Abstract
Background
Clefts of the lip and/or palate (cleft lip/palate) are notable for their complex etiology. The WNT pathway regulates multiple developmental processes including craniofacial development and may play a role in cleft lip/palate and other defects of craniofacial development such as tooth agenesis. Variations in WNT genes have been recently associated with cleft lip/palate in humans. In addition, two WNT genes, Wnt3 and Wnt9B, are located in the clf1 cleft locus in mice.
Methods
We investigated 13 SNPs located in WNT3A, WNT5A, WNT8A, WNT11, WNT3 and WNT9B genes, for association with cleft lip/palate subphenotypes in 500 cleft cases and 500 unrelated controls. Genotyping of selected polymorphisms was carried out using Taqman assays. PLINK 1.06 software was used to test for differences in allele frequencies of each polymorphism between affected and unaffected individuals. Haplotype analysis was also performed.
Results
Individuals carrying variant alleles in WNT3 presented an increased risk for cleft lip/palate (P=0.0003; OR=1.61 95% C.I: 1.29 -2.02) in the population studied.
Conclusion
Our results continue to support a role for WNT genes in the pathogenesis of cleft lip/palate. Although much remains to be learned about the function of individual WNT genes during craniofacial development, additional studies should focus in the identification of potentially functional variants in these genes as contributors to human clefting.
Keywords: WNT pathway, polymorphisms, cleft lip/palate, tooth agenesis, subphenotype
INTRODUCTION
Nonsyndromic oral facial clefts are notable for their complex etiology with interaction of genetic and environmental components (Murray, 2002). Genes involved in craniofacial development are plausible candidates for oral clefts. Wnt signaling is critical for proper development of the head and face in the mouse embryo, playing important roles in various aspects of craniofacial development ranging from axis formation to survival of cranial neural crest cells to patterning of the brain (Mani et al., 2009).
Several studies support a role for the Wnt gene family in the etiology of cleft lip/palate. Wnt3 and Wnt9b genes are expressed in the developing facial ectoderm in mice (Lan et al., 2006). A Wnt9b−/− mutant mouse was described presenting a phenotype that included incomplete penetrance of cleft lip/palate (Carroll et al., 2005). Both these genes are located within the clf1 cleft locus in chromosome 11 of A/WySn cleft susceptible mouse strains (Juriloff et al., 2005; 2006). This region is syntenic to human chromosome 17q21, a region that has been associated with nonsyndromic oral facial clefts in humans (Carinci et al., 2007). A nonsense mutation (Q83X) in the WNT3 gene has also been described in a case of tetra-amelia and cleft lip/palate in a large consanguineous family (Niemann et al., 2004). Recently, variations in WNT genes have been recently associated with human nonsyndromic oral clefts. In this particular family-based study (Chiquet et al., 2008), a variety of WNT genes (WNT3A, WNT5A, WNT8A, WNT11) showed association with cleft lip/palate in either European American or Hispanic families. Gene-gene interaction between WNT3A and both WNT3 and WNT5A was also suggested.
To investigate a role for WNT genes in nonsyndromic oral facial clefts, we interrogated thirteen SNPs in six WNT genes that had been previously associated with cleft/lip palate in humans and in animal models for association with cleft subphenotypes.
MATERIAL AND METHODS
Subjects
The subjects of this study have been described in part in previous studies (Letra et al., 2007; 2009). A total of 766 individuals of Caucasian ancestry, 463 cases with nonsyndromic clefts and 303 unrelated control individuals without clefts or family history of clefting, were ascertained at the Dental Clinics of the Hospital of Rehabilitation and Craniofacial Anomalies and Bauru Dental School, both of the University of São Paulo, Bauru, SP, Brazil and at the Center for Treatment of Craniofacial Anomalies (CTAC), Rio de Janeiro, Brazil. Individuals were considered as Caucasians when there was no history of African, Native Amerindians or Japanese descent. Subjects with clefts were examined clinically and through their medical records to determine their individual cleft status. Cleft status was based on cleft completeness (comprised of primary and secondary palates entirely) or incompleteness, and on laterality (left, right, bilateral). An “unknown” cleft status indicated that either cleft type or side could not be determined, even after medical records were reviewed. The presence of tooth agenesis was assessed clinically and through radiographs by dental professionals.
The study was conducted with the consent of the participants and approved by the Research and Ethics Committee of the aforementioned institutions. In the case of children under 15 years of age, authorization was also requested from their parents or from the individual legally in charge of the child.
