Abstract
Most eukaryotes including fungi, amoebas, and animal cells assemble an actin/myosin-based contractile ring during cytokinesis. The majority of proteins implied in ring formation, maturation, and constriction are evolutionarily conserved, suggesting that common mechanisms exist among these divergent eukaryotes. Here, we review the recent advances in positioning and assembly of the actomyosin ring in the fission yeast Schizosaccharomyces pombe, the budding yeast Saccharomyces cerevisiae, and animal cells. In particular, major findings have been made recently in understanding ring formation in genetically tractable S. pombe, revealing a dynamic and robust search, capture, pull, and release mechanism.
Keywords: Cytokinesis, contractile ring, node, anillin, myosin
1. Introduction
Cytokinesis, the final step of the cell cycle, is the process that divides one cell into two daughter cells containing a full set of chromosomes and other cellular components. Studies in fungi and animal cells have revealed similar steps during cytokinesis in these evolutionarily divergent eukaryotes [1–4]. First, cells define the location for the future cleavage site using positive and negative signaling cues. Then the actin cytoskeleton is drastically reorganized to form an actomyosin-based contractile ring consisting of actin filaments, the motor protein myosin-II, and many actin- and myosin-binding proteins. The subsequent interactions of actin filaments with myosin-II lead to the sliding of actin filaments and constriction of the assembled contractile ring. Lastly, in coordination with ring constriction, targeted secretion delivers membrane fusion machinery to the cleavage furrow, allowing the separation of the two daughter cells.
The fission yeast Schizosaccharomyces pombe has emerged as one of the favorite models for cytokinesis studies. Most proteins involved in cytokinesis are evolutionarily conserved from yeast to mammals [1,2,5], so understanding basic cytokinesis mechanisms in S. pombe could be applicable to animal cells. More than 130 proteins implied in cytokinesis have been identified and characterized in fission yeast [1,3,6–8], the current challenge is to piece these proteins together to elucidate the molecular mechanisms for cytokinesis. Besides this large inventory, the ability to integrate green fluorescent protein (GFP) and its variants into the genome by homologous recombination has been critical for microscopic studies of cytokinesis proteins expressed at their endogenous levels in live cells. Using high-resolution live cell imaging, many key cytokinesis proteins are now temporally and spatially localized, and their global and local concentrations are determined [9]. Combining numerical simulation and microscopy, contractilering assembly was mathematically described [10], providing a testable model for the field. Despite significant advances, several open questions remain regarding how cells are able to position, assemble, and contract the actomyosin ring during cytokinesis. This review aims to highlight the mechanism relating to contractile-ring assembly in fission yeast and other organisms.
2. Specification of the division site
Mechanisms that determine the position of the division plane vary depending on species. In S. pombe, the division-site selection is finalized at early mitosis and depends on both the nucleus and anillin-like protein Mid1p/Dmf1p. In the budding yeast Saccharomyces cerevisiae, the division plane is determined at late G1 phase by the budding site. In animal cells, the division site is dictated by the position of the mitotic apparatus at metaphase/anaphase. In each case, precise location of the division site ensures correct segregation of the nucleus and other components to daughter cells.
2.1. Division-site selection in fission yeast
In the fission yeast S. pombe, placement of the contractile ring (4 μm in diameter) and division site always occurs at the cell center, resulting in two daughter cells with equal size. Because S. pombe cells do not grow equally from each end after cell separation, the position of the medial cortex changes during cell-cycle progression. Thus, the mechanism by which cells specify their middle must be dynamic. Nuclear position is maintained in the cell middle through opposing pushing forces generated by the interactions between microtubules and cell cortex [11], which determines the localization of the anillin-like protein Mid1p.
Mid1p is crucial to position the division plane [12] and to assemble the contractile ring normally [13,14]. Before mitosis, Mid1p exits the nucleus and binds to the plasma membrane close to the nucleus in a band of ~65 cortical dots, called nodes, by interacting with kinase Cdr2p [12,15–17]. How Mid1p is positioned at the cell equator has attracted more and more attention in recent years. Mid1p localization is regulated by both positive and negative cues. Nuclear position determines Mid1p plasma membrane localization and the division plane [18]. Thus, changing the nuclear position affects the localization of cortical Mid1p nodes [19]. The polo kinase Plo1p interacts with Mid1p and is essential for Mid1p nuclear exit as observed in plo1-1 mutant [12]. Conversely, Plo1p overexpression induces premature Mid1p cortical accumulation mainly in the hyperphosphorylated form normally found in mitotic cells [12]. Thus, the interactions between Plo1p and Mid1p provide the positive signaling cue for Mid1p localization. However, it is not known whether Plo1p directly phosphorylates Mid1p or which residues are phosphorylated. Negative cues also regulate Mid1p localization: Pom1p (DYRK-family kinase) and other unknown protein(s) inhibit Mid1p localization at the cell poles [20,55]. The balance of the signaling cues restricts Mid1p to the cell equator, allowing the cell to localize the division plane precisely at the center. Then Mid1p acts as a scaffolding protein to recruit other proteins for contractile-ring assembly.
