Abstract
Cells constantly probe and respond to a myriad of cues present in their local surroundings. The effects of soluble cues are relatively straightforward to manipulate, yet teasing apart how cells transduce signals from the extracellular matrix and neighboring cells has proven to be challenging due to the spatially and mechanically complex adhesive interactions. Over the years, advances in the engineering of bio-compatible materials have enabled innovative ways to study adhesion-mediated cell functions, and numerous insights have elucidated the significance of the cellular microenvironment. Here, we highlight some of the major approaches and discuss the potential for future advancement.
Introduction
Cells interact with the surrounding microenvironment by processing various chemical and physical signals. Studies of growth factors, including various cytokines and hormones, have clarified mechanisms by which cells transduce soluble extracellular signals. In contrast, the current understanding of how insoluble cues, such as adhesion to the extracellular matrix (ECM) or neighboring cells, are integrated to generate cellular functions is less clear. To understand why this disparity exists, one only needs to appreciate the relative complexity of adhesive interactions, compared to processing soluble cues.
For most growth factors, the primary mechanism for signal transduction is mediated by binding to cell-surface or nuclear receptors. Although there may be effects of nonlinear cooperativity, multivalent ligand-induced avidity or downstream feedback regulation, the basic mechanisms often can be captured using steady-state approximations to describe receptor-ligand kinetics. In this case, the main parameters that one must consider are the concentration of soluble molecules and their binding to receptors, which dictate downstream cascade signaling. In contrast, the signals mediated by cell adhesion are regulated by numerous molecular and mechanical processes, namely the ligation and clustering of integrins, changes in adhesion dynamics and signaling, cytoskeleton organization, cell shape and polarity, and the generation of myosin-mediated mechanical stress between cells and the ECM. Cells attach via transmembrane integrin receptors that bind to specific motifs on the matrix proteins, such as fibronectin, collagen, and vitronectin [1,2]. Upon ligand binding, the receptors are proposed to undergo activation and clustering to induce intracellular signaling events [3]. Adhesions are also linked to the actin cytoskeleton and over 150 proteins [4,5], making them major molecular hubs where mechanical forces and biochemical signals converge for various cellular functions, including tissue organization, migration, and differentiation [6–11]. The coupling to actin and signaling proteins forms a feedback loop that regulates both adhesion dynamics [12–14] and force transmission between the cell and the ECM (Box 1) [15,16]. Interestingly, the substrate parameters, such as composition, architecture and rigidity also serve as input signals to modulate the feedback mechanism. As a result, the spatial organization and mechanical properties of the matrix provide additional layers of control on the cell-ECM interaction, and one of the challenges in cell biology is to investigate this relationship systematically in vitro.
Box 1. Adhesion dynamics and interplay with the ECM.
Cellular responses to soluble cues depend largely on ligand concentration, whereas both the density and the geometric presentations of insoluble extracellular matrix ligands are important for regulating cellular functions. Cell adhesion to the microenvironment involves not only the binding of integrin receptors to the underlying ECM but also integrin clustering and activation, connection to actin, and recruitment of adaptor and signaling proteins (Fig. I). When cells make contact with a substrate, activity of the Rho GTPase Rac increases [110], and this leads to actin polymerization at the membrane [111] and small adhesion formation [112,113]. Rac stimulates protrusion by activating WAVE (Wiskott-Aldrich syndrome protein (WASP)-family verprolin-homologous protein), which in turn regulates the Arp 2/3 complex for dendritic actin nucleation [114,115]. The Rho GTPases Rac and Cdc42 also promote actin polymerization by activating PAK (p21-activated-kinase) and mDia2, and Cdc42 can directly bind to WASP proteins. In the lamellipodium, adhesions form initially as diffraction-limited foci [12,116], and their continuous cycle of assembly and disassembly (i.e. turnover) mediates further integrin binding at the leading edge. When the protrusion pauses, nascent adhesions mature by elongating along α-actinin/actin filaments, which emerge centripetally to serve as templates [12]. Adhesion maturation is also regulated by the GTPase Rho, which induces actin stress fiber formation [117,118]. This process involves myosin II, which is activated, in part, when Rho activates ROCK (Rho-associated kinase), and ROCK phosphorylates myosin light chain (MLC) and inhibits MLC phosphatase [115]. Activated myosin II bundles actin filaments and generates contractility, which indicates that tension can modulate adhesion dynamics [119,120]. The morphogenesis of adhesions regulates global cytoskeletal restructuring, cell shape, and the strength of cellular traction on the ECM. As such, it is apparent that the cell’s homeostasis with its surroundings depends ultimately on a tightly regulated coupling between integrin-mediated adhesion to the ECM, the actin cytoskeleton, and myosin-mediated forces. Based on this mechano-chemical system, it can be appreciated how the composition, structural organization, and mechanics of the ECM can all impact both cellular structure, signaling, and function. In this review, we provide a brief overview of some of the advances in the engineering of materials that contribute to our understanding of these systems.
In addition to cell-matrix adhesion, it is clear that adhesion between neighboring cells (i.e. cell-cell adhesion) regulates many cellular structures and functions. It has been historically difficult to study the impact of such interactions due to a lack of tools to control or manipulate the spatial organization of cells with respect to each other, or the cell-cell adhesions themselves. As such, there is a growing appreciation for novel technologies that advance our understanding of cell-microenvironment interactions.
