Abstract
Resistance to 5-fluorouracil (5-FU) represents a major contributor to cancer-related mortality in advanced colorectal cancer patients. Genetic variations and expression alterations in genes involved in 5-FU metabolism and effect have been shown to modulate 5-FU sensitivity in vitro, however these alterations do not fully explain clinical resistance to 5-FU-based chemotherapy. To determine if alterations of DNA copy number in genes involved in 5-FU metabolism impacted clinical resistance to 5-FU-based chemotherapy, we assessed thymidylate synthetase (TYMS) and thymidine phosphorylase (TYMP) copy number in colorectal liver metastases. DNA copy number of TYMS and TYMP was evaluated using real time quantitative PCR in frozen colorectal liver metastases procured from 62 patients who were pretreated with 5-FU-based chemotherapy prior to surgical resection (5-FU exposed) and from 51 patients who received no pretreatment (unexposed). Gain of TYMS DNA copy number was observed in 18% of the 5-FU exposed metastases, while only 4% of the unexposed metastases exhibited TYMS copy gain (p=0.036). No significant differences were noted in TYMP copy number alterations between 5-FU exposed and unexposed metastases. Median survival time was similar in 5-FU exposed patients with metastases containing TYMS amplification and those with no amplification. However, TYMS amplification was associated with shorter median survival in patients receiving post-resection chemotherapy (hazard ratio = 2.7, 95% confidence interval = 1.1 to 6.6; p=0.027). These results suggest amplification of TYMS amplification as a putative mechanism for clinical resistance to 5-FU-based chemotherapy and may have important ramifications for the post-resection chemotherapy choices for metastatic colorectal cancer.
Keywords: Colorectal Neoplasms, Neoplasm Metastasis, DNA Copy Number Variation, Fluorouracil, Thymidylate Synthase, Thymidine Phosphorylase
Introduction
5-Fluorouracil (5-FU) is the backbone of treatment for advanced colorectal cancer, as nearly all patients will receive a 5-FU-containing regimen 1, 2. 5-FU's prominence in advanced colorectal cancer treatment is largely a function of its consistent efficacy throughout its five decades of use 3. However, one of the biggest challenges for the management of advanced colorectal cancer is 5-FU treatment failure, both inherent and acquired. Indeed, the five-year survival of those with metastatic colorectal cancer is less than ten percent 4. Deaths due to clinically-resistant metastatic colorectal cancer disproportionately account for why colorectal cancer is presently the second leading cause of cancer-related mortality 5.
Numerous investigations into 5-FU resistance in metastatic colorectal cancer have focused on genes within its known pharmacokinetic and pharmacodynamic pathways. One major focus has been the enzyme, thymidylate synthetase (TYMS), a key therapeutic target of 5-FU. Colorectal cancers that do not respond to 5-FU-based chemotherapy have been shown to possess greater TYMS enzymatic activity than cancers that do respond 6. Likewise, high levels of TYMS mRNA or protein in liver metastases have also been associated with lack of clinical response to 5-FU in vivo 7-10. A recent meta-analysis of 24 studies has indicated that metastatic colorectal tumours with low expression of TYMS are more sensitive to fluoropyrimidine-based chemotherapy 11.
Similar expression studies have also been performed for thymidine phosphorylase (TYMP) because of its role in 5-FU bioactivation. Overexpression of TYMP has been linked to increased 5-FU sensitivity in vitro 12. Xenografts transfected to overexpress TYMP showed a 43% decrease in size following 5-FU administration, whereas no response to 5-FU was observed in the xenografts lacking TYMP-transfection 13. In one small study, an increase in TYMP expression was observed in colorectal cancer metastases, which were significantly more resistant to 5-FU than their matched primary tumours 14. More recently, low TYMP expression was observed to be predictive of response to 5-FU based chemotherapy in metastatic colorectal cancer patients 15. Conversely, increased expression of TYMP measured by immunohistochemistry was associated with prolonged survival in metastatic colorectal cancer patients treated with capecitabine plus irinotecan 16. Thus the role TYMP plays in resistance to chemotherapy remains to be clarified.
