Abstract
Most pharmacogenomics knowledge is contained in the text of published studies, and is thus not available for automated computation. Natural Language Processing (NLP) techniques for extracting relationships in specific domains often rely on hand-built rules and domain specific ontologies to achieve good performance. In a new and evolving field such as pharmacogenomics (PGx), rules and ontologies may not be available. Recent progress in syntactic NLP parsing in the context of a large corpus of pharmacogenomics text provides new opportunities for automated relationship extraction. We describe an ontology of PGx relationships built starting from a lexicon of key pharmacogenomic entities and a syntactic parse of more than 87 million sentences from 17 million Medline abstracts. We used the syntactic structure of PGx statements to systematically extract commonly occurring relationships and to map them to a common schema. Our extracted relationships have a 70 to 87.7% precision and involve not only key PGx entities such as genes, drugs, and phenotypes (e.g., VKORC1, warfarin, clotting disorder), but also critical entities that are frequently modified by these key entities (e.g., VKORC1 polymorphism, warfarin response, clotting disorder treatment). The result of our analysis is a network of 40,000 relationships between more than 200 entity types with clear semantics. This network is used to guide the curation of PGx knowledge and provide a computable resource for knowledge discovery.
Keywords: Relationship Extraction, Pharmacogenomics, Natural Language Processing, Ontology, Knowledge Acquisition, Data Integration, Biological Network, Text Mining, Information Extraction
1. Introduction
Most biological knowledge exists in published scientific text. In order to support the creation of databases and to enable the discovery of new relationships, there is great interest in extracting relationships automatically. Several successful efforts use manually created rules to define patterns of relationships between entities. These approaches are efficient when used in domains that are of limited scope, such as protein–protein interactions or protein transport. However, the complexity and diversity of the semantics used to describe relationships in broad or evolving domains, such as pharmacogenomics (PGx), are harder to capture. Thus, no general set of rules exists for extracting the relationships relevant to such fields, and creating/maintaining them manually would be tedious and time consuming.
Syntactic sentence parsers can identify the subject, object and type of relationships using grammatical rules. General statistical parsing techniques have recently emerged, and there are several general-purpose parsers that yield reasonable results when applied to scientific text. These parsers depend on the need for good domain-specific lexicons of key entities, since named-entity recognition for particular fields in science can be difficult. We consider named entity recognition as the process of identifying members of the lexicon within the text, amidst other words. With such lexicons, there is an opportunity to use syntactic sentence parsers to identify rich rule sets automatically.. These rule sets take advantage of sentence structure and grammar to extract more precise information. In addition, these rule sets can be organized in an ontology that allows normalization of relationships and inference over them.
Pharmacogenomics (PGx) is the study of how individual genomic variations influence drug-response phenotypes. PGx knowledge exists for the most part in the scientific literature in sentences that mention relationships. We can represent a large fraction of this knowledge as binary relationships R (a, b), where a, and b are subjects and objects related by a relationship of type R. Sometimes, a and b are instances of a gene (e.g., VKORC1 gene), drug (e.g., warfarin), or phenotype (e.g., clotting disorder). As we shall demonstrate later, very often a and b are entities that are modified by genes (e.g., VKORC1 polymorphism), drugs (e.g., warfarin dose) or phenotypes (e.g., clotting disorder treatment). R is a type of relation described by words such as “inhibits”, “transports”, or “treats” and their synonyms. Thus, although the three key entities in PGx (genes, drugs, and phenotypes) can be target nouns for relation extraction, they are more often indicators of latent PGx knowledge, as they modify other concepts to create a second set of entities required to precisely describe PGx relationships. We call these modified entities in contrast with the key entities that modify and expand them. These modified entities can be any biomedical entity, such as a gene variation, drug effect, or disease treatment. For example, the gene entity VKORC1 (a key entity) is used as a modifier of the concept polymorphism in “VKORC1 polymorphisms affect warfarin response,” indicating that VKORC1 polymorphism is a critical (composite) PGx entity. This sentence also indicates that a modified entity, warfarin response, will be important as well.
In this paper we present a method for using a syntactical parser to identify recurrent binary relationships that express PGx knowledge. Many of these relationships use genes, drugs and phenotypes as modifiers of other entities. We organized these relationships and the associated entities in an ontology that maps diverse sentence structures and vocabularies to a common semantics. We processed 87 million sentences using this ontology to capture and normalize more than 40,000 specific PGx relationships. These relationships are summarized in the form of a semantic network (i.e., a network where entities (nodes) and relationships (edges) are associated with the semantics defined in our ontology). We anticipate that they will be useful to assist database curation and as a foundation for knowledge discovery and data mining.
2. Related Work
Our work is partially motivated by our efforts building the Pharmacogenomic Knowledge Base, PharmGKB (http://www.pharmgkb.org/) [1]. PharmGKB aims to catalog all knowledge of how human genetic variation impacts drug response phenotypes, and is a manually curated database that summarizes published gene–drug–phenotype relationships. The rapidly increasing size of the pharmacogenomic literature threatens to overwhelm the PharmGKB curators. Automatic approaches using NLP techniques are therefore promising. Methods based on co-occurrence assume that entities occurring together in a sentence are related, but the semantics of the relationships are not typically captured. Nevertheless, these approaches efficiently identify potential relationships that can subsequently be evaluated manually. For example, the Pharmspresso system uses co-occurrence to group frequently co-mentioned genes, genomic variations, drugs, and diseases [2]. These groups are then used to assist manual curation. Li et al. used the co-occurrence of drug and disease names in MEDLINE abstracts to derive drug–disease relations and to build a disease-specific drug–protein network [3]. Blaschke et al. and Rosario et al. expanded this co-occurrence approach to extract more complete relations by searching for “tri-co-occurrence” [4, 5]. Tri-co-occurrence refers to the co-occurrence of two named entities and one type of relationship in a unique piece of text. Statistical analysis of co-occurrence can help derive semantic similarities between entities [6].