Recent progress in the engineering of specialty materials and systems for cell culture has made it possible to begin to tease apart how mechanical forces, cell-matrix adhesion, cell-cell interactions, and multicellular organization might regulate cells. Here, we will provide an overview of the major tools that are now being developed within the bioengineering community which have had, or likely will have, substantial impact on our understanding of cell adhesion and its role in cellular signaling and function. In particular, we focus on engineered surfaces to control ligand presentation and organization, elastic materials used to manipulate cellular mechanics, and novel specialty biomaterials that are being developed to provide unprecedented control over additional features of native extracellular matrices. We will also briefly comment on some of the insights gained in order to illustrate the utility of these tools. Because this overview is brief and necessarily incomplete, we will refer to other reviews for more details when necessary.
Engineered extracellular matrix surfaces and cell adhesion
Traditionally, cells are grown on tissue culture (plasma-treated) polystyrene in the presence of serum. Cellular attachment is facilitated by the adsorption of ECM proteins such as fibronectin in the serum added to cell culture media. For a more controlled surface treatment, purified matrix proteins are non-covalently adsorbed prior to cell seeding, which produces a coating of specific adhesion-promoting ligands. These proteins often include multiple binding sites for cell surface adhesion receptors and can induce physiological adhesion signaling. Because ECM proteins often are large and contain multiple binding sites for different cellular receptors as well as for binding of yet other ECM proteins, promoting singular receptor interactions is usually accomplished by using short peptide sequences such as the arginine-glycine-asparate (RGD) found in several ECM proteins [17,18]. Integrins consist of at least 18 types of α and 8 types of β subunits that form about 24 known heterodimers, and each pair can interact with various ECM proteins, such as fibronectin, collagen or vitronectin [2]. When RGD is immobilized on the surface, the number of integrin subtypes involved in the adhesive interaction is limited, and these simplified surfaces are preferred for examining more specific effects of cell adhesion without additional signaling that may be prompted by other integrin-ECM pairings. The degree of integrin binding and adhesion assembly can be also controlled by varying the density of ligands adsorbed to a substrate, and this approach has proven useful to demonstrate that the amount of cell-ECM interaction regulates apoptosis, cell shape, angiogenic morphogenesis, and migration speed [19–21].
On clarifying the role of ECM geometry on cell adhesion and spreading, the uniform coating of ligands has limitations. Cultured cells exhibit diverse adhesion morphology and cytoskeletal organization that are often different from their counterparts in vivo [22–24]. Cells remodel the adsorbed ECM and secrete endogenous matrix proteins in hours to days, dramatically changing the surface properties in the process and developing a mixed population of adhesions with different sizes, molecular compositions, subcellular distributions, and dynamics [25,26]. Such heterogeneity leads to differential signaling activity within adhesions and reorganization of the actin linkage [22,27]. Although much of the current understanding of adhesion and related cellular responses has been obtained via simple homogeneous surface coating, tools to better control and understand the relationships between adhesions, cell structure, and function are needed. In vivo ECM architecture is much more complex than in cultures, ranging from relatively consistent basement membrane to fibrillar networks. These issues call for innovative engineered surfaces with high-resolution spatial patterning and adhesive specificities to control cell-ECM interaction.
One versatile technique that has emerged to pattern ECM proteins at the adhesion-scale is based on microcontact printing (Fig. 1A). Using methods developed by the semiconductor industry to lithographically fabricate micrometer-scale circuits on silicon wafers, one can similarly generate spatially defined patterns of ECM proteins onto otherwise inert surfaces. This accessible method involves producing stamps made with an inexpensive, tissue culture-compatible silicone elastomer, poly-dimethysiloxane (PDMS) [28]. ECM protein can then be inked onto the stamps and printed onto a culture substrate, leaving behind geometric features matching the micrometer-scale features of the stamp to control where cells can adhere [29,30]. To prevent non-specific ECM protein adsorption and cell adhesion outside of the printed regions, the unpatterned regions are treated with protein-resistant coatings.
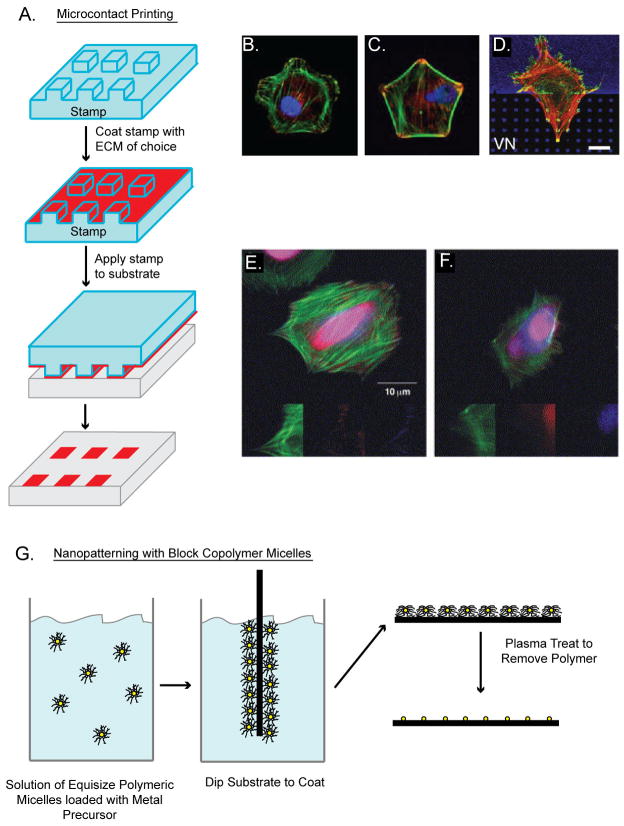
Figure 1. Methods of ECM Patterning to Control Cell Shape and Adhesions.