2.2. Division-site selection in budding yeast and animal cells
In the budding yeast Saccharomyces cerevisiae, the 1-μm diameter bud neck bridging the mother cell and the bud is the future cell-division site. Thus, the division site is determined at late G1 phase by the bud-site selection genes [21,22]. Because the mother cell is always bigger than the bud at division, cytokinesis is asymmetric in budding yeast. Five members of the septin family (Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p/Sep7p) interact with each other to form a collar-like structure at the bud neck serving to anchor the assembly of the actomyosin ring. Type II myosin Myo1p fails to localize to the bud neck in septin mutants [23–25]. By contrast, myo1 mutation does not affect septin recruitment to the division site, consistent with the model that septins are the scaffolding proteins at the division site in budding yeast.
In animal cells, the position of the mitotic apparatus at metaphase/anaphase dictates the division site. Spindle midzone or astral microtubules, or both are involved in the division site selection [2], though recent findings reveal that astral microtubules are not necessary for delivering the cytokinesis signals in sea urchin and frog embryos [26]. Accumulating evidence suggests that anillins might act as a scaffolding protein to recruit other proteins to the division site [27,28]. The spatial-temporal regulation of the division plane positioning is covered in more detail elsewhere in this series, and will not be elaborated here.
3. Assembly of an actomyosin-based contractile ring
Fission yeast, budding yeast, and animal cells all establish an actomyosin-based contractile ring during mitosis. Interestingly, although the majority of proteins implied in ring assembly are conserved among different species (Table 1), their temporal and spatial regulation and order of assembly are less conserved, probably reflecting the coordination of ring assembly with some species-specific features like cell size and cell shape. We know more about the stepwise assembly of the contractile ring in fission yeast than in other model systems. Thus, we consider three stages in the assembly process in fission yeast. The stages in budding yeast and animal cells are less distinct and so the assembly of the contractile ring is discussed as a whole.
Table 1.
Proteins involved in contractile-ring assembly and maturation.
| Generic name | S. pombe | S. cerevisiae | C. elegans | D. melanogaster |
|---|---|---|---|---|
| α-actinin | Ain1p | naa | ATN-1b | α-actinin |
| Actin | Act1p (Cps8p)c | Act1p | Actin | Actin |
| Anillin | Mid1p | na | ANI-1 | Anillin (Scraps) |
| Aurora B kinase | Ark1p (Aim1p) | Iplp | AIR-2 | Aurora B |
| Borealin | Nbl1p | Nbl1p | CSC-1d | Borr |
| Capping protein | Acp1p, Acp2p | Cap1p, Cap2p | Capping proteind | Capping proteind |
| Cdc14 phosphatase | Clp1p (Flp1p) | Cdc14p | CDC-14 | Cdc14 |
| Cofilin | Adf1p | Cof1p | UNC-60A | Twinstar |
| F-BAR domain containing protein | Cdc15p | Hof1p (Cyk2p) | TOCA-1b, TOCA-2b | Cip4b, Toca-1b, Fbp17b |
| Fimbrin | Fim1p | Sac6pb | PLST-1b | Fimbrinb |
| Formin | Cdc12p | Bni1p, Bnr1p | CYK-1 | Diaphanous |
| Hsp90 chaperone | Hsp90p (Swo1p) | Hsc82pb, Hsp82pb | HSP90 | Hsp90b |
| Inner centromere protein (INCENP) | Pic1p | Sli15p | ICP-1 | INCENP |
| IQGAP family protein | Rng2p | Iqg1p (Cyk1p) | IQGAP-related proteinb | na |
| Kinesin | Klp8pd | na | ZEN-4 | Kinesin-6 (Pavarotti) |
| Microtubule cross-linking factor | Ase1p | Ase1p | SPD-1 | Feo |
| Myosin II heavy chain | Myo2p, Myp2p | Myo1p | NMY-2 | Myosin II (Zipper) |
| Myosin II essential light chain | Cdc4p | Mlc1p | MLC-5 | Mlc-c |
| Myosin II regulatory light chain | Rlc1p | Mlc2p | MLC-4 | Spaghetti Squash |
| Phosphoinositide-dependent kinase | Pdk1p (Ppk21p) | Pkh1b, Pkh2b | PDK1b | DSTPK61b |
| Polo kinase | Plo1p | Cdc5p | PLK-1 | Polo |
| Profilin | Cdc3p | Pfy1p | PFN-1 | Chickadee |
| Rho GAP | na | na | CYK-4 | RacGAP50C |
| Rho GEF | Rgf3p, Rgf1pd, Gef2pd | Rom1p, Rom2p, Tus1p | LET-21 | Pebble |
| RhoA | Rho1p | Rho1p | RHOA | RhoA |
| ROCK | na | na | LET-502 | Rok/Drok |
| Septin | Spn1-4pd | Cdc3p, Cdc10-12p, Shs1p/Sep7p | UNC-59, UNC-61 | Peanut |
| Survivin | Bir1p (Cut17p) | Bir1p | BIR-1, BIR-2 | Faf |
| Tropomyosin | Cdc8p | Tpm1p,Tpm2p | CETMb | Tropomyosinb |
| UCS domain containing protein | Rng3p | She4p | UNC-45 | na |
na indicates no apparent known homologue involved in contractile-ring assembly.
