Abstract
Cholesterol metabolism is tightly regulated at the cellular level. In addition to classic transcriptional regulation of cholesterol metabolism (e.g., by SREBP and LXR), members of a class of non-coding RNAs termed microRNAs (miRNAs) have recently been identified to be potent post-transcriptional regulators of lipid metabolism genes, including cholesterol homeostasis. We and others have recently shown that miR-33 regulates cholesterol efflux and HDL biogenesis by downregulating the expression of the ABC transporters, ABCA1 and ABCG1. In addition to miR-33, miR-122 and miR-370 have been shown to play important roles in regulating cholesterol and fatty acid metabolism. These new data suggest important roles of microRNAs in the epigenetic regulation of cholesterol metabolism and have opened new avenues for the treatment of dyslipidemias.
Keywords: cholesterol homeostasis, microRNAs, HDL
Regulation of cellular cholesterol metabolism
Cholesterol is an essential structural component in the cell membrane of most vertebrates and a precursor in metabolic pathways, including steroid hormone and bile acid synthesis. Cholesterol homeostasis thus needs to be tightly regulated, and perturbations in lipid metabolism are associated with disease states, including atherosclerosis, metabolic syndrome, and type 2 diabetes [1].
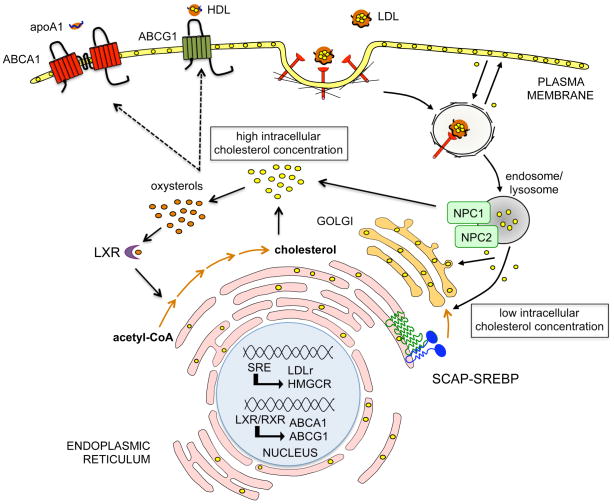
Animal cells synthesize cholesterol from acetyl-CoA through a series of more than 20 enzymatic reaction steps [2]. In addition, cells obtain cholesterol from the circulation in the form of apolipoprotein B-containing lipoproteins, especially low-density lipoproteins (LDL) [3, 4] (Figure 1). LDL particles are internalized via the LDL receptor (LDLr) on the cell surface and hydrolyzed to free cholesterol in lysosomes [3, 4]. The intracellular cholesterol concentration is tightly controlled by feedback mechanisms that operate at both transcriptional and posttranscriptional levels [5, 6]. When cells accumulate excess sterols, the activity of 3-hydroxy-3-methylglutaryl coenzyme A (HMGCoA) reductase, a rate-limiting enzyme of the biosynthetic pathway, declines by more than 90% and the cell surface expression of LDLr decreases [6]. In contrast, upon depletion of intracellular cholesterol, cells maintain a high activity of HMGCoA reductase and highly express LDLr (Figure 1). This coordinated process is regulated by ER-bound sterol regulatory element-binding proteins (SREBPs) [5, 7, 8]. The SREBP family of basic-helix-loop-helix leucine zipper (bHLH-LZ) transcription factors consists of SREBP-1a, SREBP-1c and SREBP-2 proteins that are encoded by two unique genes, SREBF-1 and SREBF-2. The SREBPs differ in their tissue-specific expression, target gene selectivity and the relative potencies of their trans-activation domains. SREBP-1c target genes are involved in fatty acid metabolism, such as fatty acid synthase (FASN) [5, 7, 8]. SREBP-2 is responsible for cholesterol related genes, such us HMGCoA reductase and LDLr. SREBP-1a targets both set of genes, but the expression of the SREBP-1c isoform predominates in vivo, particularly in the mouse liver in the fed state [5, 7, 8]. Increased SREBP activity causes cholesterol and fatty acid accumulation by downregulating the SCAP-mediated activation of the SREBP pathway by feedback inhibition.