A. Microcontact printing process. A biomolecule is absorbed to the PDMS stamp surface. The stamp is then put in contact with the substrate. B & C. Immunofluorescent images of cells in flower (B) and star (C) shapes stained for F-actin (green), vinculin (red) and nuclei (blue) reproduced from [40]. D. B16 cell expressing β 3-integrin-GFP (green) labeled for actin (red) growing on vitronectin (blue) at the border between a uniform and a patterned substratum of 1 μm2 dots. Note the redistribution of integrin receptors on the patterned substratum. Scale bars: 10 μm. Reproduced from [31]. E & F. Immunofluorescent micrographs of REF cells stained for vinculin (red), zyxin (blue) and actin (green). Cell adhering to 58 nm (E) and to 110 nm (F) nanopatterned surface for 24 h. Small inserts show each labeled protein at 2× magnification of the original images. Vinculin and zyxin can be seen to colocalize on the RGD nanodots spaced 58 nm apart but not 110 nm apart. Reproduced from [108]. G. Nanopatterning with block copolymer micelles allows the generation of substrates with a regularized pattern of gold nanoparticles. Block copolymer micelles with a polar core are used to hold a controllable amount of metal precursor in dilute solution. A substrate dipped into the solution comes out with a monolayer of micelles covering its surface. The inter-particle spacing can be controlled by using block copolymers with different lengths of blocks. The polymer is then removed by plasma treatment, leaving a quasihexagonal array of particles.
These ECM patterns can guide overall cell geometry, adhesion sizes and location, as well as organization of the actin cytoskeleton, and thus have proven to be an effective tool for studying adhesion-mediated biology [31,32]. For example, a single ECM island or an array of closely spaced dots was used to constrain or mediate cell spreading, respectively, while maintaining their total area of cell-ECM contact constant. With these substrates, it was shown that cell shape, or the area of cell spreading, rather than the amount of ECM ligand regulates apoptosis and proliferation [33]. In other words, although integrin binding initiates attachment and signaling, active cytoskeletal remodeling is critical in regulating cell function. Square ECM protein islands have been utilized to constrain the cell shape, and lamellipodia and filopodia formed at the corners, where the adhesion-mediated traction force is high [34]. In addition, cells plated on anisotropic ECM shapes, such as teardrops or arrowheads, were found to exhibit directional migration and reorientation of centrosome and Golgi, suggesting that spatial segregation of adhesions and actin can determine cell polarity [35,36]. Recently, 1-dimensional (1D) lines were used to promote elongated cell morphology and motility along the pattern, which appeared to mimic how cells adhere to and migrate on 3D fibrils [37]. Interestingly, surface patterning has also revealed the significance of cell-ECM interaction for stem cell differentiation. In addition to the importance of soluble differentiation factors, the degree of cell spreading or shape regulates cytoskeletal tension and modulates Rho GTPase signaling to guide the lineage of mesenchymal stem cells (MSC) [38–40]. Taken together, these findings indicate that physical interaction with the ECM modulates adhesive cues in cells, which in turn mediate various cellular processes.
The emergence of these geometric effects on cells has prompted a new focus on understanding how cells interact with the underlying ECM at a more fundamental level. ECM printing can be controlled over different length scales, ranging from the size of a single adhesion complex to large areas for a group of cells (Fig. 1B–D). The lower-range was explored by varying the pattern size and density to examine the effect on cell spreading, adhesion size and molecular components [31]. In this study, fibronectin dots as small as 0.1 μm2 supported cell adhesion and actin linkage, yet cell spreading was inhibited when the spacing of the dots increased to 5 μm. Application of a similar type of ECM patterning demonstrated that limiting adhesion size regulates α-smooth muscle actin-mediated contraction in myofibroblasts, which suggests that adhesion growth is part of a mechanical feedback loop that senses the microenvironment [41]. Larger-scale ECM patterns have been shown to control multicellular organization. Comparisons of single cells on ECM patches with those cultured as doublets (on twice the area) have demonstrated that cell-cell adhesion can induce planar polarity [42] and proliferation [43–45]. Alternatively, patterns containing a numbers of cells have demonstrated that cells in multicellular configuration exert greater traction stress at the corners, proliferate more [46], and, in the case of stem cells, alter their differentiation [47].
Recent advances in nanoscale technology have extended patterning techniques to test how the spatial organization of individual ECM proteins, such as fibronectin and collagen, may regulate integrin clustering and adhesion-mediated responses. One study involved functionalizing polymer stars that tethered a specific number of RGD peptides on an inert surface to control clustering density of the peptides [48]. Cells grown on the surface with at least 5 RGD peptides per star developed mature adhesions and actin stress fibers and exhibited higher migration speed, compared to those grown on stars with a single peptide, suggesting that local increases in integrin clustering are important for regulating adhesion and cytoskeletal organization. If it were known that every RGD were bound by an integrin, one could even precisely suggest that a pentameric cluster was important, but because the efficiency of binding is not yet known, we can only conclude that a cluster of five or more is sufficient for this effect.