In contrast to co-occurrence, syntactic parsing can explicitly identify relationships between two entities in text [7]. Hand-coded parsing rules can extract protein–protein interactions and protein transport relationships [8, 9]. Fundel et al. defined three general patterns of relations (specifying the semantic type of subjects and objects, and using a lexicon of association words) to identify protein–protein interactions [10]. For example their pattern “effector – relation – effectee” enables the capture of relationships of the form “protein A activates protein B”. The OpenDMAP system also uses patterns to identify protein interaction and transport [11]. Alhers et al. used vocabularies and semantic types of the UMLS (Unified Medical Language System) to specify patterns to extract gene–disease and drug–disease relationships [12]. Several groups have used extracted relationships to create networks, including molecular interaction networks [13], gene–disease networks [14], regulatory gene expression networks [15], and gene–drug–disease networks [16]. In order to be efficient, these syntactic approaches often rely on large sets of patterns and stable ontologies to guarantee performance on diverse sentence structures. Unfortunately, a systematic catalog of patterns for pharmacogenomics is not available [17, 18].
The Semantic Web community has developed methods for learning ontologies from text using unsupervised approaches [19, 20]. Most of these efforts focus on learning hierarchies of concepts. Ciaramita et al. studied unsupervised learning of relationships between concepts [21]. Their method produces a network of concepts where edges are associated with precise semantics (e.g., Virus encodes Protein). Other efforts have focused on enriching existing ontologies for NLP using Web content [22]. Cilibrasi et al. proposed a method to automatically learn the semantics of processed words, hypothesizing that semantically related words co-occur more frequently in Web pages than do unrelated words [23]. Gupta et al. used Web content to identify concept mappings for previously unrecognized words discovered while processing text [24]. We describe here our method of relationship extraction that uses (1) syntactic rules to extract relationships and (2) a learned ontology to normalize those relationships.
3. Methods
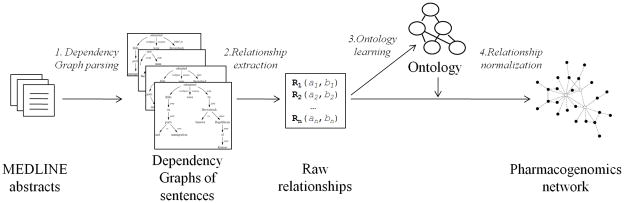
Figure 1 gives an overview of the four steps of our method, described in the following subsections. The first input is a corpus of article abstracts split into individual sentences. We benefit from previous work that made such a corpus available and also provides a convenient way to retrieve the sentences [25]. We use lexicons of PGx key entities (drugs, genes, phenotypes) from PharmGKB1 to retrieve sentences mentioning pairs of key entities.. We parse retrieved sentences with the Stanford Parser and represent the sentence using a convenient data structure called a “dependency graph” [26]. Each retrieved sentence is analyzed to extract the raw relationships between key entities themselves or other entities that they modify. After applying this procedure to many pairs of key entities, we gather the raw relationships and entities and manually map them to a much smaller set of “normalized” relationships and entities based on synonymy, arranged hierarchically in an OWL ontology2. We assume that this ontology is representative of PGx relationships mentioned in our corpus. This ontology can then be applied to all raw relationship instances in the corpus to create a very large set of normalized relationships representing the semantic content of the corpus.
Figure 1.
Overview of our method to extract pharmacogenomics (PGx) relationships from text. The method has four steps. 1. We parse the text (Medline abstracts in this work) with the Stanford Parser to yield the Dependency Graph data structure that provides the syntactical structure of each sentence. 2. We identify PGx entities and their raw relationships—“raw” because their subject, object and type use natural language terms. 3. We processed these raw relationships to build (first run only) or refine (next runs) an ontology of PGx relationships. 4. For each of the raw relationships, we map them to the ontology and express them in normalized form. Normalized relationships create a network in which nodes are PGx entities and edges are relationships, both of which are associated with a precise semantics.
3.1. Sentence Parsing of Medline into Dependency Graphs
The goal of the first step is to provide, in a format easy to process, the syntactical structure of sentences that potentially mention a PGx relationship. We focus on sentences that mention at least two PGx key entities. We used an index of individual sentence of Medline abstracts published before 2009 (17,396,436 abstracts and 87,806,828 sentences) processed by Xu et al. [25]. This index has been built on the full text of sentences with the Lucene library and can consequently be queried with any term [27]. It returns sentences that have been indexed with the query terms and also returns “parse trees” that correspond to retrieved sentences. A parse tree is a rooted tree that represents the syntactical structure of a sentence, as illustrated in Figure 2. Parse trees were previously generated by applying the Stanford Parser on every sentence. The Stanford Parser is a statistical natural language parser [26]. It uses a set of training sentences in which the grammatical function of words were manually annotated by experts to record the most likely syntactical structure of a sentence. Parse trees of sentences that mention at least two PGx key entities are subsequently transformed into Dependency Graphs (DGs) with the same Stanford Parser [27]. This DG format, described in Section 3.1.3, provides the syntactical structure of sentences that we analyze to extract relationships.
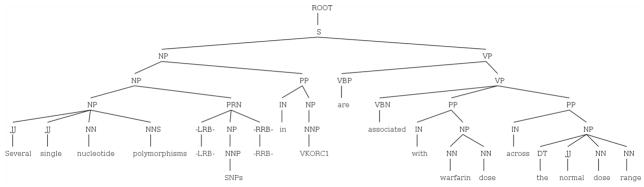
Figure 2.
Sample parse tree of the sentence “Several single nucleotide polymorphisms (SNPs) in VKORC1 are associated with warfarin dose across the normal dose range.” (PubMed ID 17161452). This parse tree is obtained when querying an index (built in previous work) with query (1) that looks for two pharmacogenomics key entities: VKORC1 (a gene) and warfarin (a drug).