Homologue known, but its relevance in contractile-ring assembly has not been determined. These proteins are also underlined.
The alternative name is in the “( )”.
The role in contractile-ring assembly needs further testing.
3.1. Contractile-ring assembly in fission yeast
3.1.1. Formation of cytokinesis nodes
As discussed in section 2.1, Mid1p is highly regulated to position the contractile ring correctly. However, it is still obscure how cells recognize the Mid1p positioning signal to initiate ring assembly. Mid1p-dependent equatorial nodes have been proposed to be the precursors of the contractile ring in wild-type cells [12,29–32]. The assembly and integrity of nodes are independent of actin filaments. More than 90 min before SPB separation, a fraction of Mid1p localizes in a broad band of ~65 nodes on the plasma membrane around the cell center [18]. Between 10 min before and 2 min after SPB separation, this broad band of Mid1p nodes are joined by conventional myosin-II (heavy chain Myo2p and its light chains Cdc4p and Rlc1p), IQGAP Rng2p, F-BAR protein Cdc15p, and the formin Cdc12p [29–32]. These studies find that Mid1p precedes other node proteins and nodes cannot form without Mid1p, reinforcing the idea that Mid1p initiates the formation of cytokinesis nodes. Of note, in absence of Mid1p, an actomyosin ring can still form, suggesting that nodes are not the only way for contractile-ring formation. However, the timing and efficiency of contractile-ring assembly and its orientation are seriously affected without nodes [33,34].
Actually, our knowledge of the node-assembly pathway is still limited, mainly due to limited information on protein interactions and localization dependence. It has been shown that the IQ domains of Myo2p interact with Cdc4p and Rlc1p [35,36]. Cdc4p also interacts with IQGAP Rng2p, probably via its IQ domains [35]. In one study, Myo2p immunoprecipitated with Mid1p when both proteins are overexpressed [37], so the authors proposed that dephosphorylated Myo2p (on S1444 residue) interacts with Mid1p to initiate ring assembly. However, a recent paper showed that Myo2p accumulates normally in nodes and assembles into a contractile ring even in a phospho-mimicking form of Myo2p [38]. Cdc15p interacts with Cdc12p [39], but their connection to the other node proteins is unknown. Unfortunately, the interactions of node proteins alone will not be enough to decipher the protein organization in nodes. To understand the architecture of a node, one solution could be to observe these proteins in vivo at nanometer resolution, as previously reported for kinetochores [40].
3.1.2. Condensation of nodes into a contractile ring
Condensation of nodes occurs from the onset of mitosis to the end of anaphase A. During this stage (zero to 10 min after spindle pole body separation), actin filaments, actin side-binding protein tropomyosin Cdc8p, and actin cross-linking protein α-actinin Ain1p join the broad band, and the nodes condenses into a compact ring at the cell equator [31]. Based on the local protein concentrations in the nodes [9], the properties of formin Cdc12p (nucleation, elongation, and anchoring the barbed end of an actin filament) [41,42] and Myo2p (gliding actin filaments to produce force) [43], and careful analysis of node movements by high temporal and spatial resolution microscopy, a mathematical model named search, capture, pull, and release (SCPR) was proposed to explain the formation of the contractile ring [10]. This model proposes that in each node, about two actin filaments are nucleated with their barbed ends anchored by Cdc12p dimers. These elongating filaments explore the neighboring space in random directions (search), because a formin dimer can successively nucleate/elongate actin filaments in different directions after each filament disassembles or breaks [44]. When a growing filament is within the capture radius (100 nm) of another node, Myo2p captures the filament and walks along the filament towards the node where the barbed end of the actin filament is anchored, producing forces that pull nodes closer to each other. Node motions are transient, because connecting actin filaments can break or dissociate from Myo2p (called release). The assumptions that Cdc12p nucleates randomly oriented actin filaments and that connections between nodes are transient are consistent with in vivo observations that nodes make many starts, stops, and changes of direction during condensation. Repeated cycles of these dynamic and transient interactions are sufficient to condense the nodes into a contractile ring in ~10 min [10].
The SCPR model represents an alternative to the spot/leading cable model [45–47]. These two models disagree mainly on three key events: formin Cdc12p origin and distribution, orientation of actin filaments, and the importance of myosin-II motor activity. In the spot/leading cable model, one Cdc12p spot moves to the equatorial plane at the onset of mitosis to form a unique actin nucleation site named the aster underneath the plasma membrane [45,48,49]. From this aster, one or two leading actin cables grow parallel to the plasma membrane but perpendicular to the cell's long axis and wrap around the cell circumference to form a contractile ring [39,45,46,49]. Thus, actin filaments with the same polarity (barbed ends facing the aster) occupy each half of the ring in early anaphase B [46]. To initiate ring constriction, actin filaments are assumed to be reoriented or mixed by Myo2p or formin Cdc12p to have filaments with opposite directionalities more homogeneously during late anaphase B [46]. However, the Myo2p motor activity is not required during ring formation according to the spot/leading cable model.