A separate nanolithography approach involves depositing metal particles of 1–15 nm on a polyethylene glycol-treated background using diblock copolymer micelles [49] (Fig. 1E–G). Each metal particle (e.g. gold) can be linked to an RGD at a tunable separation distance (up to 200 nm), and the authors found that cell spreading, leading edge dynamics and adhesion maturation are optimal at the lateral RGD spacing of < 58 nm [49–51]. This demonstrates that the proximity between neighboring ligand-receptor pairs is important for the adhesive function of integrins. More in-depth adhesion signaling is yet to be explored, but the ligation of individual integrins is now recognized as a major parameter in surface engineering to control ECM-mediated cell functions.
Elastic substrates and mechanotransduction
As cells attach and spread onto a substrate, they generate traction against the matrix. Although this phenomenon was reported thirty years ago [52], it has only recently become clear that these mechanical forces are fundamental regulators of cell adhesion and function. Alterations in the density of collagen gels or fibrin gels have been suggested to impact cell function, and this effect is largely attributed to the changes in the spatial density of ligands presented to cells, even though changing the matrix densities also impacts the mechanics of the scaffolds. To decouple these parameters, polyacrylamide (PA) gel was adopted as a substrate for cell adhesion studies, which allowed an elegant way to control substrate rigidity without affecting ligand density.
PA is a well behaved linear elastic material whose rigidity can be easily manipulated by varying the concentration of acrylamide and bis-acrylamide. By functionalizing the gel surface with immobilized matrix proteins, Pelham and Wang were able to show that substrate stiffness modulates cell adhesion, spreading, and migration [53]. Building upon this initial observation, later studies followed to show that rigidity could regulate higher order cell functions. For example, cell proliferation and survival decreased on relatively soft substrates (4.7 kPa), compared to stiffer substrates (14 kPa) [54]. On collagen-coated PA gels, elongated myotubes displayed actin and myosin striations only on the stiffness that rendered native muscle [55]. Similarly, mesenchymal stem cells cultured on substrates that match the stiffness of brain (0.1–1 kPa), muscle (8–17 kPa), or cartilage (25–40 kPa) differentiated specifically to the cells of that respective tissue, indicating that progenitor cells can determine the mechanical property of the ECM and differentiate accordingly [56]. Also, development of a malignant metastatic phenotype is linked to a hardened microenvironment, which indicates that substrates matched with pathological stiffness can drive disease-related processes [57,58]. Taken together, these data show that cells are capable of sensing a wide range of substrate rigidity and respond accordingly for various cell functions. The mechanisms by which the cell senses stiffness remain to be elucidated, but the mechanotransduction system appears to utilize integrin-mediated adhesions as main force-sensing structures that are capable of integrating bi-directional mechanical loads at the surface level [6,14,59].
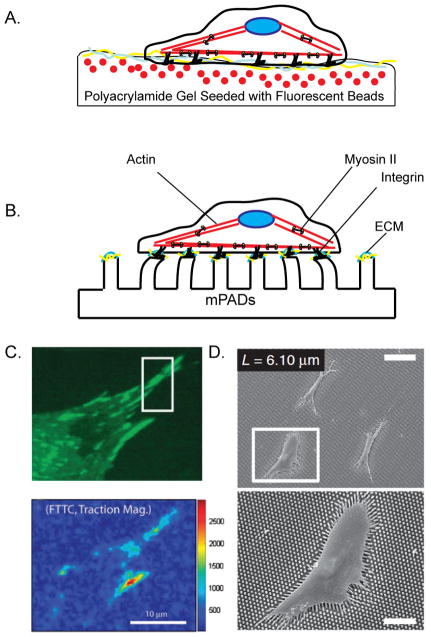
Due to its linear elastic property, PA gel provides a well defined system to measure the forces that are generated by attached cells (Fig. 2). The traction stress applied on the ECM has become a focus of great interest given the importance of cell-generated forces in sensing stiffness and in modulating adhesion dynamics. Tractions generated by cells cause the substrate material to deform, which can be readily observed by placing fiduciary markers (e.g. fluorescent beads) into the gels [60]. The deformations can then be used to estimate the distribution of forces across the cell-substrate interface [61]. These approaches have been critical in describing the forces at adhesions and their role in modulating the structure and dynamics of the adhesions and associated cytoskeletal elements [15,16,62,63]. Improvements in resolution have been achieved by using fluorescent beads of two different colors and by more rigorous computational algorithms [64,65]. Also, gel deformation and traction underneath a migrating cell can be measured in 3D by combining a digital volume correlation with volumetric confocal image stacks [66,67]. In addition to soft gels, cantilever-based substrates have been developed to measure cellular traction. Instead of relying on heavy computation for the analysis, a substrate that consists of discrete micro-cantilevers can measure local force changes. As a cell crawls over a substrate, the cantilevers lying in the plane would deflect and register the distribution of adhesion-mediated tractions [68]. More recent progression of the technology now uses vertically arrayed polymer cantilevers (microposts) that bend laterally when the cells that attach and spread across their tips exert forces [69,70]. In summary, both the gel-based and micropost-based approaches have begun providing critical insights into how ECM rigidity, mechanical forces (e.g. contractility, tension), and cellular structures (e.g. adhesions, actin) are interlinked to form a dynamic feedback loop that is central for cells to adapt and respond to adhesive cues from the microenvironment.
Figure 2. Elastic Substrates to Study Traction Forces.