3.1.1. Querying the Sentence Index Using Seeds
From the corpus, we consider only sentences with pairs of PGx key entities, (i.e., one gene and one drug, or one gene and one phenotype). For this initial work we did not focus on drug–phenotype pairs because they are numerous and the majority of these pairs are not of PGx interest. For example, to retrieve sentences that potentially mention a relationship between the gene VKORC1 and the drug warfarin, the index was queried with two sets of synonyms as follows:
| (1) |
Results of these queries were sentences (and corresponding parse trees) mentioning at least two terms, one that refers to the first entity and one that refers to the second entity. Sets of synonyms used to build such queries are from the PharmGKB lexicons. For this initial work, we used 41 important genes highlighted by PharmGKB,3 as well as 3,007 drugs and 4,202 phenotypes. Drug and phenotype names listed in lexicons are not restricted to PGx. Phenotype names include disease and adverse reaction names. Querying the index with pairs of entities named in such lexicons can be considered as a task of named-entity recognition. In one retrieved sentence (and in its corresponding parse tree), we distinguish the two particular terms, called seeds, that correspond to the two recognized entities. These are called seeds because they form the basis for relationship extraction. Seeds of the parse tree shown in Figure 2 are VKORC1 and warfarin.
3.1.2. Reducing the Set and the Size of Parse Trees
In order to reduce computational complexity, we reduce the number of parse trees or parse tree fragments considered. We compare the relative positions of the two seeds in the sentences clauses.4 If the two seeds are not located in the same clause of the sentence, the parse tree is removed from consideration (the seeds are unlikely to have an extractable relationship across clauses). If the parse tree contains more than one clause, and a clause does not contain both seeds, then the clause is pruned (we keep only clauses containing more than two seeds). For example, the parse tree in Figure 2 contains only one clause with both seeds and was neither removed from consideration nor pruned.
3.1.3. Transformation of Parse Trees into Dependency Graph
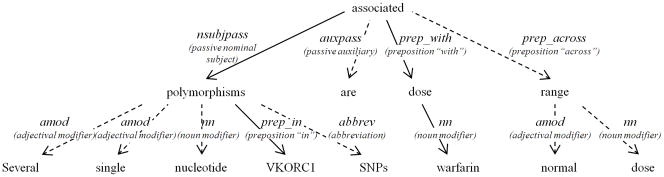
The Stanford Parser summarizes the syntactical structure of a parse tree in an easy to process format, called a Dependency Graph (DG) [28]. DGs are rooted, oriented, and labeled graphs, where nodes are words and edges are dependency relations between words (e.g., noun modifier, nominal subject). Figure 3 shows the DG built from the parse tree shown in Figure 2. DGs are easier to read and process than parse trees or other representations. Relationships between sentence words are binary, and they occur directly between “content” words (e.g., “associated” is connected directly to “dose”), rather than being mediated indirectly via less important function words (e.g., “associated” is related to “dose” via a common link to “with”). Each DG includes a root (or head) that enables easy recognition of the subject and the object of a sentence. Thus, DGs highlight semantic content and are relatively easy to understand.
Figure 3.
The Stanford Parser creates a Dependency Graph (DG) data structure from the parse tree, such as this one corresponding to the parse tree in Figure 2. Its two seeds are VKORC1 and warfarin, and its root is associated. Solid lines represent the path that connects both seeds to each other via the root. This path is used in the next step to extract the following raw relationship: associated(VKORC1_polymorphisms, warfarin_dose).
3.2. Relation Extraction
The second step of our method uses syntactic structure provided by DGs to identify raw relationships of the form R (a, b) where:
a and b are two paths (i.e., sequences of nodes) in a DG, each of which is either a single key entity (an instance of gene, drug or phenotype) or of a modified entity—an entity that is not a gene, drug or phenotype but is modified by one (e.g., an instance of gene variation, drug dose or phenotype treatment);
R is a node in the DG that connects a and b, and indicates the nature of their relationship.
In example shown Figure 3, a is “VKORC1 polymorphisms”, b is “warfarin dose” and R is “associated”. We defined an algorithm that extracts relationships from the DG that correspond to the two following patterns:
| (2) |
| (3) |
An expanded seed is a seed that matches the input key entity or that represents a modified entity in which the key entity modifies another entity. The relations are captured by verbs or nominalized versions of verbs (such as “association” that is the nominalized version of “associate”). This algorithm has three steps: seed recognition, seed expansion, and coupling of expanded seeds, described as follows.
3.2.1. Seed Recognition
Seeds are identified using the input lexicons. We use the PharmGKB lexicons for genes, drugs and phenotypes, which include a basic list of synonyms. Seeds may be a single word or a compound noun. This “seed recognition” step localizes the two seeds in the DG. When a seed is composed of one word, (e.g., thrombosis), the system uses string matching and techniques to handle plural and of capitalized forms. If a seed is composed of more than one word (e.g., venous thromboembolism), a DG for the seed itself (noted as DGseed) is created and the parsed sentence DG is examined to identify the subset of nodes matching the DGseed.
3.2.2. Seed Expansion
The DG has information that allows us to expand the seed to determine if it is being used as a key entity or a modified entity. We expand a seed by traversing edges of the DG. The method of traversal is defined by the types of dependencies that connect the seed to other concepts. Depending on these dependencies (Table 1 summarizes the decision logic), the algorithm will:
Table 1.
Summary of algorithm traversing the dependency graph from entities through root to other entities. The subject and object may be modified entities, and the dependency graph provides data types that help decide how to construct the subject, object and relationship for each graph. In particular, depending on the type associated with each edge in the graph during traversal of a DG, the seed expansion either (i) continues, (ii) ends and thereby establishes a subject or an object, or (iii) interrupts. To identify a relationship, the expansion of the two seeds has to end (one as a subject and one as an object) on a common “root” word.
| ALGORITHM ACTIONS |
||||
|---|---|---|---|---|
| (i) expand seed | (ii) end expansion |
(iii) interrupt | ||
| expanded seed is subject | expanded seed is object | |||
| DEPENDENCY TYPES | nn (noun modifier) | nsubj (nominal subject) |
dobj (direct object) iobj (indirect object) |
All others |
| prep_{for,in,into,of,on,to} (preposition) | nsubjpass (passive nominal subject) | xcomp (open clausal component) | ||
| xsubj (controlling subject) | prep_{at,as,by,for,in,into,on,than,with,within} (preposition) | |||
expand the seed (continuing traversing the DG and constructing the seed)
end the expansion by detecting a relationship type represented by a verb (e.g., activate, bind, regulate) or a nominalized form of a verb (e.g., activation, binding, regulation). The type of the dependency determines whether the seed is the subject or object in the relationship.
interrupt the expansion if neither (i) nor (ii) applies.