The underlying mechanism(s) for altered expression of genes and proteins important for drug resistance has implications for the development of strategies to overcome clinical resistance. For example, variants in the promoter region of genes are one mechanism through which expression can be influenced. A polymorphic tandem repeat sequence in the TYMS gene promoter region is associated with higher TYMS expression in tumours 17. Additionally, one study of TYMS expression in colorectal cancer patients with liver-confined metastases demonstrated an inverse association between high expressing TYMS genotypes and tumour response to 5-FU-based chemotherapy 18. It also appears that administration of 5-FU has the potential to increase TYMS expression, suggesting a potential mechanism of acquired resistance to chemotherapy 19. Alterations in DNA copy number are yet another potential mechanism for influencing gene expression. Amplification of chromosome 18p11.32, the location of the TYMS gene, has been associated with resistance to fluoropyrimidines in mouse colorectal cancer xenografts 20. TYMS gene copy number has also been associated with clinical resistance to 5-FU. In a human study, TYMS copy number gains occurred significantly more frequently in liver metastases from patients who had received 5-FU than in metastases from patients who were 5-FU naïve 21, and these copy number gains were associated with a 3.5-fold higher relative risk of death compared with normal TYMS copy number 21. Importantly, the results of this small study have not been replicated.
To gain insight into the mechanisms of clinical 5-FU resistance, we examined TYMS and TYMP copy number in resected colorectal metastases from patients exposed and not exposed to 5-FU-based chemotherapy. Gain of TYMS gene copy was associated with prior 5-FU exposure suggesting this genetic alteration as a potential mechanism of resistance to 5-FU-based chemotherapy.
Materials and Methods
Normal tissue and liver metastases were obtained from colorectal cancer patients undergoing liver resection under a University of North Carolina Institution Review Board (UNC IRB)-approved tissue banking protocol. Clinical information was retrospectively retrieved from patient records and dates of death from the medical records or the Social Security Death Index. Tissue specimen analysis and patient chart reviews was approved by the UNC IRB (IRB number 07-1525) and was performed in accordance with Health Insurance Portability and Accountability Act (HIPAA) regulations and the Helsinki Declaration. Patient samples were categorized as “5-FU exposed” if the patients received 5-FU within the 6-months preceding their liver resections; all other samples were classified as unexposed.
DNA was extracted from frozen liver metastases specimens using the Qiagen All Prep kit (Qiagen Inc, Valencia, CA, USA) according to the manufacturer's protocol. Briefly, liver metastases were lysed and homogenized via Tissuelyser (Qiagen). DNA quality was assessed based on the optical density 260/280 ratio. DNA extractions were performed at UNC Lineberger Comprehensive Cancer Center's Tissue Procurement Facility.
DNA copy number was determined by real time quantitative polymerase chain reaction (qPCR) using an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). For TYMS copy number determination, the 20 μL qPCR reaction was composed of 10 μL of TaqMan® universal PCR master mix (Applied Biosystems), 10 μL of primer probe mix containing primers and probes for FAM™-labelled TYMS (final concentration of primers = 600 nM, probe = 200 nM) and VIC™-labelled RNaseP (catalogue #: 4316844; final concentration at 1×) and 20 ng of DNA (dried in wells of the PCR plate overnight before adding the reaction mixture). These qPCR assays were performed in triplicate according to the following program: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles at 95°C for 20 seconds, and 60°C for 1 minute. The primer and probe mix for TYMS contained a forward primer – GCCTCGGTGTGCCTTTCA, reverse primer – CGTGATGTGCGCAATCATG and the TaqMan® probe – CATCGCCAGCTACGCCCTGCTC 22. The colorectal cancer cell line, H630R10 (kindly provided by Prof Patrick Johnston, Queen's University, Belfast), was used as a positive control in determination of TYMS copy number as it exhibits TYMS copy number gain 23. For TYMP copy number determination, the 20 μL qPCR reaction was composed of 10 μL of SYBR® Green universal PCR master mix (Applied Biosystems), 10 μL of PCR primers for TYMP and RNaseP (final concentration = 4 μM), and 20 ng of DNA (dried in wells of the PCR plate overnight before adding the reaction mixture). These qPCR assays were performed in duplicate, with the assays for TYMP and RNaseP for a single sample always run on the same plate according to the following program: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, 56°C for 30 seconds and 70°C for 30 seconds. The primers for TYMP were: forward primer – GTTCTCCATTGTCTCCAACCTC and reverse primer – AACTTAACGTCCACCACCAGAG 22. The SYBR® green primers for RNaseP were: forward primer - TGGGAAGGTCTGAGACTAGGG and reverse primer - CGTTCTCTGGGAACTCACCT. All primers were designed using the Primer3 program 24. The analysis of DNA copy number for each sample was performed using the comparative Ct (2-ΔΔCt) method as described previously 22, in which the threshold cycle (Ct) numbers were generated using the 7300 System SDS Software, version 1.4 (Applied Biosystems). DNA copy numbers were normalized for TYMS and TYMP to the control gene, RNaseP, because normal liver has two copies of this gene.