Recent reports help to distinguish the two models [29,38]. First, it was shown that Cdc12p localizes in >40 nodes, consistent with the SCPR model that requires Cdc12p to appear at the division site in multiple origins and to concentrate in >50% of Mid1p-myosin-II nodes [10]. Coffman et al. (2009) also shows that the Cdc12p spot does not nucleate actin filaments and disappears before accumulation of actin filaments at the division site in most cells. Moreover, cells without a Cdc12p spot do not display any cytokinesis defect [29,93]. If Cdc12p spot is not involved in ring formation under normal conditions, what is its function? This structure could serve to increase the local formin concentration after it dissolves at the cell equator. It is also possible that the spot might be involved in ring formation under stress conditions. Second, the careful analysis of Cdc12p nodes and actin filaments by high temporal and spatial resolution microscopy reveal that Cdc12p anchored actin filaments/bundles are parallel to the plasma membrane but in random orientation relative to the long axis of the cell [29]. This observation is consistent with the SCPR model but not with a previous observation by electron microscopy that actin filaments with the same polarity occupy each half of the ring in early cytokinesis [46]. This discrepancy about the orientation of actin filaments in the contractile ring remains to be solved in the future. The third key event where the two models disagree is on the importance of myosin motor activity for ring assembly. In order to investigate its role in the ring assembly process, condensation of nodes was observed in myo2-E1 and rng3-65, two mutants known to have diminished myosin motor activity at restrictive temperature [43]. Interestingly, a clear defect in node condensation was observed, indicating that myosin motor activity is required to condense the nodes together [29]. Consistent with the idea that Myo2p motor activity is critical for cytokinesis, reducing gliding activity by affecting Rng3p function or Rlc1p phosphorylation often leads to delay in contractile-ring formation [38,43].
Taken together, these data strongly support the SCPR model of contractile-ring formation where formin Cdc12p localizes at multiple nodes and nucleates actin filaments in random directions. Myosin motor activity is then required to condense nodes into a contractile ring by sliding actin filaments. However, at least two aspects of the SCPR model should be refined and further tested: 1) What are the roles of other node proteins? The SCPR model is only based on Cdc12p, myosin-II, and actin. The roles of IQGAP Rng2p and F-BAR protein Cdc15p in contractile-ring assembly remain mysterious, although Rng2p has been shown to bundle actin filaments [50]. 2) What are the roles of other actin binding proteins during node condensation? It has been observed in vivo that actin connections between nodes can be transiently stabilized [10]. The importance of this stabilization for ring assembly is unclear. Knowing more about biochemical properties of actin binding proteins like tropomyosin (Cdc8p), cofilin, fimbrin (Fim1p), and alpha-actinin-like protein (Ain1p) will help to describe actin behavior more precisely during ring assembly and to refine the SCPR model.
3.1.3. Maturation of the contractile ring
After the formation of a compact ring in fission yeast, the ring matures by recruiting additional proteins and by increasing the concentrations of some of the existing proteins. Ring maturation occurs from the beginning of anaphase B (10 minutes after SPB separation) until the initiation of ring constriction. During this prolonged stage (25 min), the ring maintains a constant diameter. The concentration of F-BAR protein Cdc15p in the ring increases about five fold [9] and new proteins like Myp2p (unconventional myosin-II), capping protein, septins, Arp2/3 complex, and presumably many other proteins are recruited to the ring or to the region adjacent to the ring.
The node protein Cdc15p is indispensable for ring maturation and stability, because cdc15 mutants are able to form an actomyosin ring that rapidly disassembles [33,51]. Ring stabilization also depends on the septation initiation network (SIN), as revealed by the observation that the ring disassembles in SIN mutants [33,52]. As we discussed in section 2.1, Mid1p is important to mark the future division site, and precedes all other cytokinesis node proteins to the division site. In mid1 null or mid1 thermosensitive mutants, no nodes are observed [31,34,37], but cells still establish an actomyosin filament or misplaced ring competent for contraction, indicating that Mid1p is not essential for ring assembly but provides the positional information. However, detailed studies reveal that the timing of ring assembly and its orientation are severely affected, and the ring also constricts more slowly [33,34]. Thus, cytokinesis nodes are not essential for ring assembly but make the process more efficient and reliable. Interestingly, it has been shown that overexpression of Cdc15p, COOH-terminal truncated formin Cdc12p, or GTPase Spg1p, a component of the SIN pathway, can induce actomyosin ring formation in interphase cells [33,53]. These observations show that another mechanism can “backup” ring formation when node formation is not possible.