Clefts subphenotypes
Individuals with oral clefts were divided by subphenotype as: All Clefts (Cleft lip + Cleft Lip and Palate + Cleft Palate), Cleft lip with or without cleft palate (CLP), and Cleft Palate only (CP). The CLP group was further divided into Bilateral CLP, Unilateral CLP, Right Unilateral CLP, and Left Unilateral CLP subgroups.
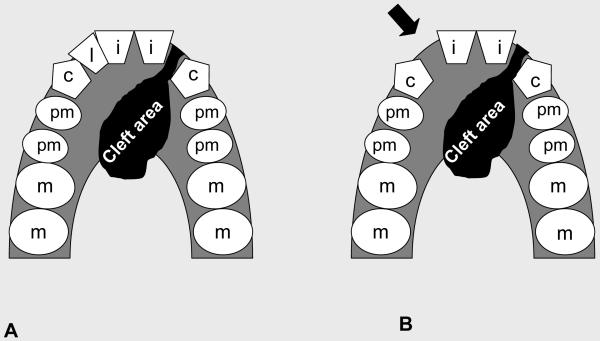
We also considered here a new dental subphenotype (unsuccessful bilateral). This trait was suggested for individuals presenting unilateral clefting and agenesis of the lateral incisor on the noncleft side (Letra et al., 2007). The absence of the incisor would represent a microform of the cleft (Figure 1). Notwithstanding, instances of tooth agenesis adjacent to the cleft area (affecting maxillary central incisors, lateral incisors, or canines) were not considered, because the absence of such teeth is likely the consequence of developmental anomalies at the cleft area.
Figure 1.
Schematic representation of unilateral cleft (A) and unilateral cleft with agenesis of the lateral incisor (arrow) on the opposite side of the cleft (B), also referred to as “unsuccessful or occult cleft” according to Letra et al. (2007).
(i=Central Incisor; l= Lateral Incisor; c= Canine; pm= Pre-molar; m= Molar)
Single Nucleotide Polymorphism (SNP) selection and genotyping
We selected thirteen SNPs spanning six WNT genes that had been previously associated with cleft/lip palate in humans (Chiquet et al., 2008) or in animal models (Juriloff et al., 2005; Juriloff et al., 2006; Lan et al., 2006) to test for association with CL/P subphenotypes in our population (Table 1).
Table 1.
Summary of candidate genes and SNPs studied
| SNP | Chr. | Gene | Base pair position |
SNP function | Base change § |
MAF |
|---|---|---|---|---|---|---|
| rs708111 | 1 | WNT3A | 226,257,988 | Promoter/regulatory | C/T | 0.47 (C) |
| rs3094912 | 1 | WNT3A | 226,276,438 | Intronic | T/A | 0.47 (A) |
| rs752107 | 1 | WNT3A | 226,313,974 | 3′ UTR | C/T* | 0.32 (T) |
| rs1745420 | 1 | WNT3A | 226,318,355 | 3′ UTR | C/G | 0.13 (G) |
| rs566926 | 3 | WNT5A | 55,495,818 | Intronic | A/C | 0.21 (A) |
| rs2040862 | 5 | WNT8A | 137,447,888 | Intronic | C/T | 0.16 (T) |
| rs1533767 | 11 | WNT11 | 75,583,448 | Silent Mutation | A/G | 0.39 (A) |
| rs142167 | 17 | WNT3 | 42,150,418 | Dowstream | A/G | 0.19(G) |
| rs199498 | 17 | WNT3 | 42,220,763 | Intronic | C/T | 0.23 (C) |
| rs111769 | 17 | WNT3 | 42,227,151 | Intronic | C/T | 0.35 (T) |
| rs9890413 | 17 | WNT3 | 42,256,448 | Upstream | A/G | 0.32 (G) |
| rs2165846 | 17 | WNT9B | 42,296,365 | Intron | A/G | 0.44 (G) |
| rs197915 | 17 | WNT9B | 42,345,521 | Upstream | A/G | 0.33 (G) |
Ancestral allele listed in bold.
not available.
MAF, minor allele frequency.
Genomic DNA was obtained from saliva samples as previously described (Menezes et al., 2008). Genotyping of selected polymorphisms was carried out using Taqman assays and reagents in an ABI 7900 automatic instrument (Applied Biosystems, Foster City, CA). We used PLINK 1.06 software (Purcell et al., 2007) to test for differences in allele frequencies of each polymorphism between affected and unaffected individuals. Haplotype analysis using “all clefts” phenotype was also performed using PLINK. We considered the P-values adjusted with Bonferroni correction as implemented in PLINK.