A. The displacement of fluorescent beads embedded within a polyacrylamide gel can be tracked beneath a migrating cell. The displacement can then be used to calculate the stress and strain fields due to the cell-generated traction forces. B. Similarly, cells can be grown on a bed of microposts (mPADs). Cell-generated traction forces deflect the posts, allowing the stress and strain fields to be calculated. C. Example of traction force microscopy. Mouse embryo fibroblasts (MEFs) marked with GFP-paxillin (green). Lower image shows a pseudo-colored map of traction magnitude calculated using Fourier-transform traction cytometry (FTTC). Units of color bar given in Pascals. White box indicates a region enlarged in a separate part of the figure (not shown). Reproduced from [65]. D. Scanning electron micrographs of human mesenchymal stem cells plated on PDMS micropost array with a post height of 6.10 μm. Lower image is a magnified version of boxed region. Scale bars are 100 μm in top image and 50 μm in lower image. Reproduced from reference [109].
It is also important to note that work from several labs now suggest that forces affect adherens junctions. Micropost-based substrates were recently adapted to show that the tugging forces across cell-cell adhesions can regulate their assembly [71]. Two other studies, one using dual micropipets [72] and the other using traditional molecular approaches to modulate myosin-mediated contractility [73], arrived at the same conclusion. These findings demonstrate that each of these tools provides a unique approach to observe a phenomenon, and, in certain situations, a synergy of different tools is needed to make a conclusive observation.
The parallels between the effects of ligand density on a substrate, micropatterned surfaces, and substrate stiffness are striking, as all of these manipulations modulate integrin clustering and adhesion assembly [74,75]. They also regulate Rho-mediated traction forces [41,69], which involve actomyosin contractility and adhesion growth. Decreasing ECM ligand density or substrate stiffness causes cells to decrease spreading as well [53,76,77], and this indicates a possibility that cell shape is coupled to adhesion-mediated mechanotransduction pathways. Interestingly, much of these changes can regulate cell proliferation and stem cell differentiation [39,56]. Together, these data suggest that the sensing mechanisms of cellular microenvironment converge to adhesions, where mechanical cues are converted to biochemical signaling events for broader cell functions. Transmission of force via adhesions may physically stretch ECM proteins, such as fibronectin [78], and contraction on the ECM has been shown to mechanically release signaling agonists, such as transforming growth factor β1 (TGFβ1), for myofibroblast differentiation [79]. Similarly, conformation-sensitive adhesion molecules, such as p130CAS [80] and vinculin [81], unfold under tension, and such a mechanism may facilitate specific protein-protein interaction or signaling (e.g. phosphorylation). Thus, a focus of future studies is likely to involve attempts to uncover the underlying cellular machinery using innovative engineered substrates.
The near future: synthetic mimetics of 3D extracellular matrix
Studying cell-environment interactions on 2D substrates has provided many useful insights into how adhesive and mechanical cues can drive cell function; however, cell-environment interactions in vivo generally occur in 3D, which provides additional contextual stimuli that could affect cell behavior. An important effect of added dimension is the altered spatial distribution and density of ECM ligands relative to functionalized 2D substrates. 2D substrates functionalized by incubation with a ECM solution results in a uniform distribution of ligand across the surface whereas 3D matrix environments are made up of a fibrous mesh with dense clusters of ligands along individual fibers [82]. The ligand distribution and fibrous architecture of 3D matrix provides additional supports for cellular interaction; having integrins bound on one face of the cell versus all around it could impact adhesion clustering, cytoskeletal organization, and the mechanical forces at the cell-ECM interface. In addition to this direct effect on adhesions, the surrounding matrix also imposes new physical constraints to cell shape [37,83,84], limits the diffusion of growth factors to cells [85], and ultimately alters cell function [82,86,87]. While no synthetic matrix can yet provide direct control over such a diverse set of ECM properties, engineered 3D matrices are now being established in order to access the biology of these functions in a more controlled manner.
Early experiments with 3D engineered matrices focused on modifying the scaffold to physically immobilize growth factors and additional peptides to add functionality. Perhaps the earliest demonstration of such engineered matrices involved the construction of a modified VEGF that contains a substrate sequence for factor XIIIa, which mediates covalent binding to fibrin gels by its transglutaminating activity during coagulation [88]. For this type of binding, cellular proteolysis of the fibrin matrix releases local VEGF slowly and allows for rapid and sustained neovascularization without systemic release of large doses of VEGF [89,90]. In addition, direct covalent binding of VEGF to the fibrin provides a vehicle for further functionalizing the matrix. In a recent follow-up to this study, researchers showed that linking fibronectin domains directly with VEGF causes dramatic synergistic signaling, suggesting that integrins and VEGF receptors may operate in close proximity endogenously [91]. Similarly, engineered matrix proteins are also being developed to mediate direct assembly and anchoring into preexisting collagen scaffolds for new functionalities. One particular strategy takes advantage of collagen mimetic peptides (CMPs), which are short peptides that can non-covalently bind native collagen molecules by entangling into the helical structure. The CMPs can then be conjugated to other bioactive components to add functionality to standard collagen matrices such as cell adhesion peptides [92], immobilizing VEGF [93] or decorating collagen scaffolds with the anti-adhesive polyethylene glycol(PEG)-CMP [94]. Such a method demonstrates the potential for engineering new functionalities into a native matrix that could direct cell migration, proliferation, and differentiation. These scaffolds offer numerous experimental options to investigate key aspects of multi-dimensional cell-matrix interactions.