3.2.3. Seed Coupling
When two expanded seeds (one subject expanded seed and one object expanded seed) each end by reaching the same verb or nominalized verb, they are associated to create a raw relationship, as subject or object depending on the dependency type. Figure 4 illustrates the expansion and subsequent coupling of seeds recognized in the DG shown in Figure 3.
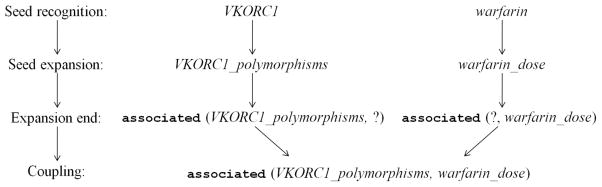
Figure 4.
Four steps of recognizing and expanding the two seeds in the example sentence shown in Figures 2 and 3. Starting with the seed entities, VKORC1 and warfarin, we use the rules provided in Table 1 to traverse the Dependency Graph in Figure 3 to recognize the subject (VKORC1_polymorphisms), object (warfarin_dose) and relationship (associated) in the Dependency Graph.
3.2.3. Evaluation of Raw Relationship Precision
We manually evaluated the precision of extracting raw relationships. We randomly selected a subset of 220 raw relationships and classified them into three categories: complete and true, incomplete and true, and false. Incomplete and true relationships are relationships that are consistent with mentioned relationships but are missing partial information. It is then required that the lack of information does not change the interpretation of the relationship. For example, if derived from the sentence “polymorphisms in VKORC1 are associated with warfarin dose.”:
associated (VKORC1_polymorphisms, warfarin_dose) would be complete and true;
associated (VKORC1_polymorphisms, warfarin) would be incomplete and true;
polymorphisms (VKORC1, warfarin_dose) would be false.
3.3. Ontology Construction
Raw relationships represent multiple equivalent ways to express a relationship. In order to simplify the analysis of the semantics, we must map many raw relationships onto a smaller, normalized set of relationships. We manually examined the raw relationships observed in the text, and grouped them into a hierarchical domain ontology of PGx relationships. We first identified the most frequent relationship types; and then merged similar ones and organized them hierarchically. We also tracked modified entities and merged these. We computed the number of raw entity and relationship types, and the number of normalized types resulting from grouping them. We describe the steps of ontology construction here. This construction is carried out only once, at the first iteration of the approach, but the ontology can be refined during subsequent iterations.
3.3.1. Identification of Relationship Types
We created four lists from the raw relationships extracted from the DGs. The lists represent (1) the most frequent types of relationships, and the most frequent modified entities modified by (2) genes, (3) drugs and (4) phenotypes as defined in our lexicons for these entities (see Figure 5). Each list is processed to remove word heterogeneity caused by captions, plurals, and conjugations. We then combined equivalent words, and computed their frequency of occurrence to produce a list sorted by frequency of use. Modified entities are the subjects or objects of relationships (i.e., a or b) grammatically modified by either a gene, a drug, or a phenotype.
Figure 5.
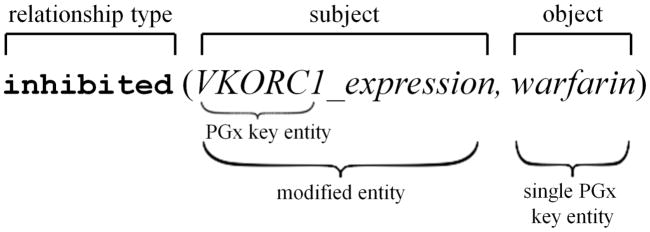
A raw relationship derived from the dependency graph has three components: relationship type, subject and object. Both subject and object can be either a single PGx key entity (e.g., warfarin) or a modified entity using the key entity as a modifier (e.g., VKORC1_expression).
3.3.2. Organization of Relationship Types and Entities in Hierarchies
We manually examined and grouped elements of each list into sets of synonyms and then organized them in role and concept hierarchies. See Figure 6 for example of roles and concepts. Relationship synonyms (e.g., decrease, reduce) represent roles in the ontology. A role is a binary relation associated with a domain and a range. It is named with one of the synonyms (e.g., decrease) and associated with labels that correspond to the other synonyms. The roles are organized in a hierarchy so that any instance of a role (e.g., inhibit) is also an instance of its parent (less precise) role (e.g., affect).
Figure 6.
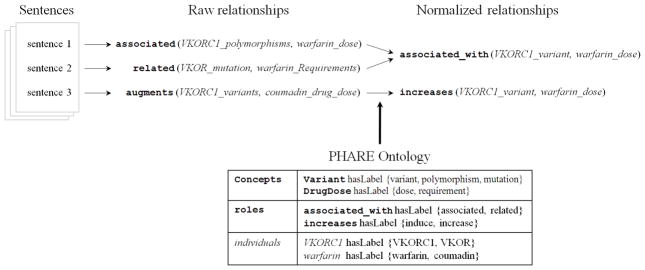
Three raw relationships normalized to two normalized expressions, using the PHARE (PHArmacogenomics RElationship) ontology of entities and relationships. The content of this ontology is described in Section 4. In this example, the first two raw relationships express the same relationship, according to the mappings in our ontology (e.g. drug dose and drug requirement are declared synonyms in the ontology). The third raw relationship maps to a more specific relationship (increases), which is a child of the more general (associated) relationship.