We used the online software program FASTSNP (Function Analysis and Selection Tool for Single Nucleotide Polymorphisms) to predict the functionality of the SNPs. FASTSNP is a web server that allows users to efficiently identify the SNPs most likely to have functional effects. It prioritizes SNPs according to putative functional effects, such as changes to the transcriptional level, pre-mRNA splicing, protein structure, etc. the prediction of functional effects is based on information extracted from external web servers. We analyzed the investigated SNPs and found that although some are located in putative regulatory regions, no information with regards to functionality is known so far.
RESULTS
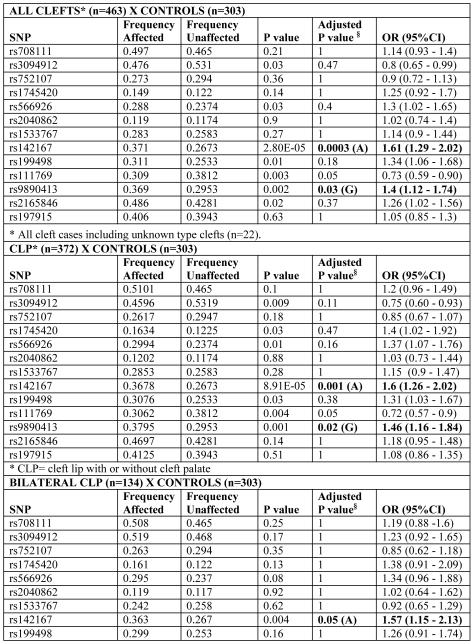
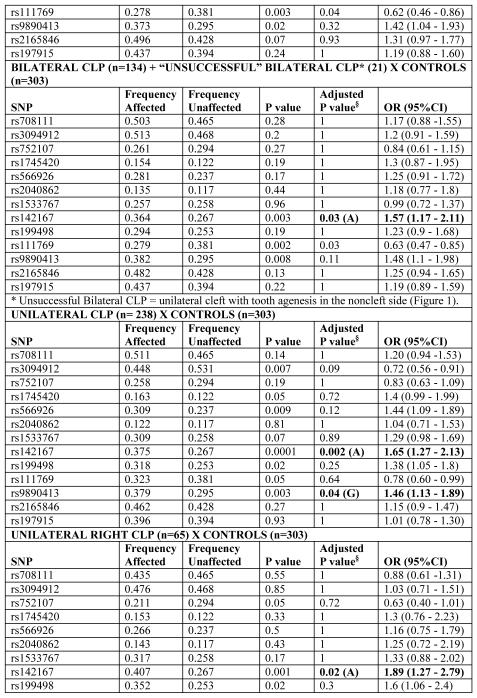
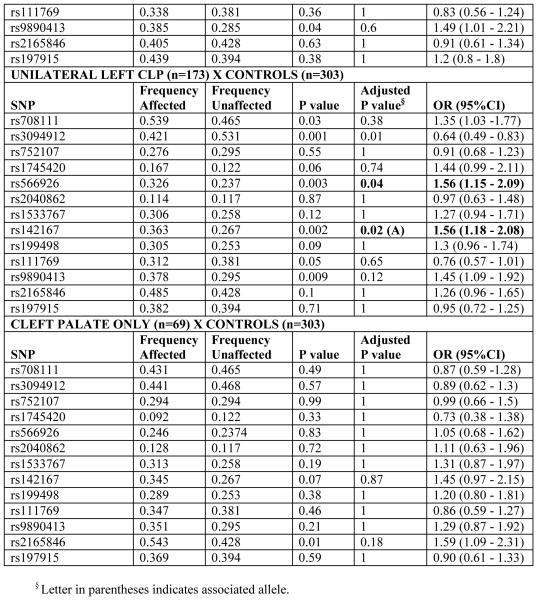
A summary of the most significant results are presented in Tables 2 and 3. Genotype and allele distributions were within Hardy-Weinberg equilibrium (data not shown).
Table 2.
Results of association tests with WNT genes in cleft lip/palate and control individuals, according to cleft subphenotype
|
|
|
Table 3.