A more de novo strategy involves starting with a completely artificial scaffold as a backbone polymer on which bioactive ligands can be tethered. Two primary examples are self-assembling peptide systems and bioinert PEG hydrogels. In the former system, the right combination of sequences and secondary structure triggers the short peptides to assemble into filamentous structures. For the gels made with PEG, they have been generated using a variety of different synthetic angles, but there are features common to all of these hydrogels. The PEG backbone acts as an inert starting material which is then polymerized covalently into a macroscopic polymer. Introducing proteins or peptides into the gels is straightforward because they are entirely synthetic, and prescribing adhesive properties and proteolytic susceptibility is achievable through inclusion of the appropriate bioactive peptide sequences that make up the hydrogels [95,96]. Hydrogels consist of hydrophilic polymers; thus, they can be used to recapitulate many processes found in natural ECM, such as diffusive transport and fluid flow. These versatile conditions enable sustained 3D culture for long-term studies, including proliferation, migration, and differentiation.
Although many of these technologies are relatively new and still being optimized, it is apparent that they will soon become an important part of the cell biology toolbox. Synthetic biomaterials are able to mimic biologically important/relevant characteristics of their natural counterparts while providing enhanced levels of control over gels biofunctionality and material properties. This added level of control provides exciting avenues to pursue studies that focus on independent cell-environment interactions while keeping other parameters constant.
Concluding remarks
We are only now beginning to appreciate the many nuances of adhesive interactions that cells are able to probe and respond to accordingly. Based on many of the examples provided here, one comes to the realization that the advent of new materials has highlighted the importance of geometric and mechanical cues in cell adhesion. Although these are only the first steps in a long journey to understand the underlying mechanisms that control these transduction pathways, their successful application is drawing more engineers into developing the next generation of tools, and more biologists to adopt and capitalize on such approaches. Along with micropatterned surfaces, there is now a robust effort in the microfabrication community towards developing microfluidic technologies for generating in vitro platforms that integrate well-defined growth factor gradients, shear stress control, or miniaturized, automated culture. Microfluidic networks designed to generate stable, linear or nonlinear gradients of soluble growth factors were pioneered almost a decade ago [97]. While popular, they required continuous flow of media over the culture and thereby introduced the confounding variable of shear stress [98]. More recent improvements have led to designs where gradients are generated without flow in the region of interest and allow the study of chemotaxis in a variety of settings [99–103]. Others are using these microfabrication approaches to examine the effects of culture miniaturization [104] and the development of high-throughput platforms for screening cell culture conditions [105]. Investigators are exploring complex combinations of purified ECM proteins and biomaterials in order to begin to understand whether complex ECM compositions and their organization elicit unique cellular responses [106,107]. These multidisciplinary efforts to elucidate how cells interact with their microenvironment are likely to continue to accelerate, as long as this growing synergy between engineering sciences and cell biology continues to unravel these important challenges.
Box Figure I. Chemical-mechano Interactions of Adhesion Maturation and Cytoskeletal Rearrangement.
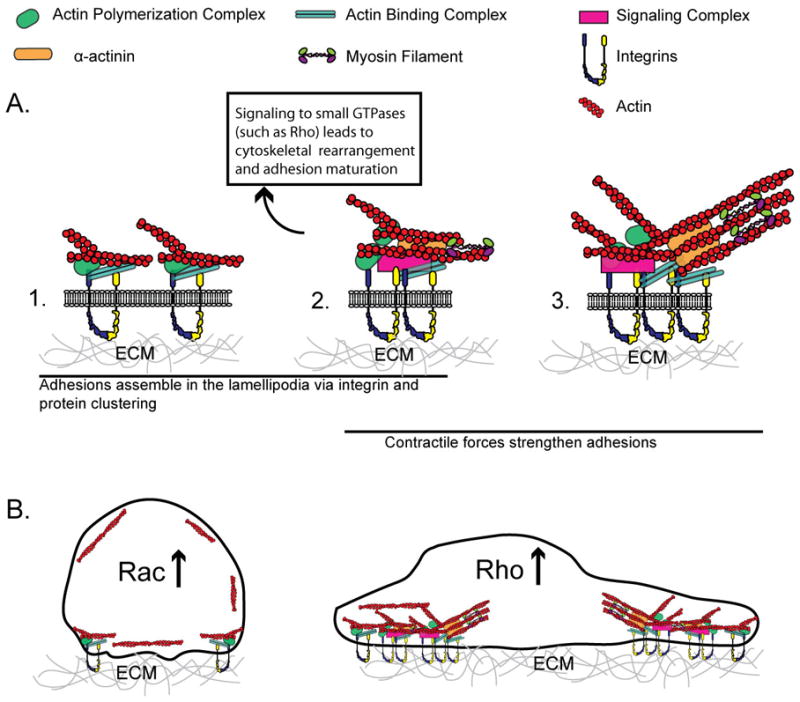
A. Schematic of adhesion maturation. (1) Nascent adhesion formation is initially driven by actin polymerization and consists of a complex of proteins linking integrins with actin. (2) Nascent adhesions elongate in response to actin-α-actinin-myosin II crosslinking. The signaling from adhesions activates small GTPases such as Rho that regulate further myosin II contractility and adhesion dynamics (3) Contractile forces generated by myosin II contribute to the further maturation of adhesions. B. During initial cell spreading, cell shape is constrained, Rac activity increases (Rho decreases), and adhesion formation is driven by actin polymerization (left). During later stages of cell spreading, the cell has spread out and flattened, forming mature adhesions and stress fibers and Rho activity is high (right).