Terms that are modified by the same kind of entity (e.g., gene) are grouped into sets of synonyms (e.g., polymorphism, mutation, variant) and lead to the creation of concepts in the ontology. A concept is named with one of the synonyms as a reference to label the group as a whole (e.g., the variant label leads to the concept name Variant) and is associated with all the other synonym labels. Distinct concepts are organized in a concept hierarchy such that any instance of a concept (e.g., Variant) is also an instance of its parent concepts (e.g., GenomicVariation). Importantly, when a new concept is created, it is associated with a description that specifies whether it is modified by genes or drugs or phenotypes. For example, the mention of a genomic variation in text can be modified by a gene name, thus the Variant concept (with alternate labels “polymorphism” and “mutation”) is associated with a description stating that instances of Variant can be modified by instances of Gene. Such a description enables “VKORC1 polymorphism” in text to be mapped to the concept Variant, since modified by a gene name (VKORC1) whereas the phrase “important polymorphism” is not mapped, since “important” is not an instance of the Gene concept. The ontology was represented in description logic and encoded in OWL using Protégé [29, 30]. The ontology was built once, examined and validated by three domain experts (curators at PharmGKB). Because we considered only the first 200 elements of each of the four lists, the ontology construction and examination took approximately 4 hours (around 1 hour per list).
3.4. Relationship Normalization
Once the ontology is built, we can use it to map most raw relationships to a common semantics. The mapping process has two steps: (1) entity names are normalized, and then (2) relationships are normalized. Normalization is a many–to-one mapping that maps multiple diverse textual statements to a common normalized form. Figure 6 illustrates the normalization process.
3.4.1. Normalization of Entity Names
To name modified entities uniformly, we implemented an algorithm that takes a modified entity name of any length and returns its normalized form according to the ontology. In the first step of the algorithm, we decompose the modified entity into its original seed and the other words in the string. The algorithm iterates over these words to construct the normalized name of the entity. The first word is the seed. Using the PharmGKB lexicons, seeds are a gene, drug, or phenotype (e.g., VKORC1, warfarin, or bleeding) and thus we can associate them with a concept Cseed. With Cseed determined, we process the next word. We search for a match between the next word and labels of concepts that are modified by Cseed, according to the ontology. If a concept matches, the processed word is associated with this new concept. In the case where no match is found, a match is searched between the processed word and labels of concepts that are modified by more general concepts (i.e., those modified by parents of Cseed). In the case where no further match is found, no concept in the ontology corresponds and then a new concept is created with the processed word as a label and with a description specifying that it is modified by Cseed (i.e., ∃ modified.Cseed). This operation is iterated for each successive word of the modified entity, each time assigning the right concept to the new processed word. When the last word is reached, the normalized version of the entity name is proposed as the concatenation of the seed plus the names of successive assigned concepts. For example, with the modified entity VKORC1_polymorphisms, VKORC1 is the seed and Cseed is the concept Gene. The next word is polymorphism, which refers to a concept modified by Gene. Polymorphism is a synonym of the concept Variant, which is thus associated with the processed word. Because there are no other words in the modified entity, the normalized name is VKORC1_variant. When the subject or the object of a raw relationship is a single PGx key entity (gene, drug, phenotype), PharmGKB lexicons provide the normalized name, which is the preferred name of the seed (e.g., VKORC1 for VKOR). Figure 7 decomposes the steps of the normalization of a modified entity made of three words.
Figure 7.
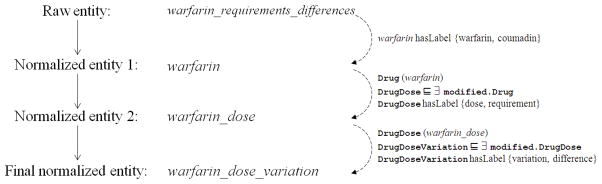
Starting with the text “differences in warfarin requirements”, we extracted the raw entity “warfarin_requirements_differences” and then apply normalization using the PHARE ontology. The first step ensures that the standard name for warfarin is used (here, Coumadin would have been mapped to warfarin, had it been used). Warfarin is the seed and the concept associated with it, noted Cseed, is Drug according to the ontology. The second step maps “requirements” to the standard ontological concept of dose, and the final step maps “differences” to the ontology concept of variation. Having learned these mappings on our initial training corpus, we can apply them broadly and prospectively to new sentences.
3.4.2. Normalization of Relationship Types
We normalize relationship types by searching for a role label that matches the raw relationship. If a label matches, the identifier of the corresponding role becomes the normalized type. For example, the type “related”, mentioned in Figure 6, matches to the role associated_with. Normalized entities and relationships are combined to form the normalized relationship. We use normalized relationships to instantiate concepts and roles from the ontology and thus to create a knowledge base of PGx relationships. Each relationship in the knowledge base is made by the instantiation of:
two concepts (e.g., Variant(vkorc1_variant) and DrugDose(warfarin_dose)) and
one role (e.g., associated_with (vkorc1_variant, warfarin_dose)).
A detailed description of the normalization algorithm is provided in Coulet et al. [31].
4. Results
We queried 87,806,828 parse trees to find a total of 295,569 sentences with pairs of PGx entities as seeds. We pruned these sentences, as described in Section 3.1.2., to extract 41,134 raw relationships, including 21,050 relationships seeded by gene–drug pairs and 20,084 by gene–phenotype pairs. Table 2 shows the relationship types and entities most frequently found in these raw relationships.5 Remarkably, we found that the 200 most frequent raw relationship types summarize 80% of extracted relationships (see Table 3). Our manual evaluation of 220 raw relationships indicated that 70% of those were complete true positives, 87.7% were complete or incomplete true positives and 12.3% were false positives. Distinction between complete and incomplete true positive is described in Section 3.2.3.
Table 2.