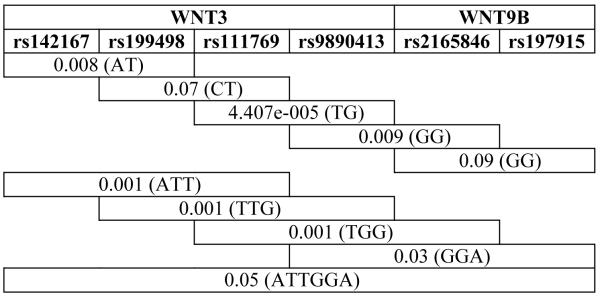
Most significant sliding window haplotype association results observed for markers in WNT3 and WNT9B genes located in chromosome 17 and cleft lip/palate in a Brazilian population
|
We found association between the WNT3 gene and the phenotypes “All Clefts” (CLP + CP) and “Cleft lip with or without cleft palate” in the studied population. SNP rs142167, located in the 5′ UTR of WNT3, showed the most significant association with the phenotypes “All clefts” [(P=0.0003; OR=1.61 (1.29-2.02)], “CLP” [P=0.001; OR=1.6 (1.26-2.02)], and “unilateral CLP” [P=0.002; OR=1.65 (1.27-2.13)]. Under a nominal value of 0.05, SNP rs9890413 in WNT3 also showed an association with the phenotypes “All clefts” [P=0.03; OR=1.4 (1.12 -1.74)] and with “CLP” and “unilateral CLP” [P=0.02; OR=1.46 (1.16-1.84) and P=0.04; OR=1.46 (1.13-1.89), respectively] (Table 2).
When analyzing the “unsuccessful bilateral” cleft subphenotype, we also observed an association for WNT3 SNP rs142167 [P=0.03; OR= 1.57 (95% CI: 1.17-2.11] (Table 2).
The results of the haplotype analysis further support the associations found for the individual SNPs (Table 3). We observed overtransmission of WNT3 haplotypes rs142167-rs199498 (P=0.008), rs111769-rs9890413 (P=4.407e-005), rs142167-rs199498-rs111769 (P=0.001), rs199498-rs111769-rs9890413 (P=0.001), and also WNT3-WNT9B haplotypes rs9890413-rs2165846 (P=0.009) and rs111769-rs9890413-rs2165846 (P=0.001) (Table 3).
DISCUSSION
The Wnt genes are involved in regulating midface development and upper lip fusion and are therefore candidates for an etiological role in nonsyndromic cleft lip with or without cleft palate. Evidences supporting Wnt genes as possible clefting loci come from the inbred A/WySn mouse strain (Juriloff et al., 2004). Wnt3 and Wnt9B are located in the clf1 region and could contribute to the clefting phenotype (Juriloff et al., 2005). In humans, variations in WNT genes have been described in cases with syndromic (Niemann et al., 2004) and nonsyndromic oral clefts (Chiquet et al., 2008).
In this study, we investigated SNPs in WNT3, WNT3A, WNT5A, WNT8A, WNT9B, and WNT11, for association with cleft lip/palate in a case-control population. Studies with cases and controls in populations characterized by significant immigration such as Brazilians deserve special caution in the interpretation of results because of possible effects of admixture. The option of using only Caucasian individuals in the genetic analyses should overcome any major stratification effects since these individuals have been shown to have major contribution of European ancestry (Lins et al., 2009). In contrast to Chiquet et al. (2008), where the authors report the association of WNT3, WNT3A, WNT7A, WNT8A, WNT9B and WNT11 with cleft lip/palate in their European American sample, we only found association with two genes. SNPs in WNT3 and WNT5A had nominal P-values of 0.05 or less, although only one SNP in WNT3 (rs142167) remained significant after Bonferroni correction. Our results corroborate with the results of Chiquet et al. (2008) for European-American families and further support the suggestion that WNT3 may be a cleft susceptibility gene. Although the SNP showing the strongest association in our study (rs142167) was not associated in the study of Chiquet et al. (2008), the fact that both studies reveal association of WNT3 with clef lip/palate warrants further investigations and reinforces a role for WNT3 in the etiology of nonsyndromic clefts. WNT3 showed association with all clefts and all cleft subphenotypes except for cleft palate alone, although our sample size with cleft palate only may be too small to draw definite conclusions. Haplotypes in WNT3 and WNT9B genes were also associated with the cleft phenotype.
The association with WNT5A was found in cases with unilateral left CL/P. We subdivided unilateral CLP into right or left because the authors believe that stratifying cleft cases into specific cleft subsets may increase homogeneity. Moreover, there may be some genetic differences in the etiology of left and right clefts. The acknowledged larger prevalence of unilateral left clefts (one third of all cleft cases) suggests that some genes may be preferentially involved in cleft side. This gene was also associated with nonsyndromic clefts in the Hispanic population described in Chiquet et al. (2008). During craniofacial development, Wnt5A signals were detected in the mesenchymal cells and around Meckel’s cartilage and in only in the mesenchyme of the elevating palatal shelves (Paiva et al., 2009).
