ATPase enzymatic cycle of myosin [42,43]. They showed that myosin binds ATP rapidly and with high affinity, whereas the binding affinities of the products (ADP and Pi) are much weaker, and that the escape of Pi from the metastable myosin–ADP–Pi complex is very slow and rate-limiting in the absence of actin. Actin binding was proposed to provide a route to lower the energy barrier of Pi release. In the early 1990s, the atomic structures of the actin monomer and the myosin head were determined [11,44,45]. These structures supported the lever theory, which states that actomyosin sliding is the consequence of lever movement Fig. 1, Box 1).
During the past twenty years, crystal structures of several myosin conformational states have been solved, each associated with different kinetic states of myosin [2,46,47]. These structures facilitated the delineation of allosteric communication pathways underlying ATP-induced actomyosin dissociation and lever priming (Box 1). Despite long-standing efforts, the actomyosin complex could not be crystallized [16]. However, a 1.4-nm resolution structure has been created by fitting the X-ray structures of actin and myosin into the cryo-electron microscopic envelope of the nucleotide-free (“rigor”) actomyosin complex [17].
The force and displacement produced by the myosin head during the powerstroke was determined in single-molecule mechanical experiments. These studies also revealed that the energy barriers of myosin enzymatic steps, especially those of nucleotide binding, ADP release, and coupled conformational changes, depend on the mechanical load applied on the lever [48-52].
In addition to the muscle myosins, numerous newly discovered forms and classes of myosin have been identified, which are specialized for a variety of cellular functions [5]. These different myosins display functionally important variations in their structural and kinetic behavior, but they all share a common mechanistic framework: their translocation along actin is associated with lever swing, and this swing is precisely linked to the actin association-dissociation cycle driven by a cyclic change of the contents of the nucleotide binding site [6]. This feature underscores the importance of a unified powerstroke model. Moreover, mechanistic knowledge of how actin mediates myosin conformational changes might enhance the understanding of the activation of diverse NTPases by track or effector proteins. Indeed, allosteric activation is an evolutionarily conserved general mechanism among motor proteins and signaling enzyme complexes including kinesin–tubulin, G protein–exchange factor (GEF) [7] and kinase–scaffold systems [8].
Futile lever swing is kinetically blocked
After rapid binding of ATP to myosin, a fast conformational equilibrium forms between down- and up-lever states (recovery step), which is followed by the hydrolysis of ATP (Fig. 1). Myosin can hydrolyze ATP only in the up-lever state and, thus, the cycle is pulled forward to lever priming [2,9]. Recent work showed that, in the absence of actin, the rate-limiting step of the cycle is the oncoming up-to-down “futile” lever swing occurring in the myosin–ADP–Pi complex, and not the subsequent release of Pi as was previously thought (Fig. 1, Box 2) [10]. Pi release is possible only from the down-lever state, and the product release steps are several orders of magnitude faster than the rate-limiting up-to-down structural transition. Thus, during steady-state enzymatic cycling in the absence of actin, myosin predominantly populates an ADP- and Pi-bound up-lever state.
The crystal structures of the up- and down-lever myosin states have been solved, and the transition between these states (the recovery step) has been investigated in detail by in silico simulations [11,12]. The nucleotide binding site and the lever are connected via the switch 2 loop of the nucleotide pocket, the relay/converter region and the central (“transducer”) β-sheet of the motor domain (Box 1). The lever swing in ATP (recovery step) and ADP–Pi states (futile lever swing), associated with switch 2 conformational changes, has been proposed to involve a seesaw-like movement of the long relay helix within the motor domain [12]. In the middle of the relay helix, a phenylalanine cluster forms a fulcrum around which the tilt might occur [12,13]. This cluster structurally connects the relay/converter and transducer regions. The latter structural element is thought to play an important role in lever swing: changes in the torsional strain of the transducer β-sheet might contribute to the free energy change associated with the powerstroke [14-16]. This model involves the idea that the conformational pathway of the lever swing in the actin-detached state is different from that during the powerstroke [2]. The futile lever swing is thought to be a reversal of the recovery step, with the important difference being that the former occurs in ADP–Pi-bound myosin whereas the latter occurs in ATP-bound myosin (Fig. 1). However, it is still unclear why the futile lever swing is four orders of magnitude slower than the recovery step [10]. Regardless of the underlying mechanism, this kinetic difference is crucial for efficient force generation.