Activation of the SIN pathway controls this backup mechanism for ring assembly [33,34]. Although tilted rings are observed in the absence of Mid1p, no rings form when both Mid1p and the SIN pathway are compromised. Conversely, tilted-ring formation in interphase cells depends on both SIN activation and Cdc15p function [51]. The model proposed by the authors is that in the absence of functional Mid1p, the SIN pathway rescues ring formation through Cdc15p, probably by regulating its phosphorylation status.
This “backup” mechanism raises two questions. First, since Mid1p determines ring localization, what kind of spatial cue is used to organize the actomyosin filament competent for contraction in the absence of Mid1p? Previously, it was proposed that Cdc15p associates and organizes lipid rafts at the division site. When Cdc15p is overexpressed, mislocalization of an abnormal lipid domain was observed [54]. One possibility is that actomyosin filament or ring formation depends on lipid rafts. The other possibility is that actin local concentration can determine the location of actomyosin filament formation as proposed previously [55]. Second, how are ring proteins recruited to form an actomyosin filament or ring? Do these proteins utilize the same interactions and temporal pathway as those of the normal ring assembly?
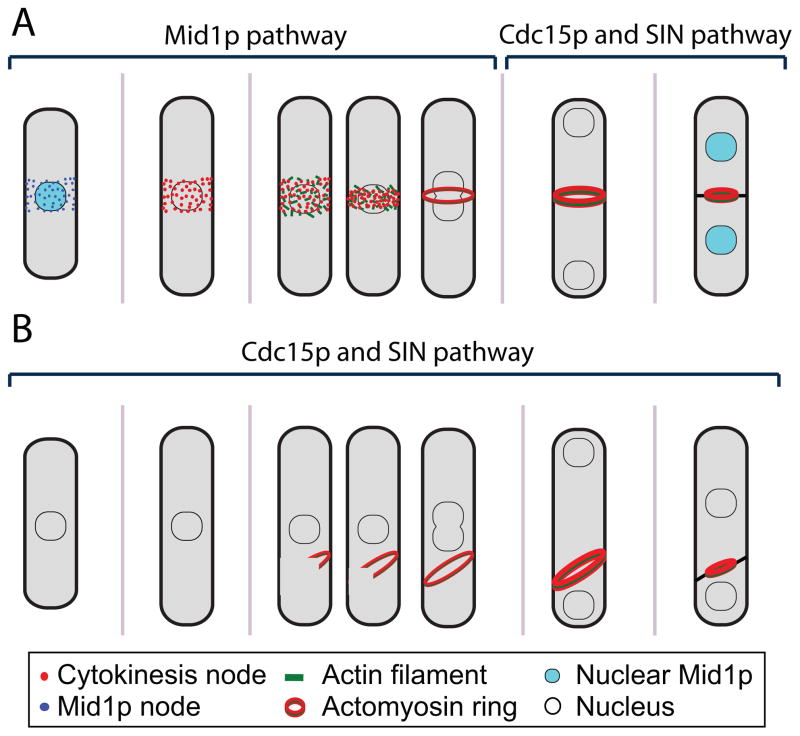
Taken together, the emerging model for actomyosin-ring formation implies two pathways: one depends on Mid1p and nodes during normal cytokinesis, and the other depends on the SIN pathway and Cdc15p (Figure 1). Plo1p, which regulates Mid1p localization and allows SIN activation, is proposed to coordinate these two pathways [33].
Figure 1.
Models for contractile-ring assembly in fission yeast. (A) Ring formation in wild-type cells. Anillin-like protein Mid1p localizes to a broad band of nodes at the equator before mitosis to specify the division site. At the onset of mitosis, Mid1p nodes become cytokinesis nodes by recruiting more proteins. Formins in the nodes nucleate actin filaments that elongate in random directions, and type II myosin Myo2p interacts with these filaments and moves nodes together to form a compact ring. F-BAR protein Cdc15p and the SIN pathway promote the maturation and constriction of the ring. During maturation, the contractile ring maintains a constant diameter and recuits more proteins. The septum forms as the ring constricts. (B) Ring formation in the absence of Mid1p depends on Cdc15p and the SIN pathway. When Mid1p function is compromised or absent, nodes cannot form and the positional cue for the contractile ring is lacking. Nevertheless, an actomyosin filament or ring competent for contraction can still form although its orientation and timing for assembly/constriction are severely affected. This actomyosin filament is also observed when Cdc15p is overexpressed or when the SIN pathway is activated during interphase.
3.2. Contractile-ring assembly in budding yeast
Unlike fission yeast, budding yeast myosin-II can form a ring without actin filaments. Myo1p, the only type II myosin in S. cerevisiae, forms a ring in the bud neck in late G1 or early S phase in a septin-dependent manner [23]. The tail of Myo1p is responsible for Myo1p localization to the bud neck [56]. The Myo1p ring remains in place until the end of anaphase, when it is joined by actin filaments to form an actomyosin ring [23]. We do not understand how Myo1p forms a ring at the budding site so early during the cell cycle or its role before cytokinesis. After Myo1p forms a septin-dependent ring, other proteins join the ring sequentially during interphase to early anaphase: formin Bnr1p, myosin essential light chain Mlc1p, F-BAR protein Hof1p, IQGAP Iqg1p/Cyk1p, and formin Bni1p [57–59]. Actin filaments appear in the ring in late anaphase. The presence of actin filaments is essential for the contractility of the Myo1p ring, but not for its formation.