The range of values for normal TYMS and TYMP were determined by performing the assays on genomic DNA isolated from cell lines from the Centre d'Etude du Polymorphisme Humain (CEPH) collection, which are EBV-transformed lymphoblastoid cells taken from healthy individuals believed to have normal TYMS and TYMP copy number. Cell lines were obtained from Coriell (Coriell Institute for Medical Research, Camden, NJ, USA). Metastases samples with copy number values more than 2 standard deviations greater than the mean CEPH copy number (2.53 for TYMS, 2.54 for TYMP) were considered to have copy number gains and those with values more than 2 standard deviations less than the mean CEPH copy number (1.31 for TYMS, 1.52 for TYMP) were considered to have copy number losses.
The differences in proportions of demographic characteristics and number of patients with copy number alterations in the 5-FU exposed and unexposed patients were compared using Fisher's exact test; Wilcoxon Rank Sum tests were used to compare continuous variables between the two groups. Based on the available sample size we had 80% power to detect a 0.15 difference between 5-FU exposed and unexposed groups. Kaplan-Meier analysis was performed to test the effect of genetic alterations on overall survival and significance was assessed using log rank tests. Overall survival data was calculated from the date of surgery to the date of death or last follow-up and Cox regression was used to calculate hazard ratios. All statistical tests were 2-sided and the significance level was set at p<0.05.
Results
Demographics
The study population consisted of 113 metastatic colorectal cancer patients who had their liver metastases surgically resected (62 exposed and 51 unexposed to 5-FU prior to surgical resection). Patient characteristics are listed in Table 1. The mean age at diagnosis in the unexposed group was 6 years greater than in the 5-FU exposed group (p=0.01); no other statistically significant differences in demographic data were observed between the two groups.
Table 1. Clinical cohort demographics.
| Characteristic | Unexposed | 5-FU Exposed | P-value |
|---|---|---|---|
| Gender | |||
| Male | 31 (61%) | 27 (44%) | 0.089 |
| Female | 20 (39%) | 35 (56%) | |
| Race | |||
| White | 36 (71%) | 48 (77%) | 0.46 |
| Black | 12 (23%) | 13 (21%) | |
| Asian/Other | 3 (6%) | 1 (2%) | |
| Age (mean) at diagnosisa | 65 (range 42 – 86) | 59 (range 38-85) | 0.01 |
| TNM staging system: Tb | |||
| 0 | 12 | 10 | 0.15 |
| 1 | 5 | 2 | |
| 2 | 7 | 5 | |
| 3 | 22 | 38 | |
| 4 | 3 | 1 | |
| TNM staging system: Nb | |||
| 0 | 27 | 25 | 0.47 |
| 1 | 14 | 17 | |
| 2 | 8 | 14 | |
| TNM staging system: Mb | |||
| 0 | 30 | 28 | 0.12 |
| 1 | 17 | 31 | |
| Initial Number of Liver Metastases | |||
| Unknown | 7 | 13 | 0.24 |
| 1 | 27 | 23 | |
| 2 | 11 | 12 | |
| >2 | 6 | 14 | |
| Range | 1-6 | 1-14 | |
| Median | 1 | 1 | |
| Post-resection chemotherapy | |||
| Yes | 22 | 34 | 0.41 |
| No | 27 | 25 | |
| Unknown | 2 | 3 | |
| Length of Neoadjuvant Chemotherapy (pretreatment in days) | |||
| Mean | 140 | NAc | |
| Median | 110 | ||
| Range | 27-564 | ||
Diagnosis refers to diagnosis of liver metastasis
TNM staging system from the AJCC Cancer Staging Handbook (2002).