Nucleotide content of the active site allosterically determines actin affinity
It has long been known that the nucleotide-free and ADP-bound forms of myosin bind actin strongly (with a Kd ≤ 1 μM), whereas myosin–nucleotide complexes in which the γ-phosphate site is occupied (i.e. ATP and ADP–Pi) have weak actin affinity (Kd > 100 μM at physiological ionic strength) [2]. This coupling can be explained by an allosteric linkage between the actin binding region and the nucleotide pocket, located in relatively distant parts of the motor domain (Box 1). The conformation of the actin binding region determines actin affinity. This region is divided by a large cleft, which must close in order for myosin to adopt a strong actin binding conformation [16,17]. The switch 1 loop of the nucleotide pocket can adopt two distinct conformations (open and closed) depending on the occupancy of the γ-phosphate site [18]. In the presence of ATP and ADP–Pi (or their analogs), the equilibrium is shifted to the closed-switch-1 state, whereas the open and closed states are almost equally populated in the presence of ADP. Crystal structures and fluorescence experiments indicate that switch 1 opening might be coupled to the closure of the acting binding cleft, thereby delineating an allosteric route that links nucleotide states to actin affinity [4,18-20]. Actin affinity thus mainly depends on the equilibrium constant of the switch 1 open-closed transition. It remains unresolved whether actin binding is regulated only by the switch 1 equilibrium or also by the position of the switch 2/relay/converter/lever region, and how the weak-to-strong actin binding transition is involved in the mechanism of the powerstroke [2,4]. Importantly, rapid product release can only occur when switch 1 or switch 2 or both are open. Both nucleotide and Pi release are much slower when switch 1 is closed, and Pi release is extremely slow in closed-switch-2 states.
Much like the uncertainty surrounding the mechanism of coupling between switch-1 and the actin cleft, the conformational equilibria of switch 2 and the relay/converter region link lever movement and Pi release, which likely happen differently in actin-detached and actin-attached states, is incompletely understood. Nonetheless, the key process of the mechanochemical cycle is the powerstroke, which consists of three coupled events: the binding of myosin to actin, the weak-to-strong actin binding transition, and the lever swing. In the next sections we describe the potential order of these events and the kinetic/thermodynamic background of the coupling mechanism.
Actin-induced acceleration of the lever swing channels the pathway towards effective powerstroke
In the absence of actin, the rate-limiting step of the myosin enzymatic cycle is the futile lever swing occurring in ADP–Pi state (Fig. 1) [10]. Actin accelerates the lever swing of ADP–Pi-bound myosin by at least two orders of magnitude. This lies in stark contrast to the recovery step occurring in ATP-bound form—actin has little effect on this process [21]. Actin activation is therefore a crucial component in driving the system through effective (actin-attached) powerstroke even though it starts in a weak-actin-affinity (ADP–Pi) state. Principally, the reaction flux is channeled into the pathways involving lever swing in actin-attached states via kinetic pathway selection (Box 3). As the actin-detached futile lever swing step is kinetically blocked, the predominant reaction flux is diverted towards actin attachment even though this pathway is thermodynamically less favorable owing to low actin affinity of the myosin–ADP–Pi complex. This model highlights the importance of non-equilibrium situations in pathway selection mechanisms applied by biochemical systems (Box 3).