The functional actomyosin ring contracts and disappears after mitotic exit. It will be interesting to know if and how actin may promote some reorganization of proteins in the ring. However, the small diameter of the ring and the presence of actin patches at the bud neck make this study challenging. It is of note that although budding yeast employs the contractile-ring mechanism for cell division, the contractile ring is not essential for cytokinesis in budding yeast [23,60].
A recent breakthrough has been achieved in S. cerevisiae with the successful isolation and partial purification of the cytokinesis machinery, which consisting of actin, myosin heavy and light chains, IQGAP, septins, and F-BAR protein Hof1p [61]. Studies of the isolated cytokinetic apparatus suggested that it may be functional in vitro. This achievement holds the potential to shed light on the structure of the assembled actomyosin ring in S. cerevisiae at the molecular level, and to apply similar methods to other genetically tractable species, like S. pombe.
3.3. Contractile-ring assembly in animal cells
Although the existence of a contractile ring based on actin filaments and myosin-II was established four decades ago in animal cells [62], much less is known about the assembly mechanism in animal cells than in S. pombe. Myosin-II and anillin are found to be enriched in node-like patches or puncta distributed over the cortex of animal cells [63–66]. Actin filaments are then assembled by formins around the cell equator area or are translocated to the cleavage furrow by cortical flow [66] or on spindle microtubule tracks [67]. In contrast, there was no detectable cortical flow of myosin-II in the rat kidney cells, suggesting equatorial de novo assembly of myosin-II [66]. The authors further pointed out that inhibition of disassembly played some role in the myosin band formation at the cell equator. Myosin II cortical flow has been previously reported in fibroblasts cells, Xenopus egg, and Dictyostelium cells [64,68,69]. It is reasoned that the inconsistency might arise from observation of myosin II movement at different recruitment stages or multiple recruitment mechanisms are adopted by different organisms, as in D. discoideum [70].
RhoA plays a central role in the contractile-ring assembly in animal cells. Centralspindlin at the equator recruits Pebble or ECT2 to activate RhoA [71–75], which then recruits the contractile machinery via phosphorylation of myosin-II RLC [76]. However, the latest study suggested that myosin RLC phosphorylation is not required for its furrow recruitment [77]. RacGAP also stabilizes the contractile ring by binding to anillin [78], which binds both myosin-II and actin.
Several other actin regulating proteins have been implied in cytokinesis. Tropomyosin, an actin side-binding and stabilizing protein, was found to be present in the cleavage furrow in metazoan cells [79]. Profilin, an actin-monomer binding protein, is essential for cytokinesis in C. elegans and D. melanogaster by regulating actin polymerization [80–82]. Formin is essential for contractile-ring assembly in C. elegans and D. melanogaster as the disruption of its function leads to the failure of furrow formation [80,83]. Formin mDia2 has been found to be essential in mammalian cell cytokinesis by scaffolding the contractile ring and help to focus it at the correct division site [83]. PSTPIP is a Cdc15-like F-BAR protein found in mammalian cells. It is located to the cleavage furrow, but its roles in cytokinesis remain to be further determined [84]. Although S. pombe and S. cerevisiae IQGAP proteins Rng2p and Cyk1p/Iqg1p are both required for contractile-ring formation, the roles of IQGAP-like proteins in cytokinesis are still poorly understood in animal cells. An IQGAP localizes to the division site in sea urchin egg [85]. The IQGAP GAPA might be involved in midbody separation but not in myosin II recruitment in Dictyostelium [86]. Actin cross-linking protein α-actinins also localize to the cleavage furrow in chick embryos [87], sea urchin eggs [88], and mammalian cells [89]. It is unclear if they are involved in contractile-ring formation. A more recent study suggests α-actinin regulates the speed of furrow ingression [90]. Despite the progresses, it is remain obscure how these proteins interact to assembly the contractile ring in animal cells.
4. Concluding remarks
The use of a contractile ring for cytokinesis can be dated back to about one billion years ago across phyla. This review discusses the assembly mechanism of the contractile ring in fission yeast, budding yeast, and animal cells, emphasizing that many proteins and mechanisms are conserved in these evolutionarily distant eukaryotes. Importantly, cells precisely mark their future division site where a set of conserved proteins like actin and myosin-II form the contractile ring, though the order of interactions is not well conserved.