NA = not applicable
TYMS
TYMS DNA copy number was obtainable from 111 samples (62 5-FU exposed and 49 unexposed). The values of TYMS copy number ranged from 1.06 to 3.80 for the 5-FU exposed samples and 1.12 to 3.28 for the unexposed samples. The median copy number was 1.89 for exposed samples and 1.74 for unexposed samples. 5-FU exposed samples exhibited a significantly greater frequency of TYMS amplification compared to unexposed samples (19% vs. 4%, p=0.036; Figure 1). TYMS loss was infrequent in both 5-FU exposed and unexposed groups (3 samples for each group).
Figure 1.
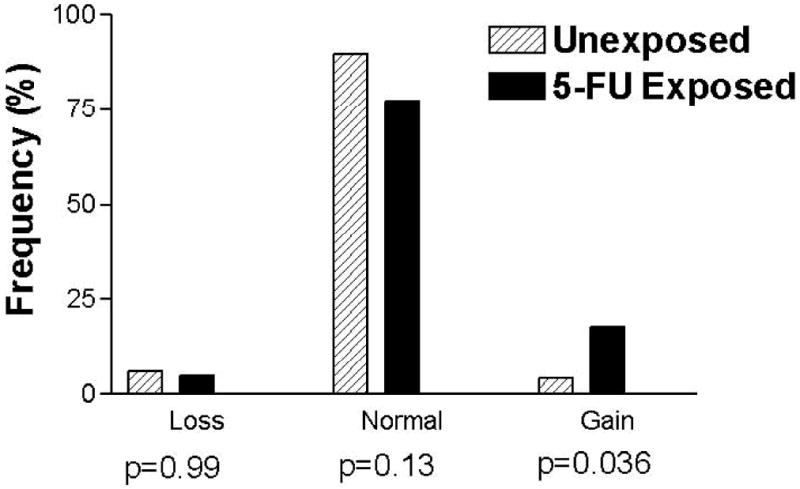
Histogram of TYMS copy number in tumours from 5-FU exposed and unexposed patients. Tumours with copy number < 1.31 were classified as loss of copy number and tumours with copy number > 2.53 were classified as gain of copy number. The p-values correspond to the Fisher's Exact Test comparing each group to the other two combined.
TYMP
TYMP DNA copy number was obtainable for 99 samples (54 5-FU exposed and 45 unexposed). The values of TYMP copy number ranged from 0.93 to 6.78 for the 5-FU exposed samples and 1.06 to 6.98 for the unexposed samples. The median copy number was 1.97 for 5-FU exposed samples and 2.00 for unexposed samples. While a greater percentage of tumours exhibited TYMP copy number alterations than TYMS copy number alterations, the prevalence of TYMP copy number alterations was similar in the two treatment groups (37% for 5-FU exposed and 42% for unexposed metastases; p=0.68). Amplification of TYMP was roughly equivalent in both treatment groups (20% for 5-FU exposed and 29% for unexposed; p=0.35), as was loss of TYMP (17% for 5-FU exposed and 13% for unexposed; p=0.78; Figure 2).
Figure 2.
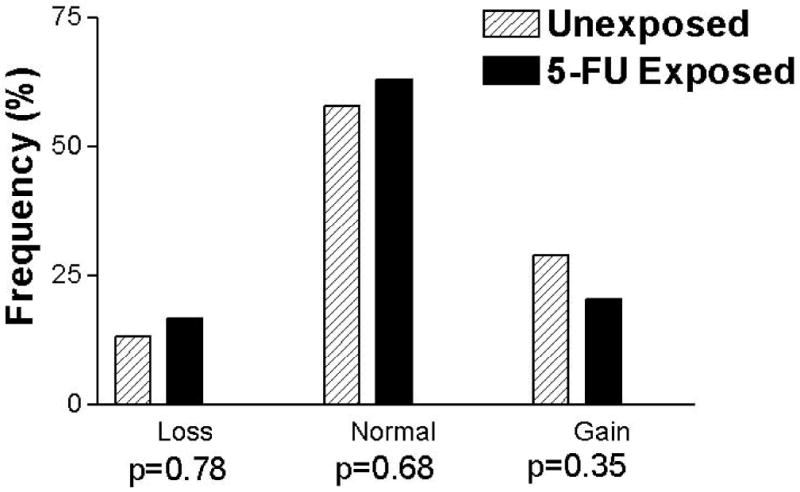
Histogram of TYMP copy number in tumours from 5-FU exposed and unexposed patients. Tumours with copy number < 1.52 were classified as loss of copy number and tumours with copy number > 2.54 were classified as gain of copy number. The p-values correspond to the Fisher's Exact Test comparing each group to the other two combined.