The 30 most frequent relationship types and entities modified by genes, drugs or phenotypes. Numbers correspond to their frequency of occurrence in 41,134 raw relationships extracted from 17,396,436 MEDLINE abstracts. Entities can be composed of one or several words.
| relationship types | entities modified by |
||
|---|---|---|---|
| genes | drugs | phenotypes | |
| 2538 associate | 1237 gene | 267 metabolism | 304 cell |
| 1017 increase | 1000 inhibitor | 229 activity | 114 line |
| 985 inhibit | 935 polymorphism | 226 administration | 101 patient |
| 825 induce | 775 expression | 213 treatment | 71 risk |
| 763 metabolize | 773 activity | 207 effect | 35 tissue |
| 666 involve | 689 mutation | 205 inhibitor | 34 specimen |
| 643 reduce | 685 genotype | 146 dose | 33 case |
| 547 catalyze | 393 inhibition | 137 concentration | 27 treatment |
| 515 cause | 329 level | 104 level | 26 rate |
| 509 affect | 245 gene_mutation | 103 substrate | 26 effect |
| 490 decrease | 232 gene_polymorphism | 90 clearance | 26 breast=cancer |
| 433 show | 227 allele | 88 antagonist | 22 incidence |
| 428 express | 162 variant | 84 channel | 21 factor |
| 392 relate | 156 enzyme | 75 inhibition | 21 resistance |
| 392 use | 138 mrna | 73 responsible | 20 sample |
| 387 correlate | 125 protein | 72 hydroxylation | 18 model |
| 385 influence | 83 channel | 70 enzyme | 16 exposure |
| 355 determine | 81 isoform | 67 oxidation | 15 type |
| 354 contribute | 78 effect | 65 gene | 15 development |
| 319 factor | 77 isozyme | 63 formation | 15 group |
| 318 mediate | 76 cell | 62 blocker | 14 % |
| 317 had | 73 deficiency | 60 metabolite | 14 activity |
| 301 found | 71 overexpression | 57 dependent | 14 mellitus |
| 299 measure | 67 substrate | 52 exposure | 13 gene |
| 287 investigate | 67 induction | 46 ratio | 13 cause |
| 284 result | 63 gene_expression | 45 consumption | 13 presence |
| 281 studied | 59 c677t | 44 due | 13 all |
| 280 detect | 58 inhibitor_use | 43 drug | 13 level |
| 274 association | 57 gene_allele | 40 response | 12 severity |
| 274 have | 55 content | 38 bioavailability | 12 study |
Table 3.
Percentages of raw relationships covered using the 100 and 200 most frequent relationship types and entities. n represents the number of distinct types or entities identified in all relationships. Thus, for example, the top 200 entities modified by genes account for 85% of all raw gene-related entities mentioned in the corpus. Similarly, the top 200 relationship types account for 80% of all raw relationships in our corpus.
| Percentage of covered raw relationships | relationship types (n=1921) | entities modified by | ||
|---|---|---|---|---|
| genes (n=1210) | drugs (n=1243) | phenotypes (n=445) | ||
| by 100 most frequent relationship types or entities | 68% | 77% | 58% | 71% |
| by 200 most frequent relationship types or entities | 80% | 85% | 71% | 84% |
We created an ontology of the 200 most frequent relationship types and modified entities called PHARE (PHArmacogenomics RElationships).6 PHARE is made of 237 concepts and 76 roles. PHARE concepts are instantiated with 26,966 distinct entities and PHARE roles are instantiated with 46,523 explicit7 relationships between pairs of entities. The number of role instantiations is greater than the number of raw relationships because we count both role and inverse role instantiations (e.g., R (a, b) and R− (b, a)). Finally one role instantiation can be supported by several sentences and one entity can be involved in several role instantiations. Table 4 presents a list of the most commonly used concepts and roles.
Table 4.
The fifteen most instantiated normalized roles (first column) and normalized concepts modified by Gene (second column), Drug (third column), and Phenotype (fourth column) in the knowledge base. Numbers to the left represent the number of instances of the role or entity in 41,134 raw relationships. The number in brackets is the number of unique instances of concepts used in these raw relationships.
| roles | concepts modified by |
||
|---|---|---|---|
| Drug | Gene | Phenotype | |
| 2981 associated_with | 6075(1083) Drug | 8040 (285) Gene | 3854 (990) Disease |
| 1580 demonstrates | 1063 (558) DrugTreatment | 2082 (210) Variant | 604 (354) PhenotypeRisk |
| 1460 increases | 606 (344) DrugDose | 2702 (187) Expression | 211 (163)DiseaseExacerbation |
| 1428 reduces | 302 (160) DrugEffect | 826 (134) GenomicVariation | 171 (183) DiseaseSeverity |
| 1420 studies | 263 (195) DrugMetabolism | 644 (64) Enzyme | 117 (95) Symptom |
| 986 inhibits | 199 (135) DrugActivity | 520 (185) GeneProductFunction | 99 (77) DiseaseCause |
| 924 influences | 130 (107) DrugElimination | 192 (103) GeneProductSynthesis | 57 (38) DiseaseSensitivity |
| 894 causes | 137 (90) DrugTransformation | 592 (103) Repression | 49 (53) PhenotypeMechanism |
| 841 includes | 101 (61) Hydroxylation | 317 (79) Overexpression | 49 (36) Phenotype |
| 707 metabolizes | 93 (75) DrugAnalysis | 285 (81) GeneProductActivity | 44 (38) DiseaseEffect |
| 698 uses | 88 (81)DrugPharmacokinetics | 169 (67) Protein | 29 (33) DiseaseRelief |
| 655 induces | 87 (72) DrugMetabolite | 128 (81) GeneAnalysis | 27 (35) PhenotypeAnalysis |
| 488 produces | 80 (62) DrugInhibitor | 88 (75) GenomicRegion | 25 (24) DiseaseDuration |
| 464 affects | 67 (62) DrugDoseVariation | 73 (44) GeneProduct | 9 (10) DiseaseSurvival |
| 449 determines | 59 (59) DrugAnalog | 57 (50) GeneProductActivityChange | 5 (13) DiseaseAbsence |
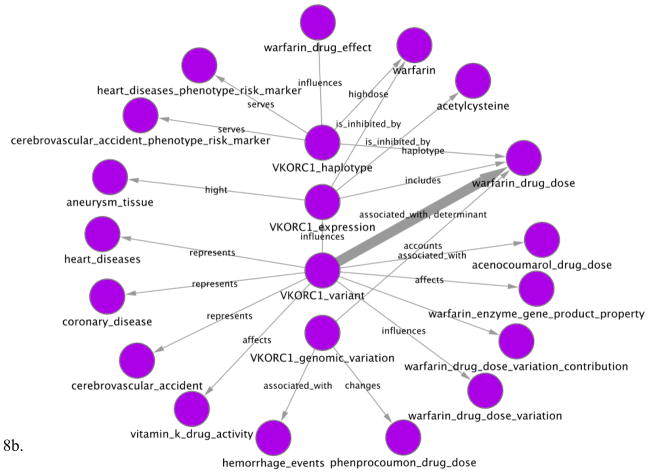
We used the resulting knowledge base to create PGx networks where nodes are PGx entities and edges are normalized relationships between these entities. Of course, we mapped these entities and relationships to common semantics as defined in the knowledge base, and thus they are semantic networks. Figures 8a and 8b show such semantic networks related to the VKORC1 gene. Figure 9 summarizes the number of entities in each entity class and the number of relationships between these and other entity types.