Figure 1.
Regulation of cellular cholesterol metabolism. Animal cells synthesize cholesterol from acetyl-CoA. In addition, cells obtain cholesterol from the circulation in the form of apolipoprotein B-containing lipoproteins, especially LDL. The circulating LDL particles carrying cholesterol and cholesterol esters are internalized through LDLr and transported to sorting endosomes. LDL particles are subsequently transported to late endosomes (LE) and lysosomes (LY), while LDL receptors are recycled to the plasma membrane. Free cholesterol egress from LE/LY in a process mediated by Niemann-Pick type C 1 and 2 proteins (NPC1 and NPC2). Under low intracellar cholesterol concentration, the SCAP-SREBP complex moves to the Golgi complex where the SREBP is processed to its nuclear form. The nuclear SREBP turns on genes involved in cholesterol biosynthesis (e.g. HMGCR) and cholesterol uptake (LDLr). Conversely, in response to cellular cholesterol excess, the oxysterols generated bind and activate the liver X receptors (LXRs), which heterodimerize with retinoid X receptors (RXR) and activate the expression of the ATP transporters (ABCA1 and ABCG1). ABCA1 and ABCG1 promote cholesterol efflux via apoA1 and HDL respectively and help to maintain intracellular cholesterol homeostasis.
The liver X receptor (LXR) nuclear hormone receptors also contribute to cholesterol homeostasis by activating the transcription of genes involved in the response to cholesterol excess, including ABCA1, ABCG1 and ABCG5/8 [9–11]. These transporters promote cellular cholesterol efflux to HDL and its associated apolipoprotein, apo-A1, a critical step in the initiation of reverse cholesterol transport to the liver for excretion [12, 13] (Figure 1), whereas ABCG5/8 promote cholesterol excretion into bile [14]. ABCA1 is also primarily responsible for initiating HDL formation in the liver, a finding brought to light when the mutation in Tangier disease, a condition characterized by low plasma HDL, was mapped to the ABCA1 gene [15–18]. As HDL levels are inversely correlated with atherosclerosis susceptibility, these discoveries have set off an enormous amount of research on the regulation, mechanism of action, and suitability of ABCA1 as a target to increase HDL levels for the treatment of atherosclerosis.
Mechanisms of action and biogenesis of miRNAs
As noted, microRNAs have newly recognized influences on cholesterol homeostasis. MicroRNAs (miRNAs) constitute a large family of small (~22 nucleotide [nt]) noncoding RNA molecules, single stranded in the mature form, that are important post-transcriptional regulators of gene expression [19–21] in metazoan animals, plants and protozoa. Since their discovery in C. elegans [22–24], hundreds of miRNAs have been identified as a result of the cloning and sequencing of size-fractionated RNAs, deep sequencing technologies, and computational prediction methods [25].
miRNAs typically control the expression of their target genes by imperfect base pairing to the 3′ untranslated regions (3′UTR) of messenger RNAs (mRNAs) thereby inducing repression of the target mRNA. This inhibitory effect can occur by either transcript destabilization, translational inhibition, or both [21, 26, 27]; however, recent studies suggest that miRNAs may also repress mRNA targets by binding to other regions including 5′ UTRs or protein-coding exons [28–31]. Bioinformatic predictions and experimental approaches indicate that a single miRNA may target more than a hundred mRNAs [32]. Indeed, human miRNAs are predicted to control the activity of more than 60% of all protein-coding genes [26, 32, 33]. miRNAs have now been implicated in the control of a wide range of biological functions including development, differentiation, metabolism, growth, proliferation and apoptosis [19, 20, 34–39].