As one of our hypotheses was that TYMS amplification would result in poorer response to 5-FU we compared overall survival in patients with TYMS gains (n=11) to patients with normal or low TYMS copy (n=50). No difference in survival was demonstrated between patients with normal/low TYMS copy number and those with TYMS gain (Figure 3A: Median survival for normal/low copy = 2.52 years, TYMS gain = 2.11 years, p=0.13). However, for the subset of patients treated with chemotherapy after surgical resection of their metastases for which we have TYMS copy number data (n=54), TYMS gain (n=8) was associated with a poorer median survival post-resection compared to patients with normal or low TYMS copy number (n=46) (Figure 3B: 2.11 years vs. 3.61 years, p=0.027). This difference represented a 2.7-fold higher hazard of death compared to patients without TYMS copy number gain (95% CI = 1.1 to 6.6). There were no associations between TYMP copy number alterations and survival for either the whole patient population or the treated and untreated subgroups (data not shown).
Figure 3.
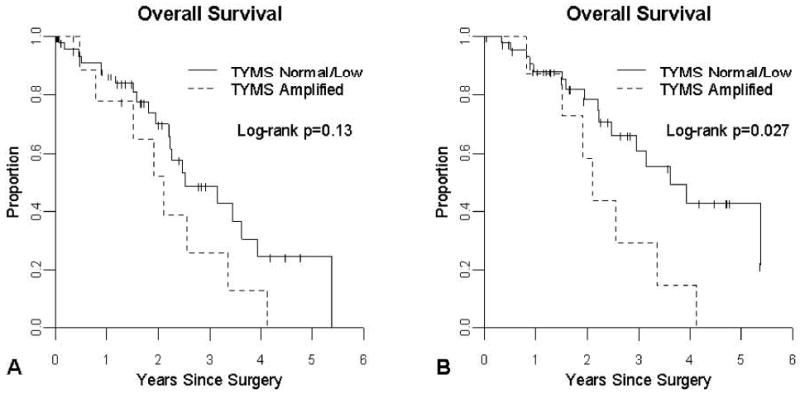
Effect of TYMS amplification on overall survival in metastatic colorectal cancer patients receiving chemotherapy. 3A: Kaplan-Meier analysis of overall survival in patients with metastatic colorectal cancer receiving neoadjuvant chemotherapy prior to surgical resection of their metastases (median survival time for TYMS amplification = 2.11 years, median survival for normal/low copy = 2.52 years). 3B: Kaplan-Meier analysis of overall survival in patients with metastatic colorectal cancer receiving adjuvant chemotherapy following surgical resection of their metastases (median survival time for TYMS amplification = 2.11 years, median survival for normal/low copy = 3.61 years).
Discussion
Aneuploidy is a common phenomenon in cancer which can have a pharmacologic impact if the altered chromosomal regions contain genes important for modulating drug response. Trisomies are common in acute lymphoblastic leukaemia cells 25 and chromosomal gains that encompass the pharmacologically important genes thiopurine S-methyltransferase and gamma-glutamyl hydrolase have been associated with higher activity of the encoded proteins in leukemic cells, which would be expected to impact the clinical response to mercaptopurine 26. In breast cancer, amplification of v-erb-b2 erythroblastic leukaemia viral oncogene homolog 2 (ERBB2 - also known as HER-2/neu) is found in a subset of tumours, which has prognostic importance 27 and is associated with decreased response to tamoxifen 28. In non-small-cell lung cancer, amplification of epidermal growth factor receptor (EGFR) is associated with better response to the EGFR tyrosine kinase inhibitor, gefitinib 29. Thus one mechanism for drug resistance appears to include copy number alterations of pharmacologically important genes in cancer tissue which can impact drug response 26.