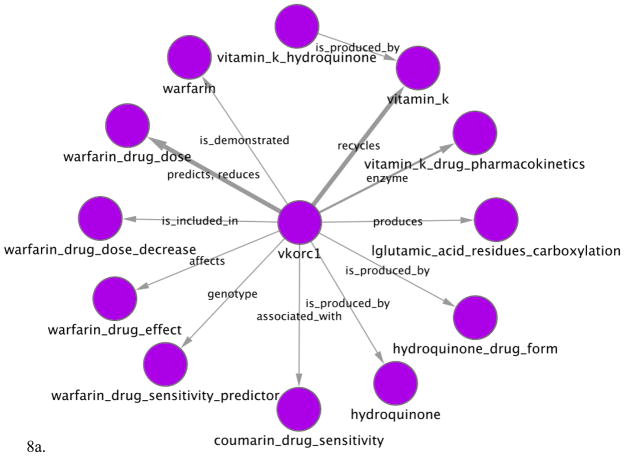
Figure 8.
Two semantic networks extracted for the VKORC1 gene. (a) Displays Pharmacogenomics (PGx) relationships extracted from sentences that contain VKORC1 or one of its synonyms as a key entity. Thus, for example, it shows that VKORC1 predicts warfarin drug dose. (b) Displays PGx relationships for entities that are modified by VKORC1 (e.g. VKORC1_haplotype, VKORC1_variant). Thus, for example, VKORC1 haplotypes influence warfarin drug effect. Each node represents a PGx key or modified entity, e.g. warfarin or warfarin_drug_effect. Edges represent relationships between these entities that are mentioned in MEDLINE abstracts. When several sentences mention a relationship between the same two entities, the edge is wider and is labeled with the most frequent types of relationship. Networks have been generated using Cytoscape v2.6.3 (http://www.cytoscape.org).
Figure 9.
A summary of the Pharmacogenomics (PGx) Concept Network. Nodes represent concepts frequently appearing in PGx relationships. Their size is dependent on the number of instantiated PGx entities. Edges represent relationships between instances of two concepts. Their width is dependent on their number. This network has been built from the knowledge base of 41,134 relationships extracted from the text of Medline abstracts. Thus, for example, there are many statements in the PGx literature relating drugs to genes, and genes to diseases. There are somewhat less relating drug metabolism specifically to genomic variation. This network has been generated using Cytoscape v2.6.3 (http://www.cytoscape.org).
5. Discussion
The two main advantages of our method are: (1) the identification of both PGx key entities (genes, drugs and phenotypes), as well as crucial and novel PGx entities modified by genes, drugs and phenotypes, and (2) the association of extracted relationships with a normalized semantics, captured in a manually built ontology. The syntactical structure of sentences allows us to use our key entity lexicons to bootstrap the discovery and normalization of the modified entities critical to PGx and the ontology allows us to record these entities and recognize them under very general textual conditions.
Our method is flexible because it uses syntactical patterns that are much more general than specific rules (e.g., x inhibits y). It is precise because it is based on the detection of relationships in natural language text, and does not depend upon simple co-occurrence of two recognized entities. A drawback of syntactical parsing approaches compared to co-occurrence is lower recall. In our work, low recall is attenuated by large size of the corpus, which gives us multiple opportunities to recognize a relationship. We may further improve precision by using full text. Our recognition of named entities in sentences is based on string matching plus normalization techniques. At this time, we capture, but do not use qualifiers that modulate the relationship itself such as negation, adverbs (e.g., not, highly, hypothetically). One improvement of our approach would be to consider subcategorization frames in particular for ditransitive or caused-motion verbs (such as to transform for instance) that are reporting several relationships between one subject and multiple objects (e.g., x transforms y in z).
We created and validated our ontology manually, and were fortunate that the language used to describe PGx relationships degenerates to a small core of unique concepts. Other efforts for detecting synonyms use resources such as WordNet8, but this is not applicable to technical biological domains. Instead, we used domain experts to create acceptable synonym mappings. The decision to group words can be approximate, and some grouped words are not exact synonyms, such as SNP and allele. These similar words have been grouped to limit the number of distinct concepts in the ontology. The approach described in this paper is completely applicable to other domains. The main drawback of such domain change is the human effort that will be required to develop an ontology adapted to the domain if none is available.
5. Conclusion
We have described a new method that uses the syntactical structure of sentences to extract biomedical relationships from text. We use key pharmacogenomic entities (genes, drugs, phenotypes) to bootstrap a process whereby other entities that are modified by these concepts are identified and stored in an ontology. The relationships used in pharmacogenomics literature are also captured and normalized, yielding a core set of 41,134 relationships that capture approximately 80% of extracted relationships in the text. Our ontology allows us to label automatically any parsed sentence that provides a relationship between the key entities or the derivative modified entities—totaling more than 200 total entity types. We created a knowledge base of relationships from 17 million MEDLINE abstracts containing 87 million sentences. This knowledge base allows us to create semantically rich summaries of the relationships between genes, drugs and phenotypes. By going beyond classic entity recognition for gene, drug and phenotype, and by not requiring prior enumeration of relationship types, we have created a novel accurate and extensible approach to processing PGx text. To the best of our knowledge, our work is the first to use reasoning with an OWL ontology to integrate heterogeneous relationships extracted from text.
Acknowledgments
This work was supported by the NIH Roadmap National Centers of Biomedical Computing grant to the National Center for Biomedical Ontologies (U54HG004028) and by the PharmGKB GM61374, as well as LM-05652, with computing cluster support from NSF CNS-0619926.