Among all the model systems, the fission yeast S. pombe provides the leading advances on the contractile-ring assembly, but some critical questions should be addressed in the future. First, while Mid1p is critical for cytokinesis node formation, it remains unclear how Mid1p is able to recognize the plasma membrane. Recently, studies in bacteria have shown cortical proteins can recognize membrane curvature [91,92]. Whether Mid1p localization depends on membrane curvature should be addressed in the future. Second, it is unknown whether formin Cdc12p dimers nucleate actin filaments at each node and how this protein is activated. Unlike other formins, no evidence suggesting that Rho GTPases or an inhibitory DAD-like domain regulate cdc12p activity [93]. Third, to gain insight into the cytokinesis process, understanding how node proteins interact with each other is critical. Fourth, the mechanism and protein(s) allowing ring “nucleation” in the absence of Mid1p remains unknown. Fifth, it is unknown whether the “search, capture, pull, and release” model for contractile-ring assembly in fission yeast is applicable to animal cells. Although some similarities exist, like formation of dynamic node-like structures for myosin and anillin, more studies using quantitative live cell observations, biochemical characterization, and mathematical modeling will be essential to understand the molecular mechanism of cytokinesis in other model systems.
Acknowledgments
We thank Valerie Coffman and Tom Pollard for critical reading of the manuscript. We regret that we cannot cite all the papers on contractile-ring assembly due to the space limitation. The Wu laboratory is supported by American Heart Association, Great Rivers Affiliate; Basil O’Connor Starter Scholar Research Award from the March of Dimes Foundation; and National Institute of Health (NIH) GM086546.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–18. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–60. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–55. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe BA, Gould KL. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–8. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–9. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 6.Feierbach B, Chang F. Cytokinesis and the contractile ring in fission yeast. Curr Opin Microbiol. 2001;4:713–9. doi: 10.1016/s1369-5274(01)00273-9. [DOI] [PubMed] [Google Scholar]
- 7.Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–78. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan S, Wachtler V, Balasubramanian M. Cytokinesis in fission yeast: a story of rings, rafts and walls. Trends Genet. 2003;19:403–8. doi: 10.1016/S0168-9525(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 9.Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–4. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 10.Vavylonis D, Wu J-Q, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 11.Tran PT, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bähler J, Steever AB, Wheatley S, Wang Y-l, Pringle JR, Gould KL, et al. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–16. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang F, Wollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–42. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- 14.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–19. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 15.Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, et al. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol. 2009;19:961–6. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol. 2004;24:10621–35. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–60. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 18.Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–73. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci USA. 2005;102:8228–32. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padte NN, Martin SG, Howard M, Chang F. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol. 2006;16:2480–7. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 21.Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–44. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 22.Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000;113:365–75. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- 23.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, et al. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–12. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–66. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roh D-H, Bowers B, Schmidt M, Cabib E. The septation apparatus, an autonomous system in budding yeast. Mol Biol Cell. 2002;13:2747–59. doi: 10.1091/mbc.E02-03-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Dassow G, Verbrugghe KJ, Miller AL, Sider JR, Bement WM. Action at a distance during cytokinesis. J Cell Biol. 2009;187:831–45. doi: 10.1083/jcb.200907090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–6. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 28.D'Avino PP. How to scaffold the contractile ring for a safe cytokinesis - lessons from anillin-related proteins. J Cell Sci. 2009;122:1071–9. doi: 10.1242/jcs.034785. [DOI] [PubMed] [Google Scholar]
- 29.Coffman VC, Nile AH, Lee IJ, Liu H, Wu J-Q. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motegi F, Nakano K, Mabuchi I. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J Cell Sci. 2000;113:1813–25. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- 31.Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–34. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 32.Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, et al. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–16. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–88. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Souza VM, Naqvi NI, Wang H, Balasubramanian MK. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct Funct. 2001;26:555–65. doi: 10.1247/csf.26.555. [DOI] [PubMed] [Google Scholar]
- 36.Naqvi NI, Wong KC, Tang X, Balasubramanian MK. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat Cell Biol. 2000;2:855–8. doi: 10.1038/35041107. [DOI] [PubMed] [Google Scholar]
- 37.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–95. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sladewski TE, Previs MJ, Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol Biol Cell. 2009;20:3941–52. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–62. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–9. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–87. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–30. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–25. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–33. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 45.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–82. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–71. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra M, Oliferenko S. Cytokinesis: catch and drag. Curr Biol. 2008;18:R247–50. doi: 10.1016/j.cub.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 48.Arai R, Mabuchi I. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2002;115:887–98. doi: 10.1242/jcs.115.5.887. [DOI] [PubMed] [Google Scholar]