In a similar context, we considered that individuals with unilateral clefts who also present tooth agenesis of the lateral incisor on the noncleft side may in fact have a bilateral cleft, and the absence of the lateral incisor represents a microform of the cleft, namely an unsuccessful bilateral cleft (Letra et al., 2007). Comparison of this cleft subphenotype with controls also revealed association with WNT3. WNT intercellular signaling molecules have been implicated in the regulation of murine tooth development, where Wnt3 shows specific expression in the enamel knot at the cap stage. The importance of Wnt3 during odontogenesis is further highlighted by the progressive loss of ameloblasts from postnatal incisor teeth in transgenic mice (Millar et al., 2003).
One may argue that most investigated SNPs are in intronic or intergenic regions and do not seem to alter transcription factor binding sites or have any other potentially damaging effect. Overall, in silico analysis predicted that none of the SNPs tested would abolish the protein domain, although some SNP alleles could arguably have a deleterious effect. For example, WNT5A SNP rs566926, associated with unilateral left CLP, is located at a transcription factor binding site with intronic enhancer function for which the A allele creates two additional binding sites for SOX5. Both these transcription factors are involved in the regulation of embryonic development and in the determination of the cell fate. The encoded protein may also play a role in chondrogenesis (Woods et al., 2007), which is a critical step in palatogenesis. We should also consider that these SNPs, although putatively neutral, may also be in linkage disequilibrium with an etiologic variant which could explain the results observed here.
In summary, our results continue to support the involvement of WNT genes in human clefting. Although much remains to be learned about the function of individual WNT genes during craniofacial development, additional studies should focus in the identification of potentially functional variants in these genes as contributors to human clefting.
ACKNOWLEDGMENTS
The authors are indebted to the participants of the study. We thank the staff at CTAC for their help with sample collection: Marcelo Couto, Leonardo Garcia, Anália Pereira, Bruna Cruz, Bernardo Ribeiro. Many thanks to XJ Wang for valuable assistance with PLINK, Ashley Rowles and Ryan Rylands for their technical support.
AK was partially supported by the Carnegie Mellon’s Undergraduate Research Office (URO). This work was supported by NIH grants 1K99DE018413-01A1 (to RM); 1K99DE018954 (to AL); and UL1 RR024153. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
REFERENCES
- Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71:1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, et al. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, Hecht JT. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet. 2008;17:2212–2218. doi: 10.1093/hmg/ddn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Dewell SL. A digenic cause of cleft lip in a A-strain mice and definition of candidate genes for the two loci. Birth Defects Res A Clin Mol Teratol. 2004;70:509–518. doi: 10.1002/bdra.20041. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Dewell SL, Brown CJ, Mager DL, Gagnier L, Mah DG. Investigations of the genomic region that contains the clf1 mutation, a casual gene in multifactorial cleft lip and palate in mice. Birth Defects Res A Clin Mol Teratol. 2005;73:103–113. doi: 10.1002/bdra.20106. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Menezes R, Granjeiro JM, Vieira AR. Defining subphenotypes for oral clefts based on dental development. J Dent Res. 2007;86:986–991. doi: 10.1177/154405910708601013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Menezes R, Granjeiro JM, Vieira AR. AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts. Birth Defects Res A Clin Mol Teratol. 2009;85:169–173. doi: 10.1002/bdra.20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, Grattapaglia D, Pereira RW. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol. 2010;22:187–192. doi: 10.1002/ajhb.20976. [DOI] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R. Visualizing canonical Wnt signaling during mouse craniofacial development. Dev Dyn. 2009;239:354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Letra A, Ruff J, Granjeiro JM, Vieira AR. Studies of genes in the FGF signaling pathway and oral clefts with or without dental anomalies. Am J Med Genet A. 2008;164A:1614–1617. doi: 10.1002/ajmg.a.32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE, Koyama E, Reddy ST, Andl T, Gaddapara T, Piddington R, Gibson CW. Over- and ectopic expression of Wnt3 causes progressive loss of ameloblasts in postnatal mouse incisor teeth. Connect Tissue Res. 2003;44:124–129. [PubMed] [Google Scholar]
- Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- Niemann S, Zhao C, Pascu F, Stahl U, Aulepp U, Niswander L, Weber JL, Muller U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am J Hum Genet. 2004;74:558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva KB, Silva-Valenzuela MD, Massironi SM, Ko GM, Siqueira FM, Nunes FD. Differential Shh, Bmp and Wnt gene expressions during craniofacial development in mice. Acta Histochem. 2009 doi: 10.1016/j.acthis.2009.05.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Wang G, Dupuis H, Shao Z, Beier F. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J Biol Chem. 2007;282:23500–23508. doi: 10.1074/jbc.M700680200. [DOI] [PubMed] [Google Scholar]