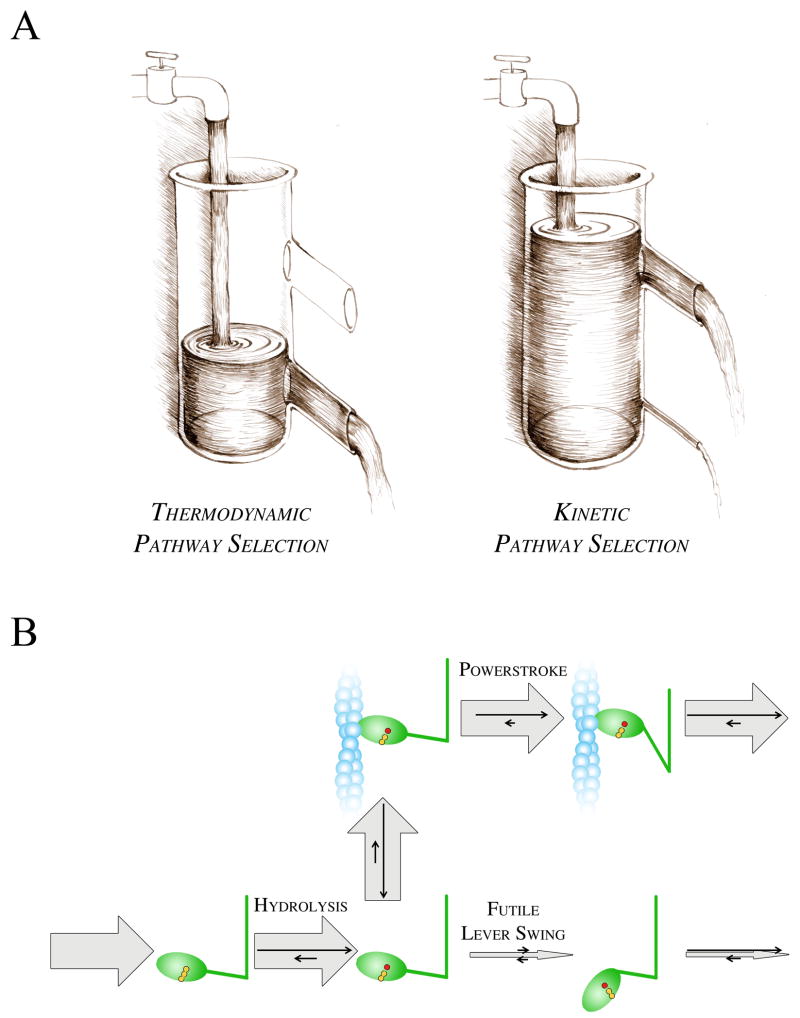
Box 3. Kinetic and thermodynamic pathway selection.
Biochemical reactions can happen in parallel pathways. In certain conditions, one of these pathways must be selected for efficient biological function. Theoretically, pathway selection can occur in a thermodynamic or a kinetic manner. Thermodynamic pathway selection occurs when the system follows the lowest free energy path (Fig. IA, i). Kinetic pathway selection can occur only in non-equilibrium situations when the flux through the lowest free energy route is hindered to the extent that the system will use an alternative escape route through a higher free energy path (Fig. IA, ii). In kinetic pathway selection, reaction rates prior to the “branching” intermediate (represented by water accumulated in the vessel) must be greater than that of the hindered step.
Kinetic pathway selection is a commonly used switch mechanism in biochemical systems where the reaction must be channeled into the efficient pathway instead of futile ones. In signal transduction systems, certain pathways are also selected in this manner. In the actomyosin system, actin activates the rate-limiting step of the myosin ATPase cycle, i.e. the post-hydrolytic futile lever swing (Fig. IB). Because the flux through the futile lever swing is greatly suppressed in the absence of actin, the main flux will proceed through the much faster actin-attached pathway despite the low actin affinity of myosin. In a representative example, the rate-limiting lever swing is accelerated by actin by 100 times and the actomyosin dissociation constant is Kd = 100 μM. In these conditions, as much as 91 % of the reaction flux will proceed through the actin-attached pathway even at a low actin concentration of 10 μM, although only 9 % of myosin heads would bind to actin in an equilibrium situation.