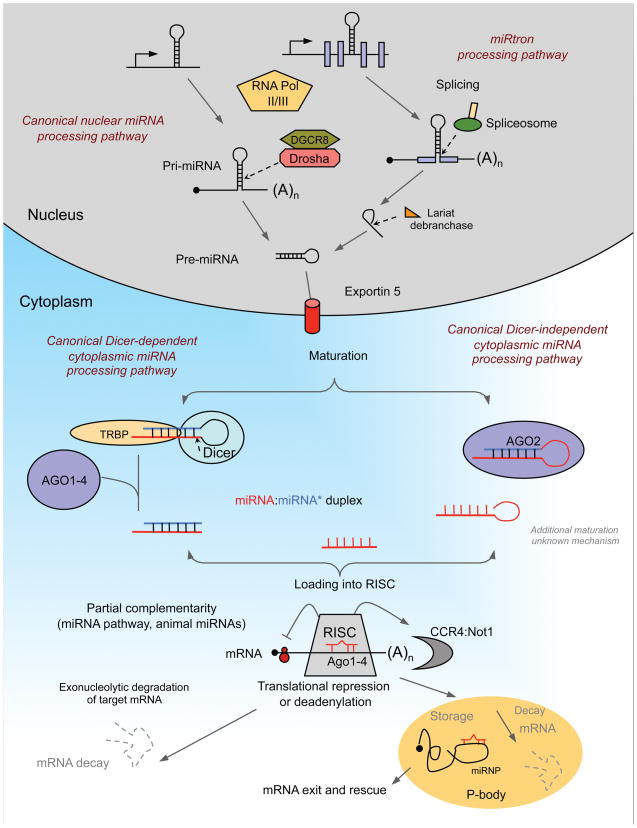
miRNAs are located throughout the genome within intronic and exonic portions of protein-coding genes, as well as intergenic regions. As illustrated in Figure 2, the production of the functional, ~22 nt mature miRNA involves multiple processing steps. Most animal miRNAs are transcribed by RNA polymerase II as long primary transcripts, generating a stem-loop containing primary miRNA (pri-miRNA) [40] The pri-miRNA is processed within the nucleus by the microprocessor complex, which consists of a ribonuclease III (RNase III) called Drosha [41], a RNA-binding protein DGCR8/Pasha [42] and a variety of co-factors [DEAD box helicases p68 and p72, and heterogeneous nuclear ribonucleoproteins) [43] thought to promote the fidelity, specificity, and/or activity of Drosha cleavage. In most mammalian miRNAs, Drosha processing occurs co-transcriptionally, i.e. before splicing of host RNA (canonical pathway). However, a subset of intronic miRNAs called “miRtrons” can circumvent the Drosha pathway and are made by splicing and debranching of short hairpin introns [44–46]. The product of Drosha cleavage is a 70–100 nucleotide hairpin-shaped precursor, the pre-miRNA, that is transported to the cytoplasm via an Exportin-5- and Ran-GTP-dependent mechanism [47]. This pre-miRNA is further processed to produce a ~22 nt long miRNA duplex (miRNA:miRNA*) by another RNaseIII enzyme, Dicer [48–50], or alternatively through cleavage by Ago2, an Argonaute protein that is part of the complex that aligns the miRNA and messenger RNA [51–53]. The miRNA duplex produced via either Dicer or Ago2 processing is then unwound into the mature single-stranded form (guide strand) and its complementary strand (passenger strand or miRNA*). This single stranded RNA associates with members of the Ago protein family within the effector ribonucleoprotein complex RISC (RNA-induced silencing complex) [43, 54–56]. As part of the RISC, the mature miRNA guides the complex to its RNA target by Watson-Crick base-pairing interactions regulating protein synthesis [21]. In animals, most miRNAs appear to pair imperfectly within the 3′ UTR of their mRNA targets, and repress translation and/or decay of the mRNA [57]. The mechanistic details of protein synthesis inhibition by miRNAs are not well understood, but may include sequestration from ribosomes by relocation into P bodies, blockage of translational initiation, translational repression after initiation or target deadenylation coupled to transcript degradation [57, 58].
Figure 2.