Footnotes
OWL (Web Ontology Language): http://www.w3.org/TR/owl-features/
A clause is a group of terms of a sentence. Some sentences contain several independent clauses. For example the sentence “I am a doctor, and my wife is a lawyer” has two independent ones.
Complete lists are available at http://www.stanford.edu/~coulet/material/entity_lists/.
PHARE is available at http://www.stanford.edu/~coulet/material/ontology/phare.owl
Those are considered explicit in contrast with inferred instantiations that can be considered implicit.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klein T, Chang J, Cho M, Easton K, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver D, Rubin D, Shafa F, Stuart J, Altman R. Integrating genotype and phenotype information: An overview of the PharmGKB project. The Pharmacogenomics Journal. 1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 2.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 10(S-2) doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Zhu X, Chen JY. Building disease-specific drug–protein connectivity maps from molecular interaction networks and pubmed abstracts. PLoS Comput Biol. 2009;5(7):e1000450. doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic Extraction of Biological Information from Scientific Text: Protein–Protein Interactions. ISMB; 1999. pp. 60–67. [PubMed] [Google Scholar]
- 5.Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. ACL; 2004. pp. 430–437. [Google Scholar]
- 6.Cohen T, Widdows D. Empirical distributional semantics: Methods and biomedical applications. Journal of Biomedical Informatics. 2009;42(2):390–405. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wermter J, Hahn U. You can’t beat frequency (unless you use linguistic knowledge) - a qualitative evaluation of association measures for collocation and term extraction. ACL; 2006. [Google Scholar]
- 8.Hirschman L, Krallinger M, Wilbur J, Valencia A, editors. Genome Biology. Vol. 9. 2008. The BioCreative II - Critical Assessment for Information Extraction in Biology Challenge. [Google Scholar]
- 9.Tsujii J, editor. Proceedings of the BioNLP 2009 Workshop Companion Volume for Shared Task. 2009. [Google Scholar]
- 10.Fundel K, Kuffner R, Zimmer R. Relex - relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 11.Hunter L, Lu Z, Firby JWB, Jr, Johnson H, Ogren P, Cohen K. BMC Bioinformatics. 78. Vol. 9. OpenDMAP: An open-source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Rindflesch TC. Extracting semantic predications from medline citations for pharmacogenomics. Pacific Symposium on Biocomputing. 2007:209–220. [PubMed] [Google Scholar]
- 13.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. Genies: a natural-language processing system for the extraction of molecular pathways from journal articles. ISMB (Supplement of Bioinformatics) 2001:74–82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 14.Rindflesch TC, Libbus B, Hristovski D, Aronson AR, Kilicoglu H. Semantic relations asserting the etiology of genetic diseases. AMIA Annu Symp Proc, 2003; 2003. pp. 554–558. [PMC free article] [PubMed] [Google Scholar]
- 15.Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from medline. Bioinformatics. 2006;22(6):645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- 16.Tari L, Hakenberg J, Gonzalez G, Baral C. Querying parse tree database of medline text to synthesize user-specific biomolecular networks. Pacific Symposium on Biocomputing. 2009:87–98. [PubMed] [Google Scholar]
- 17.Dumontier M, Villanueva-Rosales N. Towards Pharmacogenomics Knowledge Discovery on the Semantic Web. Briefings in Bioinformatics. 2009;10(2):153–163. doi: 10.1093/bib/bbn056. [DOI] [PubMed] [Google Scholar]
- 18.Coulet A, Smail-Tabbone M, Napoli A, Devignes MD. Suggested Ontology for Pharmacogenomics (SO-Pharm): Modular Construction and Preliminary Testing. KSinBIT. 2006;LNCS 4277:648–57. [Google Scholar]
- 19.Aussenac-Gilles N, Soergel D. Text analysis for ontology and terminology engineering. Applied Ontology. 2005;1(1):35–46. [Google Scholar]
- 20.Buitelaar P, Cimiano P, Magnini B. Vol. 123 of Frontiers in Artificial Intelligence. IOS Press; 2005. Ontology Learning from Text: Methods, Evaluation and Applications. [Google Scholar]
- 21.Ciaramita M, Gangemi A, Ratsch E, Saric J, Rojas I. IJCAI. 2005. Unsupervised learning of semantic relations between concepts of a molecular biology ontology; pp. 659–664. [Google Scholar]
- 22. [accessed 02/11/2010];Ontology Development Information Extraction (ODIE) project. http://www.bioontology.org/ODIE-project.
- 23.Cilibrasi R, Vitányi PMB. Kolmogorov Complexity and Applications. 2006. Automatic Meaning Discovery Using Google. [Google Scholar]
- 24.Gupta A, Oates T. Using Ontologies and the Web to Learn Lexical Semantics. IJCAI. 2007:1618–1623. [Google Scholar]
- 25.Xu R, Supekar K, Morgan A, Das A, Garber A. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. AMIA Annu Symp Proc 2008; 2008. pp. 820–824. [PMC free article] [PubMed] [Google Scholar]
- 26.Klein D, Manning CD. Accurate unlexicalized parsing. ACL; 2003. pp. 423–430. [Google Scholar]
- 27.Agichtein E, Gravano L. Snowball: extracting relations from large plaintext collections. ACM DL; 2000. pp. 85–94. [Google Scholar]
- 28.de Marneffe MC, Manning CD. COLING Workshop on Cross-framework and Cross-domain Parser Evaluation. 2008. The stanford typed dependencies representation. [Google Scholar]
- 29.Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF. The Description Logic Handbook. Cambridge University Press; 2003. [Google Scholar]
- 30.Knublauch H, Fergerson RW, Fridman Noy N, Musen MA. The Protégé OWL Plugin: An Open Development Environment for Semantic Web Applications. ISWC; 2004. pp. 229–243. [Google Scholar]
- 31.Coulet A, Altman RB, Musen MA, Shah NH. Integrating heterogeneous relationships extracted from natural language sentences. Proceedings of the Bio-ontologies SIG, ISMB; 2010. [Google Scholar]