Box 3, Figure I. Pathway selection mechanisms.
A, Water flow analogy of pathway selection. Heights represent free energies, whereas the magnitude of rate constants is symbolized by the width of outlets (escape routes). If the lower outlet is wide enough to release all the water flowing in, then all flux will be conveyed through this outlet (i). Conversely, if the influx rate remains unchanged but the lower outlet is narrow, water will accumulate in the vessel up to the point where the main flux will be conveyed by the wider upper outlet (ii). B, Pathway selection in actomyosin. Relative magnitudes of fluxes and rate constants are represented by thick gray and thin black arrows, respectively.
Coupling between actin binding, cleft movement and lever swing during the powerstroke
An efficient mechanical step can only take place if lever swing occurs when the myosin head is bound to actin. However, there are several pathways that can produce an efficient powerstroke. The different pathways determine different energetic profiles for the powerstroke, which could greatly influence the mechanical performance of the system. Thus, it is fundamentally important to determine the kinetic fluxes of the powerstroke through different parallel reaction pathways.
The powerstroke can be defined as involving three key events including actin binding, a structural change leading to strong actin binding (linked to cleft closure), and the lever swing. The process starts in an actin-detached, up-lever, weak-actin-binding (open-cleft) form of myosin, whereas it ends in a strongly actin-bound (closed cleft), down-lever state (Fig. 2). Results on muscle fibers, from several laboratories, show that force generation (and thus the major lever swing) occurs before Pi release, and thus all substeps (Fig. 2) could occur in an ADP–Pi-bound form of the myosin head [22-25]. The possible modes of coupling between substeps define three possible effective powerstroke pathways whereby the lever swing occurs in the actin-attached form of myosin. The other pathways lead to futile lever swing (Fig. 2). Thus, it is important to assess which pathways are the most probable and how the probability of futile pathways is kept relatively low. The powerstroke events (Fig. 2) are followed by nucleotide exchange, which is practically irreversible owing to the tight binding of the next ATP molecule; thus it pulls the system towards the next mechanochemical cycle.
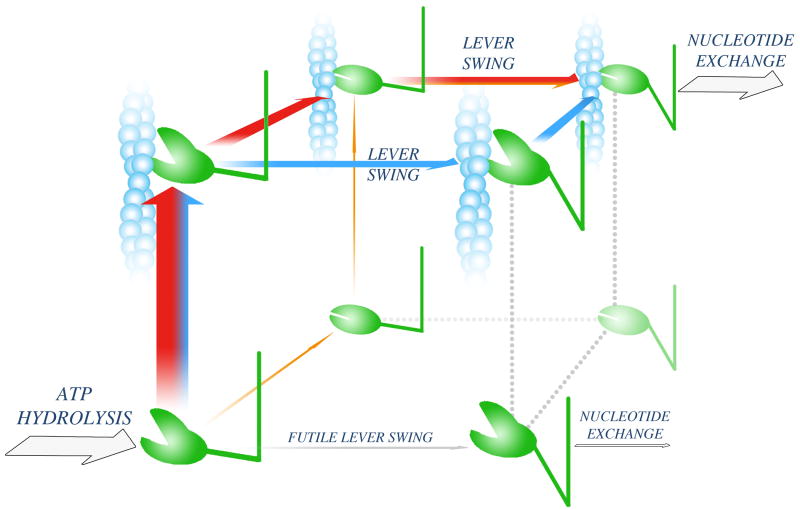
Figure 2. Powerstroke pathways.