miRNA biogenesis and function. miRNAs are transcribed in the nucleus into primary transcripts (pri-miRNAs). They are transcribed from independent miRNA genes, from polycistronic transcripts or from introns of protein-coding genes. Pri-miRNAs are then processed in two steps in the nucleus and cytoplasm, catalyzed by the RNase III type endonuclease Drosha and Dicer, respectively. These enzymes function in complexes with dsRNA-binding domains proteins, DGCR8 and TRBP for Drosha and Dicer, respectively. In the canonical pathway (illustrated here) Drosha-DGCR8 processes the transcript to a stem-loop-structured precursor (pre-miRNA). A subset of miRNAs, called miRtrons, also derived from introns, is processed into pre-miRNAs by the spliceosome and the debranching enzyme. Both canonical miRNAs and miRtrons are exported to the cytoplasm via Exportin 5, where they are further processed by Dicer-TRBP to yield ≈ 20-bp miRNA duplexes. The typical Dicer cleavage product features 5′ phosphate groups and two-nucleotide overhangs at the 3′ ends. One strand is selected to function as mature miRNA and loaded into the RISC, while the partner miRNA* strand is preferentially degraded. In contrast, the precursor of miR-451 is recognized directly by Ago2. The unusual structure of the precursor (short stem, miRNA sequence spans the loop) promotes binding and cleavage by Ago2 after the 30th nucleotide. Therefore, miR-451 is produced independently of Dicer. The miRNA is further matured by so far unknown mechanisms. The mature miRNA produced by these two mechanisms leads to translational repression or mRNA degradation. The key components of the RISC are components of the Argonaute family (Ago 1–4). Animal miRNAs usually show only partial complementarity to the target mRNA promoting translational repression (initiation and post initiation steps) or deadenylation coupled to exonucleolytic degradation of target mRNA. mRNAs repressed by deadenylation or at the translation-initiation step are moved to P-bodies for either degradation or storage.
Identifying functionally important miRNA target genes is crucial for understanding the impact of specific miRNAs on cellular function. This has been challenging, as miRNAs usually have imperfect complementarity with their targets [21]. In mammals, the most consistent requirement of miRNA:target interaction, although not always essential, is a contiguous and perfect base pairing of the miRNA nt 2–8, representing the “seed” region [59]. In many cases, the seed seems to determine this recognition; in other cases, additional determinants are required, such as reasonable complementarity to the miRNA 3′ half to stabilize the interaction (nt 13–16). In addition, some other features of the 3′ UTR sequences surrounding the target site, such as AU-rich sequences, positioning within the 3′ UTR at least 50 nt from the stop codon and away from the center of long UTRs, may boost miRNA efficacy [21]. Interestingly, another class of miRNA target sites has recently been described. The new “centered sites” lack both perfect seed pairing and 3′-compensatory pairing and instead have 11–12 contiguous Watson-Crick pairs to the center of the miRNA [60]. The existence of multiple public miRNA target prediction algorithms has greatly facilitated the rapid identification of miRNA target genes; however, these require validation.
miRNAs and lipid metabolism
The first microRNA that regulates lipid metabolism, miR-122, was identified as the most highly expressed miRNA in the adult liver, where it accounts for 70% of all miRNAs [61]. Several studies of miR-122 suggest its importance in establishing gene expression patterns critical to the differentiated state of the liver. Notably, inhibition of this miRNA in vivo has pronounced effects on cholesterol and fatty acid metabolism. Using antisense strategies, several groups have reported that antagonism of miR-122 in the liver results in sustained decreases in plasma cholesterol levels in both mice and non-human primates [62–66]. Mice treated with antisense oligonucleotides (ASO) to miR-122 showed 25–35% reductions in total cholesterol, and this reflected decreases in both the LDL and HDL lipoprotein fractions [63, 65]. In African green monkeys, efficient silencing of miR-122 in the liver was achieved by only 3 doses of anti-miR-122 and caused sustained decreases in total plasma cholesterol without any apparent liver toxicity or histopathological changes [62]. Furthermore, prolonged antagonism of miR-122 in chimpanzees, achieved by weekly injections of anti-miR-122 for 12 weeks, also reduced plasma cholesterol by 30%; however, unlike in mice, these changes were predominantly observed in the LDL fraction [66]. Interestingly, the reduction in cholesterol levels persisted for several weeks after the end of treatment suggesting that miR-122 ASO treatment has prolonged effects on hepatic gene expression and cholesterol metabolism.