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (EB00262, EB08396, HL73305, HL90747, GM74048), the RESBIO Technology Resource for Polymeric Biomaterials, and the Center for Engineering Cells and Regeneration of the University of Pennsylvania.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buck CA, Horwitz AF. Cell surface receptors for extracellular matrix molecules. Annu Rev Cell Biol. 1987;3:179–205. doi: 10.1146/annurev.cb.03.110187.001143. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110 (6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat Cell Biol. 2002;4 (4):E65–68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 4.Zaidel-Bar R, et al. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9 (8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burridge K, et al. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 6.Chen CS. Mechanotransduction - a field pulling together? J Cell Sci. 2008;121 (Pt 20):3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 7.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84 (3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 8.Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213 (3):565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 9.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84 (3):345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 10.Discher DE, et al. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324 (5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7 (4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 12.Choi CK, et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10 (9):1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6 (2):154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 14.Geiger B, et al. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10 (1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 15.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3 (5):466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 16.Beningo KA, et al. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153 (4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc Natl Acad Sci U S A. 1984;81 (19):5985–5988. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 19.Ingber DE, Folkman J. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J Cell Biol. 1989;109 (1):317–330. doi: 10.1083/jcb.109.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palecek SP, et al. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385 (6616):537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 21.Re F, et al. Inhibition of anchorage-dependent cell spreading triggers apoptosis in cultured human endothelial cells. J Cell Biol. 1994;127 (2):537–546. doi: 10.1083/jcb.127.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiger B, et al. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2 (11):793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 23.Vicente-Manzanares M, et al. Integrins in cell migration--the actin connection. J Cell Sci. 2009;122 (Pt 2):199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardel ML, et al. Mechanical Integration of Actin and Adhesion Dynamics in Cell Migration. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz BZ, et al. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol Biol Cell. 2000;11 (3):1047–1060. doi: 10.1091/mbc.11.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamir E, et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2 (4):191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 27.Calderwood DA, et al. Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J Biol Chem. 2000;275 (30):22607–22610. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- 28.Whitesides GM, et al. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 29.Mrksich M, Whitesides GM. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Annu Rev Biophys Biomol Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 30.Singhvi R, et al. Engineering cell shape and function. Science. 1994;264 (5159):696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 31.Lehnert D, et al. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J Cell Sci. 2004;117 (Pt 1):41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 32.Thery M, et al. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Motil Cytoskeleton. 2006;63 (6):341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 33.Chen CS, et al. Geometric control of cell life and death. Science. 1997;276 (5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 34.Parker KK, et al. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. Faseb J. 2002;16 (10):1195–1204. doi: 10.1096/fj.02-0038com. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X, et al. Directing cell migration with asymmetric micropatterns. Proc Natl Acad Sci U S A. 2005;102 (4):975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thery M, et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci U S A. 2006;103 (52):19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle AD, et al. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184 (4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, et al. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28 (3):564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBeath R, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6 (4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 40.Kilian KA, et al. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107 (11):4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goffin JM, et al. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J Cell Biol. 2006;172 (2):259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desai RA, et al. Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci. 2009;122 (Pt 7):905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu WF, et al. E-cadherin engagement stimulates proliferation via Rac1. J Cell Biol. 2006;173 (3):431–441. doi: 10.1083/jcb.200510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;514 (2–3):238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- 45.Nelson CM, Chen CS. VE-cadherin simultaneously stimulates and inhibits cell proliferation by altering cytoskeletal structure and tension. J Cell Sci. 2003;116 (Pt 17):3571–3581. doi: 10.1242/jcs.00680. [DOI] [PubMed] [Google Scholar]
- 46.Nelson CM, et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A. 2005;102 (33):11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26 (11):2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maheshwari G, et al. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113 (Pt 10):1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 49.Arnold M, et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 2004;5 (3):383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 50.Cavalcanti-Adam EA, et al. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. 2007;92 (8):2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold M, et al. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8 (7):2063–2069. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris AK, et al. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208 (4440):177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 53.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94 (25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang HB, et al. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000;279 (5):C1345–1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 55.Engler AJ, et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166 (6):877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126 (4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 57.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139 (5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8 (3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Ingber D. Integrins as mechanochemical transducers. Curr Opin Cell Biol. 1991;3 (5):841–848. doi: 10.1016/0955-0674(91)90058-7. [DOI] [PubMed] [Google Scholar]