Following ATP hydrolysis, the myosin head adopts a weak actin-binding (open-cleft), up-lever state (lower left corner). Three possible pathways leading to an effective powerstroke are shown as orange, red and blue arrows. On the orange pathway, cleft closure is followed by association to actin and subsequent lever swing. The other two pathways (red and blue) start with actin attachment. On the red pathway, cleft closure precedes lever swing. On the blue pathway, lever swing occurs while the cleft is still open, and the pathway is completed with cleft closure. Experimental data indicate that the flux through the orange pathway is limited, whereas the red and blue pathways might both convey significant fluxes. A futile lever swing, which would lead to an ATP-wasting cycle (grey arrow), is kinetically blocked (Box 3).
The starting state of the powerstroke process is formed rapidly after ATP binding and hydrolysis (Fig. 1). In all three possible effective pathways, actin binding must precede the lever swing (Fig. 2). The crucial role of actin activation is in preventing the flux through the pathway starting with a futile lever swing (grey arrow in Fig. 2), given that the rate constant of the latter step is at least 100 times lower than those of the lever swing in the actin-attached forms of myosin (see also Box 3).
Closure of the cleft in the first step denotes the first possible pathway for an effective powerstroke (thin orange arrow in Fig. 2). During this step, the myosin head adopts a strong actin-binding state, which can then attach to actin and perform the lever swing. However, the flux through this pathway is very low because the open-closed transition of the cleft in the actin-detached myosin-products complex, the first step in this sequence, is very unfavorable (K < 0.01) [18].
The remaining two effective powerstroke pathways (marked in red and blue in Fig. 2) both start with actin attachment, first leading to a weakly attached up-lever actomyosin complex. From this state, the strengthening of the actomyosin interaction and the lever swing can occur in two different sequences. On the pathway represented by red arrows, a rapid interconversion between open- and closed-cleft (weakly and strongly actin-bound) states is followed by the lever swing. Alternatively, the lever swing might occur while the myosin head is still weakly attached to actin and the subsequent strengthening of the actomyosin interaction could serve a stabilizing (“locking”) role for the post-powerstroke intermediate (blue arrows), provided that cleft closure occurs more rapidly than actin detachment [28]. Based on available kinetic and thermodynamic data, both the red and blue pathways could convey significant fluxes [10,25]. Thus, the effective powerstroke probably proceeds through parallel reaction pathways. It is important to note that, due to the fact that many events take place at similar timescales, the powerstroke intermediates never reach internal equilibration.
Importantly, the equilibrium constant of the interconversion between weak and strong actin binding states was determined in the absence of a mechanical load exerted on the lever. It will be important to determine whether this equilibrium depends on external load. Notably, recent findings indicate that a high actin affinity up-lever myosin state is stabilized in the presence of ADP and blebbistatin, a widely-used myosin inhibitor [29-31]. This complex might resemble a previously inaccessible powerstroke intermediate in which the weak-to-strong actin binding transition has taken place but the lever swing has not occurred.
Concluding remarks and future perspectives
In summary, the powerstroke is initiated by the binding of a low actin-affinity (open-cleft) state of myosin to actin. Because the flux involving the futile lever swing in actin-detached myosin is limited due to kinetic reasons, myosin binds to actin before the lever swing despite the fact that this pathway is thermodynamically unfavorable. Thus, allosteric activation by actin provides a kinetic type of pathway selection, which determines a high ratio of fluxes through efficient versus futile pathways. Importantly, this activation mechanism is not required for motor function per se, but it determines the energetic efficiency of the mechanochemical system. The degree of actin activation is generally lower in myosin isoforms that, under some conditions, primarily exert load-bearing functions as opposed to rapid contraction. These myosins include smooth muscle and non-muscle myosin II, and other myosins from classes I, V and VI functioning as load-dependent anchors or processive transporters [6]. In such isoforms, in addition to the powerstroke mechanism discussed herein, an additional lever swing occurring in ADP-bound actomyosin greatly contributes to load-sensitive tuning of actomyosin mechanical function [32].