Microarray analyses of the livers of miR-122 ASO treated mice showed upregulation in the mRNAs of up to 400 genes, of which approximately 70% had at least one 6-nt seed match for miR-122 in their 3′UTR [62, 63, 65]. While these analyses revealed that miR-122 actively represses a large number of mRNAs in the liver, the gene pathways regulating cholesterol biosynthesis were not foremost among those, and it remains unclear which direct gene targets of miR-122 cause the changes in circulating cholesterol. However, the gene expression profiling approaches used are limited in that they only identify miRNA effects on mRNA stability. As a large majority of miRNAs regulate their targets by translational inhibition, such changes would not have been detected using this method. Despite this caveat, several genes of interest were noted among the mRNAs altered by anti-miR-122, including FASN, ACC1 and ACC2, which are involved in fatty-acid synthesis and oxidation (Table 1). Hepatocytes from anti-miR-122 treated mice showed reduced fatty acid synthesis and increased fatty acid oxidation rates, and accordingly, antagonism of miR-122 in a mouse model of diet-induced obesity significantly improved hepatic steatosis and reduced levels of triglyceride accumulation [63].
Table 1.
Cholesterol and lipid-related genes regulated by miR-33, miR-122 and miR-370
| miRNA | Putative Target gene | Biological Function | Tissue/cell type | Ref |
|---|---|---|---|---|
| miR-33 | ABCA1 | Cholesterol efflux, HDL biogenesis | Human hepatoma cell lines (HepG2, Huh7, Hep3B), mouse liver, mouse hepatic cell line (Hepa), J774, human endothelial cell lline (EAhy926), Primary mouse and human macrophages, Hepa, THP-1, Hep3B | [67] |
| [68] | ||||
| [69] | ||||
| ABCG1 | Cholesterol efflux | |||
| NPC1 | Intracellular cholesterol transport | |||
| miR-122 | Aldoa | Primary mouse hepatocytes | [60] | |
| Gysl | [61] | |||
| Ccngl | ||||
| P4hal | ||||
| Ndrg3 | ||||
| Tmed3 | ||||
| Hfe2 | ||||
| Slc35a4 | ||||
| miR-370 | CPT1a | Fatty acid oxidation | Human hepatoma cell line (HepG2) | [66] |
The lack of toxicity in primate studies suggests that miR-122 inhibition in humans may be feasible, and this is being actively investigated for the treatment of chronic hepatitis C virus infection [67, 68]. Interestingly, the 5′ noncoding region of the HCV genome contains two highly conserved miR-122 binding sites that are essential for accumulation of viral RNA in hepatic cells. Notably, antagonism of miR-122 in chronically HCV-infected chimpanzees markedly suppresses viremia, suggesting that it may be an effective therapeutic [66]. In addition to reducing viral replication, antagonism of miR-122 likely also reduces HCV propagation by decreasing the expression of enzymes involved in lipid metabolism, which can enhance HCV replication in cell culture models. The continued study of miR-122 in this disease model is likely to yield important insights, not only into the potential of miR-122 antagonists as anti-HCV therapies, but also into its effects on genes involved in hepatic lipid metabolism.
A second microRNA, miR-370, was recently identified to have similar effects on lipid metabolism [68]. However, whereas miR-122 promotes changes in lipogenesis directly, miR-370 was shown to act primarily by modifying the expression of miR-122 [68] Transfection of the human hepatic cell line HEPG2 with miR-370 upregulates expression of miR-122 and conversely, miR-370 inhibition decreases miR-122 levels. Like miR-122, transfection of hepatic cells with miR-370 upregulates the expression of lipogenic genes, including SREBP1c and DGAT2. In addition, miR-370 directly regulates the expression of carnitine palmitoyl transferase 1a (Cpt1a), an enzyme that promotes fatty acid β-oxidation. These findings highlight the complex regulatory role that microRNAs are likely to play in modulating lipid metabolism, through both direct and indirect effects on target gene expression.