Previously, copy number gains of the gene TYMS were shown to be inversely correlated with survival in a small study of patients with colorectal metastases following exposure to 5-FU 21. Our analysis of gene copy number in a larger population indicates a significant association between exposure to 5-FU-based chemotherapy and amplification of the TYMS gene in surgically resected metastatic colorectal lesions, consistent with the aforementioned results. While Wang and colleagues noted TYMS gains in only tumours exposed to 5-FU 21, our results suggest that amplification of TYMS in metastatic colorectal cancer can be found in the absence of 5-FU exposure, although it is possible patients received a 5-FU-based chemotherapy regimen for their primary disease before the onset of metastasis. It is unknown whether exposure to 5-FU induces amplification of the chromosomal region containing the TYMS gene or if 5-FU chemotherapy preferentially targets cancer cells without the amplification, resulting in the survival of resistant cells with TYMS amplification. However, it has been shown that expression of TYMS increases after bolus exposure to 5-FU in vivo 19. Additional work needs to be conducted to determine the mechanism for the observed amplification of TYMS in colorectal liver metastases following exposure to 5-FU.
The importance of TYMS amplification for metastatic colorectal cancer patients was illustrated by previous results that indicated TYMS amplification resulted in shorter survival times in patients pretreated with 5-FU 21. Our results do not strongly support this hypothesis, although our patients with tumours exhibiting TYMS amplification exhibited a trend toward shorter (almost 5 months) survival that did not reach statistical significance (Figure 3A). However, it should be noted that the patient cohort examined in the study by Wang and colleagues were treated only with 5-FU, as this was the standard of care therapy at the time of that study 21, whereas the patients examined in this study were treated with neoadjuvant chemotherapy regimens containing 5-FU plus oxaliplatin, irinotecan and/or bevacizumab. Since TYMS amplification is not significantly associated with shorter survival time in patients pre-treated with 5-FU-based chemotherapy, this suggests that the use of additional chemotherapeutic agents in the neoadjuvant setting lessens the impact of TYMS amplification on clinical resistance. On the other hand, there may be some bias in our patient groups, in terms of both age and comorbidity, which favour some patients to receive pre-operative chemotherapy, which has the potential to mask any associations with survival.
However, in those patients treated with adjuvant chemotherapy following surgical resection of their metastases, median survival time was 1 ½ years shorter in patients with tumours containing TYMS amplification than those with normal or lower TYMS copy number (Figure 3B). This suggests that tumours exhibiting TYMS amplification respond more poorly to chemotherapy. Alternatively, TYMS amplification could be associated with more aggressive tumour behaviour. Since the number of patients with tumours exhibiting TYMS amplification is small, additional studies need to be conducted to confirm the importance of TYMS amplification in metastatic colorectal cancer. If validated, TYMS copy number alteration could potentially serve as a biomarker for clinical resistance to 5-FU based adjuvant chemotherapy in metastatic colorectal cancer patients, at which point, TYMS copy number gain in resected tumours could be used to indicate which patients should receive an adjuvant chemotherapy regimen devoid of 5-FU due to resistance.
The cause of 5-FU resistance in metastatic colorectal cancer patients is believed to be multifactorial 6, 30-33. TYMS amplification clearly does not fully explain resistance to 5-FU based chemotherapy, as some patients without gain of TYMS also had short survival times. A pathway-based approach to interrogate all the genes and proteins that are postulated to be involved in 5-FU metabolism and efficacy is a more likely strategy to identify the relevant mechanisms underlying clinical resistance to 5-FU. While our data do not support a role for TYMP copy number alterations in resistance to 5-FU based chemotherapy, there are still many other genes that could logically be investigated. In addition, the problem of clinical resistance has become more complicated by the additional chemotherapeutic agents that are utilized with 5-FU in metastatic colorectal cancer patients. Detailed investigation of the role played by genes relevant to each of the chemotherapy agents used will provide a more complete picture of the mechanism(s) underlying clinical resistance to complex drug regimens. In addition, since our knowledge of the mechanisms through which these drugs exert their anticancer effects is incomplete, an unbiased approach, such as through genome-wide profiling, may be required to fully elucidate the mechanisms underlying clinical resistance in metastatic colorectal cancer patients.