Activation mechanisms similar to that described for actomyosin can be found in various NTPase-effector systems exerting motor and signaling functions, including microtubule-based motors, GEFs and kinase–scaffold protein complexes [8,33-35]. In the large family of P-loop NTPases, conserved switch loops within the active site sense the nucleotide state and transmit conformational changes associated with nucleotide binding, hydrolysis or product dissociation to the partner binding interface. Similarities between the organization of functional domains and active site chemistry suggest that the cores of these NTPases have diverged from a common ancestral nucleotide-binding motif, whereas later insertions enabled functional specialization [34]. The primary common mechanistic principle applied during the functioning of these diverse enzymes is the allosteric activation of the NTPase which occurs in a NTPase–nucleotide–partner ternary complex [7]. In many cases, nucleotide exchange is an intrinsically slow process, which is accelerated by the partner, often through interactions with a Mg2+ ion associated with the bound nucleotide. Specific modulations to this common mechanism precisely determine the mechanochemical activities or signaling properties of individual NTPases (Box 4). Much like the actomyosin system, which has been characterized in greatest detail, kinetic pathway selection probably plays an important role in delineating physiologically-functional biochemical pathways.
Box 4. Outstanding questions.
How is effective force generation initiated in motor enzyme systems instead of futile enzymatic cycling?
What is the biological role of the allosteric activation of motor and signaling enzymes by track or effector proteins?
Acknowledgments
AMC is supported by the European Research Council. MK is supported by the Fogarty International Center and the National Heart, Lung and Blood Institute (Grant 1 R01TW007241), and Hungarian Scientific Research Fund (OTKA) grants K71915 and NNF78783. We thank Andrew G. Szent-Györgyi and Clive R. Bagshaw for insightful discussions. We also thank Zoltán Simon and Tamás Cserna for figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollard TD. Reflections on a quarter century of research on contractile systems. Trends Biochem Sci. 2000;25:607–611. doi: 10.1016/s0968-0004(00)01719-9. [DOI] [PubMed] [Google Scholar]
- 2.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith DA, et al. Towards a unified theory of muscle contraction. I: foundations. Ann Biomed Eng. 2008;36:1624–1640. doi: 10.1007/s10439-008-9536-6. [DOI] [PubMed] [Google Scholar]
- 4.Sweeney HL, Houdusse A. Structural and Functional Insights into the Myosin Motor Mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 5.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Goody RS, Hofmann-Goody W. Exchange factors, effectors, GAPs and motor proteins: common thermodynamic and kinetic principles for different functions. Eur Biophys J. 2002;31:268–274. doi: 10.1007/s00249-002-0225-3. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 9.Bauer CB, et al. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- 10.Gyimesi M, et al. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J Biol Chem. 2008;283:8153–8163. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- 11.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4- Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 12.Fischer S, et al. Structural mechanism of the recovery stroke in the myosin molecular motor. Proc Natl Acad Sci U S A. 2005;102:6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kintses B, et al. Experimental investigation of the seesaw mechanism of the relay region that moves the myosin lever arm. J Biol Chem. 2008;283:34121–34128. doi: 10.1074/jbc.M805848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Coureux PD, et al. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes KC, et al. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes KC, et al. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 18.Kintses B, et al. Reversible movement of switch 1 loop of myosin determines actin interaction. EMBO J. 2007;26:265–274. doi: 10.1038/sj.emboj.7601482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yengo CM, et al. Actin-induced closure of the actin-binding cleft of smooth muscle myosin. J Biol Chem. 2002;277:24114–24119. doi: 10.1074/jbc.M111253200. [DOI] [PubMed] [Google Scholar]
- 20.Conibear PB, et al. Myosin cleft movement and its coupling to actomyosin dissociation. Nat Struct Biol. 2003;10:831–835. doi: 10.1038/nsb986. [DOI] [PubMed] [Google Scholar]
- 21.Conibear PB, et al. The effect of F-actin on the relay helix position of myosin II, as revealed by tryptophan fluorescence, and its implications for mechanochemical coupling. Biochemistry. 2004;43:15404–15417. doi: 10.1021/bi048338j. [DOI] [PubMed] [Google Scholar]