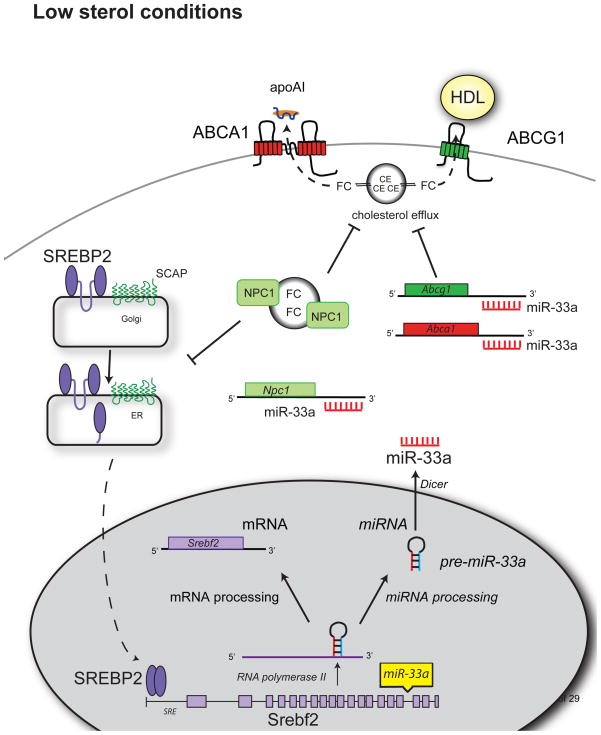
Most recently, three independent studies demonstrated that miR-33a, an intronic microRNA located within the SREBF-2 gene, plays important roles in the homeostatic regulation of cholesterol metabolism [69–71]. These reports showed that miR-33a targets genes involved in cholesterol trafficking, including ABCA1, ABCG1 and NPC1. Interestingly, each group uncovered the role of miR-33a in regulating cholesterol homeostasis by independent means. In our report, miR-33a was identified through genome-wide profiling of miRNAs whose expression was altered by cellular cholesterol content [69]. We found that during states of cholesterol depletion, miR-33a expression rises, and conversely miR-33a levels decrease during states of cholesterol enrichment in both macrophages and hepatocytes. Notably, miR-33a was also regulated in the livers of mice when dietary cholesterol was altered. Marquart et al and Najafi-Shoushtari et al each identified miR-33a using in silico bioinformatic screening and noted its intriguing presence within intron 16 of SREBF-2 (Figure 3) [70, 71]. The sequence and genomic location of miR-33a is highly conserved from Drosophila to humans, and its intronic location within SREBF-2 suggests that these two transcripts may be co-regulated. Indeed, we found that miR-33a and SREBF-2 are co-transcribed during states of cholesterol depletion [69], and expression levels of miR-33a and SREBF-2 were also comparable across a variety of tissues [71].
Figure 3.
miR-33 Reduces Cholesterol Transport And Efflux Under Low Sterol Conditions. When sterol conditions are low, SREBP-2 is cleaved in the Golgi and translocates to the nucleus, where it acts as a transcription factor to regulate genes containing a sterol response element (SRE), including SREBP-2 itself. miR-33a is located within intron 16 of SREBP-2, and is co-transcribed along with its host transcript. Once processed by the miRNA system, mature miR-33a can bind the 3′UTR of its target genes, such as ABCA1, ABCG1 and NPC1 repressing their expression. This results in reduced cholesterol efflux to apoA1 and HDL.
Interestingly, target prediction algorithms place ABCA1 at the top of the list for predicted target genes of miR-33a, and all three studies demonstrate that miR-33a strongly represses the expression of this cholesterol transporter under a variety of conditions. The 3′UTR of ABCA1 contains three highly conserved consensus-binding sites for miR-33a, and mutational analysis of these sites showed that they cumulatively contribute to miR-33-dependent repression of ABCA1 [69]. As a consequence of this targeting, miR-33a expression limits the efflux of cholesterol to apoA1, the building block of HDL, in both macrophages and hepatocytes. Conversely, antagonism of endogenous miR-33 using anti-sense oligonucleotides upregulates ABCA1 expression both in vitro and in vivo, and promotes the efflux of cholesterol to apoA1, the first step in reverse cholesterol transport to the liver (Figure 3). Interestingly, a second sterol transporter, ABCG1, which promotes the efflux of cholesterol to pre-lipidated apoA1 (HDL), is also targeted by miR-33a [69]. Despite the presence of a miR-33a binding site in the 3′UTR of human ABCG1, the targeting of ABCG1 by miR-33a only appears to be of significant consequence in mouse cells [69, 70]. In mouse macrophages and hepatocytes, expression of miR-33 reduces ABCG1 protein and decreases cholesterol efflux to HDL [69]. These species-specific differences in the post-transcriptional regulation of ABCG1 may have contributed to previous discordant findings of ABCG1 regulation in mouse and human cells. Together, these findings establish that during sterol-limited states, miR-33a is coincidentally generated with SREBF-2 transcription and works to increase cellular cholesterol levels by limiting cholesterol export through the downregulation of ABCA1 and ABCG1 (Figure 3).