In conclusion, we found neoadjuvant treatment with 5-FU-based chemotherapy was associated with gain of TYMS gene copy number. In addition, among patients treated with chemotherapy following surgical resection of their metastases, those with tumours containing TYMS amplification exhibited significantly shorter overall survival time. These data, along with previously published results, suggest that TYMS amplification may be involved in clinical resistance to 5-FU-based chemotherapy in metastatic colorectal cancer patients. If validated in larger clinical trials, these results suggest a prognostic importance for TYMS copy number gain in metastatic colorectal cancer patients, which can assist in the selection of the chemotherapy regimens most likely to be of clinical benefit.
Acknowledgments
We wish to thank the UNC Tissue Procurement Facility for their assistance in this study. This research was supported in part by a GI SPORE grant from the NCI (P50-CA106991), the NIH Pharmacogenetics Research Network (U01 GM63340), and the Lineberger Comprehensive Cancer Center (P30-CA016086).
Financial Support: This research was supported by a GI SPORE grant from the NCI (P50-CA106991) [JTA], the NIH Pharmacogenetics Research Network (U01 GM63340) [HLM], and the Lineberger Comprehensive Cancer Center (P30-CA016086).
Footnotes
Conflict of Interest Statement: The authors declare that they have no conflicts of interest that could inappropriately influence this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bleiberg H. Role of chemotherapy for advanced colorectal cancer: new opportunities. Semin Oncol. 1996;23:42–50. [PubMed] [Google Scholar]
- 2.de Gramont A, Vignoud J, Tournigand C, et al. Oxaliplatin with high-dose leucovorin and 5-fluorouracil 48-hour continuous infusion in pretreated metastatic colorectal cancer. Eur J Cancer. 1997;33:214–9. doi: 10.1016/s0959-8049(96)00370-x. [DOI] [PubMed] [Google Scholar]
- 3.Moertel C. Chemotherapy for colorectal cancer. N Engl J Med. 1994;330:1136–42. doi: 10.1056/NEJM199404213301608. [DOI] [PubMed] [Google Scholar]
- 4.Sanoff HK, Sargent DJ, Campbell ME, et al. Five-year data and prognostic factor analysis of oxaliplatin and irinotecan combinations for advanced colorectal cancer: N9741. J Clin Oncol. 2008;26:5721–7. doi: 10.1200/JCO.2008.17.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Cancer Society. Cancer Facts & Figures 2009. 2009 [Google Scholar]
- 6.Etienne M, Chazal M, Laurent-Puig P, et al. Prognostic value of tumoral thymidylate synthase and p53 in metastatic colorectal cancer patients receiving fluorouracil-based chemotherapy: phenotypic and genotypic analyses. J Clin Oncol. 2002;20:2832–43. doi: 10.1200/JCO.2002.09.091. [DOI] [PubMed] [Google Scholar]
- 7.Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumours. Cancer Res. 1995;55:1407–12. [PubMed] [Google Scholar]
- 8.Leichman CG, Lenz HJ, Leichman L, et al. Quantitation of intratumoral thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol. 1997;15:3223–9. doi: 10.1200/JCO.1997.15.10.3223. [DOI] [PubMed] [Google Scholar]
- 9.Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 10.Corsi DC, Ciaparrone M, Zannoni G, et al. Predictive value of thymidylate synthase expression in resected metastases of colorectal cancer. Eur J Cancer. 2002;38:527–34. doi: 10.1016/s0959-8049(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 11.Qiu LX, Tang QY, Bai JL, et al. Predictive value of thymidylate synthase expression in advanced colorectal cancer patients receiving fluoropyrimidine-based chemotherapy: Evidence from 24 studies. Int J Cancer. 2008;123:2384–9. doi: 10.1002/ijc.23822. [DOI] [PubMed] [Google Scholar]
- 12.Evrard A, Cuq P, Ciccolini J, Vian L, Cano J. Increased cytotoxicity and bystander effect of 5-fluorouracil and 5-deoxy-5-fluorouridine in human colorectal cancer cells transfected with thymidine phosphorylase. Br J Cancer. 1999;80:1726–33. doi: 10.1038/sj.bjc.6690589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccolini J, Cuq P, Evrard A, et al. Combination of thymidine phosphorylase gene transfer and deoxyinosine treatment greatly enhances 5-fluorouracil antitumor activity in vitro and in vivo. Mol Cancer Ther. 2001;1:133–9. [PubMed] [Google Scholar]