- 60.Pelham RJ, Jr, Wang Y. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol Biol Cell. 1999;10 (4):935–945. doi: 10.1091/mbc.10.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munevar S, et al. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80 (4):1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aratyn-Schaus Y, Gardel ML. Transient frictional slip between integrin and the ECM in focal adhesions under myosin II tension. Curr Biol. 2010;20 (13):1145–1153. doi: 10.1016/j.cub.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardel ML, et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183 (6):999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butler JP, et al. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282 (3):C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 65.Sabass B, et al. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94 (1):207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hur SS, et al. Live Cells Exert 3-Dimensional Traction Forces on Their Substrata. Cell Mol Bioeng. 2009;2 (3):425–436. doi: 10.1007/s12195-009-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maskarinec SA, et al. Quantifying cellular traction forces in three dimensions. Proc Natl Acad Sci U S A. 2009;106 (52):22108–22113. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci U S A. 1997;94 (17):9114–9118. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan JL, et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100 (4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.du Roure O, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci U S A. 2005;102 (7):2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z, et al. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107 (22):9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez-Rico C, et al. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. J Cell Sci. 2010;123 (Pt 5):712–722. doi: 10.1242/jcs.047878. [DOI] [PubMed] [Google Scholar]
- 73.Brevier J, et al. The asymmetric self-assembly mechanism of adherens junctions: a cellular push-pull unit. Phys Biol. 2008;5 (1):016005. doi: 10.1088/1478-3975/5/1/016005. [DOI] [PubMed] [Google Scholar]
- 74.Cavalcanti-Adam EA, et al. Cell adhesion and response to synthetic nanopatterned environments by steering receptor clustering and spatial location. Hfsp J. 2008;2 (5):276–285. doi: 10.2976/1.2976662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CS, et al. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307 (2):355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 76.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86 (1 Pt 1):617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ingber DE. Fibronectin controls capillary endothelial cell growth by modulating cell shape. Proc Natl Acad Sci U S A. 1990;87 (9):3579–3583. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith ML, et al. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5 (10):e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wipff PJ, et al. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J Cell Biol. 2007;179 (6):1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127 (5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466 (7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17 (5):524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 83.Cukierman E, et al. Taking cell-matrix adhesions to the third dimension. Science. 2001;294 (5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 84.Ochsner M, et al. Dimensionality controls cytoskeleton assembly and metabolism of fibroblast cells in response to rigidity and shape. PLoS One. 2010;5 (3):e9445. doi: 10.1371/journal.pone.0009445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raghavan S, et al. Decoupling diffusional from dimensional control of signaling in 3D culture reveals a role for myosin in tubulogenesis. J Cell Sci. 2010;123 (Pt 17):2877–2883. doi: 10.1242/jcs.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beningo KA, et al. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci U S A. 2004;101 (52):18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meshel AS, et al. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat Cell Biol. 2005;7 (2):157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- 88.Zisch AH, et al. Covalently conjugated VEGF--fibrin matrices for endothelialization. J Control Release. 2001;72 (1–3):101–113. doi: 10.1016/s0168-3659(01)00266-8. [DOI] [PubMed] [Google Scholar]
- 89.Ehrbar M, et al. Cell-demanded liberation of VEGF121 from fibrin implants induces local and controlled blood vessel growth. Circ Res. 2004;94 (8):1124–1132. doi: 10.1161/01.RES.0000126411.29641.08. [DOI] [PubMed] [Google Scholar]
- 90.Ehrbar M, et al. The role of actively released fibrin-conjugated VEGF for VEGF receptor 2 gene activation and the enhancement of angiogenesis. Biomaterials. 2008;29 (11):1720–1729. doi: 10.1016/j.biomaterials.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 91.Martino MM, et al. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30 (6):1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamazaki CM, et al. A collagen-mimetic triple helical supramolecule that evokes integrin-dependent cell responses. Biomaterials. 2010;31 (7):1925–1934. doi: 10.1016/j.biomaterials.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 93.Wang AY, et al. Immobilization of growth factors on collagen scaffolds mediated by polyanionic collagen mimetic peptides and its effect on endothelial cell morphogenesis. Biomacromolecules. 2008;9 (10):2929–2936. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- 94.Wang AY, et al. Facile modification of collagen directed by collagen mimetic peptides. J Am Chem Soc. 2005;127 (12):4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 95.Gobin AS, West JL. Cell migration through defined, synthetic ECM analogs. Faseb J. 2002;16 (7):751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 96.Lutolf MP, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100 (9):5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dertinger SKW, et al. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73 (6):1240–1246. [Google Scholar]
- 98.Walker GM, et al. Effects of flow and diffusion on chemotaxis studies in a microfabricated gradient generator. Lab Chip. 2005;5 (6):611–618. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Atencia J, et al. The microfluidic palette: a diffusive gradient generator with spatio-temporal control. Lab Chip. 2009;9 (18):2707–2714. doi: 10.1039/b902113b. [DOI] [PubMed] [Google Scholar]
- 100.Saadi W, et al. Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber. Biomed Microdevices. 2007;9 (5):627–635. doi: 10.1007/s10544-007-9051-9. [DOI] [PubMed] [Google Scholar]
- 101.Irimia D, et al. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7 (12):1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Breckenridge MT, et al. A microfluidic imaging chamber for the direct observation of chemotactic transmigration. Biomed Microdevices. 2010;12 (3):543–553. doi: 10.1007/s10544-010-9411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park ES, et al. Continuously perfused, non-cross-contaminating microfluidic chamber array for studying cellular responses to orthogonal combinations of matrix and soluble signals. Lab Chip. 2010;10 (5):571–580. doi: 10.1039/b919294h. [DOI] [PubMed] [Google Scholar]
- 104.Meyvantsson I, Beebe DJ. Cell culture models in microfluidic systems. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 105.Melin J, Quake SR. Microfluidic large-scale integration: the evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 106.Flaim CJ, et al. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2 (2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 107.Lutolf MP, et al. Designing materials to direct stem-cell fate. Nature. 2009;462 (7272):433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cavalcanti-Adam EA, et al. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur J Cell Biol. 2006;85 (3–4):219–224. doi: 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 109.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7 (9):733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.del Pozo MA, et al. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. Embo J. 2000;19 (9):2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ridley AJ, et al. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70 (3):401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 112.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81 (1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 113.Rottner K, et al. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol. 1999;9 (12):640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 114.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112 (4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 115.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116 (2):167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 116.Alexandrova AY, et al. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS One. 2008;3 (9):e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133 (6):1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70 (3):389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 119.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128 (3):561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vicente-Manzanares M, et al. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176 (5):573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]