- 49.Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr Biol. 1999;9:849–52. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- 50.Takaine M, Numata O, Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–31. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:3254–66. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, et al. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–75. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yonetani A, Chang F. Regulation of cytokinesis by the formin cdc12p. Curr Biol. 2010;23:561–6. doi: 10.1016/j.cub.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeda T, Kawate T, Chang F. Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast. Nat Cell Biol. 2004;6:1142–4. doi: 10.1038/ncb1189. [DOI] [PubMed] [Google Scholar]
- 55.Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–70. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lister I, Tolliday N, Li R. Characterization of the minimum domain required for targeting budding yeast myosin II to the site of cell division. BMC Biology. 2006;4:19. doi: 10.1186/1741-7007-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyne J, Yosuf H, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113:4533–43. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- 58.Luo J, Vallen EA, Dravis C, Tcheperegine SE, Drees B, Bi E. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J Cell Biol. 2004;165:843–55. doi: 10.1083/jcb.200401040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol. 2000;10:727–30. doi: 10.1016/s0960-9822(00)00539-x. [DOI] [PubMed] [Google Scholar]
- 60.Tolliday N, Pitcher M, Li R. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell. 2003;14:798–809. doi: 10.1091/mbc.E02-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young BA, Buser C, Drubin DG. Isolation and partial purification of the Saccharomyces cerevisiae cytokinetic apparatus. Cytoskeleton. 2010;67:13–22. doi: 10.1002/cm.20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroeder TE. The contractile ring: II. Determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J Cell Biol. 1972;53:419–34. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107:3077–90. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- 64.Noguchi T, Arai R, Motegi F, Nakano K, Mabuchi I. Contractile ring formation in Xenopus egg and fission yeast. Cell Struct Funct. 2001;26:545–54. doi: 10.1247/csf.26.545. [DOI] [PubMed] [Google Scholar]
- 65.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 66.Zhou M, Wang YL. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell. 2008;19:318–26. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alsop GB, Chen W, Foss M, Tseng KF, Zhang D. Redistribution of actin during assembly and reassembly of the contractile ring in grasshopper spermatocytes. PLoS ONE. 2009;4:e4892. doi: 10.1371/journal.pone.0004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeBiasio RL, LaRocca GM, Post PL, Taylor DL. Myosin II transport, organization, and phosphorylation: evidence for cortical flow/solation-contraction coupling during cytokinesis and cell locomotion. Mol Biol Cell. 1996;7:1259–82. doi: 10.1091/mbc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yumura S. Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J Cell Biol. 2001;154:137–46. doi: 10.1083/jcb.200011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yumura S, Ueda M, Sako Y, Kitanishi-Yumura T, Yanagida T. Multiple mechanisms for accumulation of myosin II filaments at the equator during cytokinesis. Traffic. 2008;9:2089–99. doi: 10.1111/j.1600-0854.2008.00837.x. [DOI] [PubMed] [Google Scholar]
- 71.Kamijo K, Ohara N, Abe M, Uchimura T, Hosoya H, Lee J-S, et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishimura Y, Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J Cell Sci. 2006;119:104–14. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- 73.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 74.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–82. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao WM, Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc Natl Acad Sci USA. 2005;102:13158–63. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dean SO, Spudich JA. Rho kinase's role in myosin recruitment to the equatorial cortex of mitotic Drosophila S2 cells is for myosin regulatory light chain phosphorylation. PLoS ONE. 2006;1:e131. doi: 10.1371/journal.pone.0000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–83. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gregory SL, Ebrahimi S, Milverton J, Jones WM, Bejsovec A, Saint R. Cell division requires a direct link between microtubule-bound RacGAP and anillin in the contractile ring. Curr Biol. 2008;18:25–9. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 79.Clayton L, Johnson MH. Tropomyosin in preimplantation mouse development: identification, expression, and organization during cell division and polarization. Exp Cell Res. 1998;238:450–64. doi: 10.1006/excr.1997.3854. [DOI] [PubMed] [Google Scholar]
- 80.Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, et al. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Severson AF, Baillie DL, Bowerman B. A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–75. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 82.Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin–dependent processes during Drosophila development. Development. 1994;120:717–28. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- 83.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–38. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, et al. PSTPIP: A tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol. 1997;138:845–60. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nishimura Y, Mabuchi I. An IQGAP-like protein is involved in actin assembly together with Cdc42 in the sea urchin egg. Cell Motil Cytoskeleton. 2003;56:207–18. doi: 10.1002/cm.10146. [DOI] [PubMed] [Google Scholar]
- 86.Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol. 1997;137:891–8. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujiwara K, Porter ME, Pollard TD. Alpha-actinin localization in the cleavage furrow during cytokinesis. J Cell Biol. 1978;79:268–75. doi: 10.1083/jcb.79.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mabuchi I, Hamaguchi Y, Kobayashi T, Hosoya H, Tsukita S. Alpha-actinin from sea urchin eggs: biochemical properties, interaction with actin, and distribution in the cell during fertilization and cleavage. J Cell Biol. 1985;100:375–83. doi: 10.1083/jcb.100.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanger JM, Mittal B, Pochapin MB, Sanger JW. Stress fiber and cleavage furrow formation in living cells microinjected with fluorescently labeled α-actinin. Cell Motil Cytoskel. 1987;7:209–20. doi: 10.1002/cm.970070304. [DOI] [PubMed] [Google Scholar]
- 90.Mukhina S, Wang YL, Murata-Hori M. Alpha-actinin is required for tightly regulated remodeling of the actin cortical network during cytokinesis. Dev Cell. 2007;13:554–65. doi: 10.1016/j.devcel.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–7. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–5. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–19. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]