- 14.Okumura K, Shiomi H, Mekata E, et al. Correlation between chemosensitivity and mRNA expression level of 5-fluorouracil-related metabolic enzymes during liver metastasis of colorectal cancer. Oncol Rep. 2006;15:875–82. [PubMed] [Google Scholar]
- 15.Gustavsson B, Kaiser C, Carlsson G, et al. Molecular determinants of efficacy for 5-FU-based treatments in advanced colorectal cancer: mRNA expression for 18 chemotherapy-related genes. Int J Cancer. 2008;124:1220–6. doi: 10.1002/ijc.23852. [DOI] [PubMed] [Google Scholar]
- 16.Meropol NJ, Gold PJ, Diasio RB, et al. Thymidine Phosphorylase Expression Is Associated With Response to Capecitabine Plus Irinotecan in Patients With Metastatic Colorectal Cancer. J Clin Oncol. 2006;24:4069–77. doi: 10.1200/JCO.2005.05.2084. [DOI] [PubMed] [Google Scholar]
- 17.Marsh S. Thymidylate synthase pharmacogenetics. Invest New Drugs. 2005;23:533–7. doi: 10.1007/s10637-005-4021-7. [DOI] [PubMed] [Google Scholar]
- 18.Graziano F, Ruzzo A, Loupakis F, et al. Liver-only metastatic colorectal cancer patients and thymidylate synthase polymorphisms for predicting response to 5-fluorouracil-based chemotherapy. Br J Cancer. 2008;99:716–21. doi: 10.1038/sj.bjc.6604555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauritz R, van Groeningen CJ, Smid K, Jansen G, Pinedo HM, Peters GJ. Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA expression after administration of 5-fluorouracil to patients with colorectal cancer. Int J Cancer. 2007;120:2609–12. doi: 10.1002/ijc.22626. [DOI] [PubMed] [Google Scholar]
- 20.Ooyama A, Okayama Y, Takechi T, Sugimoto Y, Oka T, Fukushima M. Genome-wide screening of loci associated with drug resistance to 5-fluorouracil-based drugs. Cancer Sci. 2007;98:577–83. doi: 10.1111/j.1349-7006.2007.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang TL, Diaz LA, Romans K, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci U S A. 2004;101:3089–94. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Miller R, Zhang W, et al. Copy-number analysis of topoisomerase and thymidylate synthase genes in frozen and FFPE DNAs of colorectal cancers. Pharmacogenomics. 2008;9:1459–66. doi: 10.2217/14622416.9.10.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rooney PH, Stevenson DA, Marsh S, et al. Comparative genomic hybridization analysis of chromosomal alterations induced by the development of resistance to thymidylate synthase inhibitors. Cancer Res. 1998;58:5042–5. [PubMed] [Google Scholar]
- 24.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pui CH, Relling MV, Downing JR. Acute Lymphoblastic Leukemia. N Engl J Med. 2004;350:1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Q, Yang W, Raimondi SC, Pui CH, Relling MV, Evans WE. Karyotypic abnormalities create discordance of germline genotype and cancer cell phenotypes. Nat Genet. 2005;37:878–82. doi: 10.1038/ng1612. [DOI] [PubMed] [Google Scholar]
- 27.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 28.Arpino G, Green SJ, Allred DC, et al. HER-2 Amplification, HER-1 Expression, and Tamoxifen Response in Estrogen Receptor-Positive Metastatic Breast Cancer. Clin Cancer Res. 2004;10:5670–6. doi: 10.1158/1078-0432.CCR-04-0110. [DOI] [PubMed] [Google Scholar]
- 29.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 30.Ahnen D, Feigl P, Quan G, et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group study. Cancer Res. 1998;58:1149–58. [PubMed] [Google Scholar]
- 31.Etienne MC, Formento JL, Chazal M, et al. Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil-based treatment in advanced colorectal cancer patients. Pharmacogenetics. 2004;14:785–92. doi: 10.1097/00008571-200412000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, Huang K, Cheng Y, et al. P53 overexpression predicts poor chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV colorectal cancers after palliative bowel resection. Int J Cancer. 2002;97:451–7. doi: 10.1002/ijc.1637. [DOI] [PubMed] [Google Scholar]
- 33.Peters G, Backus H, Freemantle S, et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587:194–205. doi: 10.1016/s0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]