- 22.Dantzig JA, et al. Reversal of the cross-bridge force-generating transition by photogeneration of phosphate in rabbit psoas muscle fibres. J Physiol. 1992;451:247–278. doi: 10.1113/jphysiol.1992.sp019163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai M, Halvorson HR. Two step mechanism of phosphate release and the mechanism of force generation in chemically skinned fibers of rabbit psoas muscle. Biophys J. 1991;59:329–342. doi: 10.1016/S0006-3495(91)82227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleep J, et al. The ATP hydrolysis and phosphate release steps control the time course of force development in rabbit skeletal muscle. J Physiol. 2005;563:671–687. doi: 10.1113/jphysiol.2004.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caremani M, et al. Effect of inorganic phosphate on the force and number of myosin cross-bridges during the isometric contraction of permeabilized muscle fibers from rabbit psoas. Biophys J. 2008;95:5798–5808. doi: 10.1529/biophysj.108.130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis JS, Rodgers ME. Indirect coupling of phosphate release to de novo tension generation during muscle contraction. Proc Natl Acad Sci U S A. 1995;92:10482–10486. doi: 10.1073/pnas.92.23.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis JS, Epstein ND. Mechanistic role of movement and strain sensitivity in muscle contraction. Proc Natl Acad Sci U S A. 2009;106:6140–6145. doi: 10.1073/pnas.0812487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferenczi MA, et al. The “roll and lock” mechanism of force generation in muscle. Structure. 2005;13:131–141. doi: 10.1016/j.str.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Zhao FQ, et al. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu S, et al. Stabilization of helical order in the thick filaments by blebbistatin: further evidence of coexisting multiple conformations of myosin. Biophys J. 2009;96:3673–3681. doi: 10.1016/j.bpj.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takacs B, et al. Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke. Proc Natl Acad Sci U S A. 2010;107:6799–6804. doi: 10.1073/pnas.0907585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyitrai M, Geeves MA. Adenosine diphosphate and strain sensitivity in myosin motors. Philos Trans R Soc Lond B Biol Sci. 2004;359:1867–1877. doi: 10.1098/rstb.2004.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vale RD. Switches, latches, and amplifiers: common themes of G proteins and molecular motors. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kull FJ, et al. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil. 1998;19:877–886. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- 35.Thomas C, et al. Structural evidence for a common intermediate in small G protein-GEF reactions. Mol Cell. 2007;25:141–149. doi: 10.1016/j.molcel.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Uyeda TQ, et al. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci U S A. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banga I, et al. In: Studies from the Institute of Medical Chemistry. Karger S, Gergely R, editors. University Szeged; 1942. [Google Scholar]
- 38.Biro NA, Szent-Gyorgyi AG. The effect of actin and physico-chemical changes on the myosin ATP-ase system, and on washed muscle. Hung Acta Physiol. 1949;2:120–133. [PubMed] [Google Scholar]
- 39.HUXLEY H, HANSON J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 40.HUXLEY AF, NIEDERGERKE R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 41.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 42.Bagshaw CR, et al. The magnesium ion-dependent adenosine triphosphatase of myosin. Two-step processes of adenosine triphosphate association and adenosine diphosphate dissociation. Biochem J. 1974;141:351–364. doi: 10.1042/bj1410351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagshaw CR, Trentham DR. The characterization of myosin-product complexes and of product-release steps during the magnesium ion-dependent adenosine triphosphatase reaction. Biochem J. 1974;141:331–349. doi: 10.1042/bj1410331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W, et al. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 45.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 46.Houdusse A, et al. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coureux PD, et al. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 48.Debold EP, et al. Slip sliding away: load-dependence of velocity generated by skeletal muscle Myosin molecules in the laser trap. Biophys J. 2005;89:L34–L36. doi: 10.1529/biophysj.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovacs M, et al. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci U S A. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altman D, et al. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 51.Veigel C, et al. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 52.Veigel C, et al. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]