It has been known for some time that SREBP2 activation downregulates the expression of ABCA1, an LXR target gene. However, until now, the exact mechanism of this downregulation remained incompletely understood. The discovery that miR-33a is expressed when SREBP-2 is induced, and limits sterol export by targeting ABCA1, provides the missing link in our understanding of this pathway and suggests that the SREBP and the LXR axes of cholesterol control are elegantly connected.
Our study also identified the lysosomal cholesterol transport protein NPC1 as a target of miR-33a [69]. Interestingly, the human NPC1 gene is strongly repressed by miR-33a owing to the presence of two miR-33a binding sites, whereas mouse NPC1 contains only one miR-33a binding site resulting in a lower degree of targeting in this species. While it may appear paradoxical that during times of cholesterol depletion, miR-33a limits intracellular cholesterol mobilization by blocking NPC1 expression, NPC1 is known to work in concert with ABCA1 to promote cholesterol efflux and to play an important role in the lysosomal export of cholesterol to the mitochondria, where CYP27 action generates 27-hydroxycholesterol, a potent agonist of LXR [72]. Another possibility is that miR-33a inhibits NPC1 expression in an effort to reduce cholesterol transport to the ER, providing a feedback loop to increase SREBP-2 activation and increase cholesterol synthesis. Thus by reducing NPC1 expression, miR-33a likely limits the availability of LXR ligands and activation of LXR target genes, including ABCA1, as well as prevents cholesterol transport to the ER, until the cholesterol needs of the cell are met.
Manipulating the expression of miR-33a in vivo, whether through locked-nucleic acids or viral delivery of sense or antisense oligonucleotides, significantly altered HDL cholesterol levels in mice [69–71]. Our study and that of Marquart et al., showed that delivery of miR-33 in vivo decreases hepatic ABCA1 expression and reduces circulating HDL [69–70]. Conversely, anti-sense inhibition of miR-33 increased HDL levels [69–71]. Notably, hepatic gene expression profiling showed that, other than the specific upregulation of miR-33a target genes ABCA1 and ABCG1, inhibition of miR-33a results in few detectable changes in lipid metabolism-related genes [69]. Similar results were seen in macrophages treated with anti-miR-33a, showing that anti-miR-33a specifically relieves the repression of miR-33a target genes [69]. Interestingly, another member of the miR-33 family, miR-33b, is found within intron 17 of human SREBP-1c [71]. Unlike miR-33a however, the sequence of miR-33b is only present in humans and is not conserved in mice or other small mammals. Moreover, the mature sequences of miR-33a and miR-33b differ by only 2 nucleotides, resulting in both miR-33a and miR-33b having overlapping gene targets, including ABCA1. Therefore under conditions in which SREBP-1 is activated to promote fatty acid synthesis (for example, by LXR or insulin), miR-33b may coordinately downregulate ABCA1 expression and cholesterol efflux, and hence reduce HDL biogenesis. This may be particularly relevant in the setting of metabolic syndrome, which is characterized by increased triglycerides, and plasma levels of VLDL, and reduced plasma HDL. Although it has yet to be investigated, miR-33b may contribute to these pathological alterations in lipid metabolism in insulin resistant states.
Summary and concluding remarks
Despite the promise of these early studies, our understanding of the role of miRNAs in regulating lipid metabolism is still quite limited. The liver, a major site of lipid metabolism, is the organ most amenable to the delivery of oligonucleotides following systemic injection, thus facilitating future studies of miRNA regulation of lipid metabolism in vivo. Clinical translation of these findings for the use of microRNA therapeutics in the treatment of hyperlipidemia and atherosclerosis is likely to be in the distant future (Box 1). However, the limited treatment duration of anti-miR-122 in HCV infection and the positive safety profile in preclinical studies suggest that this type of therapy may be established sooner for HCV patients at high risk for developing HCV-related morbidity and mortality. Indeed phase I clinical trials are currently underway to investigate the use of anti-miR-122 in the treatment of chronic HCV infection.
Clinical Implications
miR-33a represses expression of genes involved in cholesterol transport
ABCA1: transporter that mediates cholesterol efflux to lipid-poor apoA1 (nascent HDL) during HDL biogenesis and reverse cholesterol transport
ABCG1: transporter that mediates cholesterol efflux to HDL
NPC1: lysosomal cholesterol transport protein
Inhibitors of miR-33a alter cholesterol metabolism
-
Increase cellular cholesterol efflux to apoA1 in both:
hepatocyte - site of HDL biogenesis
macrophage – a key player in atherosclerotic plaque
Raise circulating HDL by 25–50%
HDL raising ability similar in magnitude to niacin, currently the most widely used HDL targeted therapeutic
Future directions to development of miR-33 antagonists
Testing the safety and pharmacokinetic profile of miR-33 anti-sense oligonucleotides (ASOs) in animal models, including non-human primates
Establishing that miR-33 ASOs have a disease modifying effect in animal models cholesterol-associated diseases such as atherosclerosis
Acknowledgments
The Moore Lab is supported by the National Institute of Health (R01AG02055), the Suárez lab is supported by grant from American Heart Association (SDG-0835481N) and the Fernández-Hernando lab is supported by grants from American Heart Association (SDG-0835585D) and National Institute of Health (1P30HL101270-01). We thank Dr. Edward A. Fisher for providing critical feedback. We apologize to those whose work could not be cited owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 2.Bloch K. Summing up. Annu Rev Biochem. 1987;56:1–19. doi: 10.1146/annurev.bi.56.070187.000245. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. Receptor-mediated control of cholesterol metabolism. Science. 1976;191:150–154. doi: 10.1126/science.174194. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 5.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 7.Horton JD, et al. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton JD, Shimomura I. Sterol regulatory element-binding proteins: activators of cholesterol and fatty acid biosynthesis. Curr Opin Lipidol. 1999;10:143–150. doi: 10.1097/00041433-199904000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 10.Peet DJ, et al. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 11.Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 12.Tall AR, et al. Regulation and mechanisms of macrophage cholesterol efflux. J Clin Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tall AR, et al. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Yu L, et al. Disruption of Abcg5 and Abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proc Natl Acad Sci U S A. 2002;99:16237–16242. doi: 10.1073/pnas.252582399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks-Wilson A, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 16.Frikke-Schmidt R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis. 2010;208:305–316. doi: 10.1016/j.atherosclerosis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Oram JF, Vaughan AM. ATP-Binding cassette cholesterol transporters and cardiovascular disease. Circ Res. 2006;99:1031–1043. doi: 10.1161/01.RES.0000250171.54048.5c. [DOI] [PubMed] [Google Scholar]
- 18.Rust S, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 23.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 24.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 25.Berezikov E, et al. Approaches to microRNA discovery. Nat Genet. 2006;38(Suppl):S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 26.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forman JJ, Coller HA. The code within the code: MicroRNAs target coding regions. Cell Cycle. 2010;9 doi: 10.4161/cc.9.8.11202. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lytle JR, et al. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orom UA, et al. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 32.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 35.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 36.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim VN, et al. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 42.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 44.Kim YK, Kim VN. Processing of intronic microRNAs. Embo J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamura K, et al. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruby JG, et al. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund E, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 48.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y, et al. The role of PACT in the RNA silencing pathway. Embo J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheloufi S, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasmussen KD, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory RI, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 55.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Pillai RS, et al. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 59.Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Shin C, et al. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 62.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 63.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 66.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Branch AD, Rice CM. Antisense gets a grip on miR-122 in chimpanzees. Sci Transl Med. 2010;2:13ps11. doi: 10.1126/scitranslmed.3000605. [DOI] [PubMed] [Google Scholar]
- 68.Iliopoulos D, et al. MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marquart TJ, et al. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science. 2010 doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu X, et al. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J Biol Chem. 2001;276:38378–38387. doi: 10.1074/jbc.M105805200. [DOI] [PubMed] [Google Scholar]