Abstract
Much of the experimental data used to construct mathematical models of molecular networks are derived from in vitro measurements. However, there is increasing evidence that in vitro measurements fail to capture both the complexity and the individuality found in single, living cells. These limitations can be overcome by live cell microscopy which is evolving to enable in vivo biochemistry. Here we survey the current capabilities of live cell microscopy and illustrate how a number of different imaging approaches could be applied to analyze a specific molecular network. We argue that incorporation of such quantitative live cell imaging methods is critical for the progress of systems biology.
Keywords: FRAP, Mathematical models, Optical imaging, Molecular interaction networks, FRET
Introduction
Quantitative modeling of molecular networks has attracted increasing interest over the past decade. This reflects a natural evolution in biomedical research. Most previous work has focused on identifying the key molecules comprising a regulatory network and the interactions of these molecules with each other. Now, with the sequencing of many genomes and the accumulation of biochemical data for many pathways, the focus of attention is shifting to the question of how the molecular network operates as a system to perform its biological function.
Most network models have been generated from in vitro data. This has recently changed as several modeling studies have been fueled instead by quantitative in vivo data generated by live cell microscopy [1–4]. Even in these studies, however, only a limited range of the available live-cell imaging techniques have been used. Thus, imaging approaches are currently a niche methodology in systems biology research.
Here we describe how live cell imaging methods might be more broadly harnessed to extract crucial information on molecular interaction networks. We use a sample molecular network for NF-κB to specifically illustrate how critical molecular processes can be probed by microscopy. We conclude with a discussion of current challenges and future prospects for live cell imaging applications that are relevant for systems cell biology.
Network models and experimental data
Over the past decade, a growing number of laboratories have developed mathematical and dynamical models of molecular networks [5, 6]. These run the gamut from bacterial chemotaxis pathways [7] to cell cycle networks in yeasts [8–10] to an assortment of higher eukaryotic networks. Among the latter are various conserved developmental pathways that have been studied quantitatively in model organisms such as Drosophila, zebrafish, and Xenopus [11–13]. Mammalian molecular systems have also been modeled including signal transduction pathways such as ERK, EGFR, JAK-STAT, MAPK [14–17] and gene regulatory networks such as p53, NF-κB, Rb/E2F, Hes [18–21]. Virtually all of these networks regulate key cellular processes critical for development, stress response, or disease.
A common feature of these network models is that they rely on experimental data for both their construction and validation. The data play a formative role in compiling the list of molecules in a network and identifying connections between the molecules. Once the molecular network is defined, experimental data are also used to provide estimates for the model's free parameters. These include the strengths of interactions between molecules, typically given by chemical rate constants, and the initial concentrations for each of the molecules in the network. After these parameter estimates and initial concentrations are obtained, the model can be used to predict how the concentrations of each molecule change over time. Moreover, more comprehensive analyses of the model can reveal systems properties and key reaction steps that control the network behavior. Some of these predictions can be tested by acquiring new experimental data.
Limitations of population averages in most current experimental data
Most of the experimental data that have been used to date to either develop or test network models have been averaged measurements from a large population of cells. This approach has some serious limitations [22], as demonstrated by a growing body of recent work.
It is now well established that individual cells, even from a genetic clone, display a remarkable degree of heterogeneity. This variability is typically not eliminated when cells are synchronized in some way (for example by cell cycle or hormone induction). Rather the variability arises from other intrinsic or extrinsic sources [23, 24]. This variability is invisible to population average measurements.
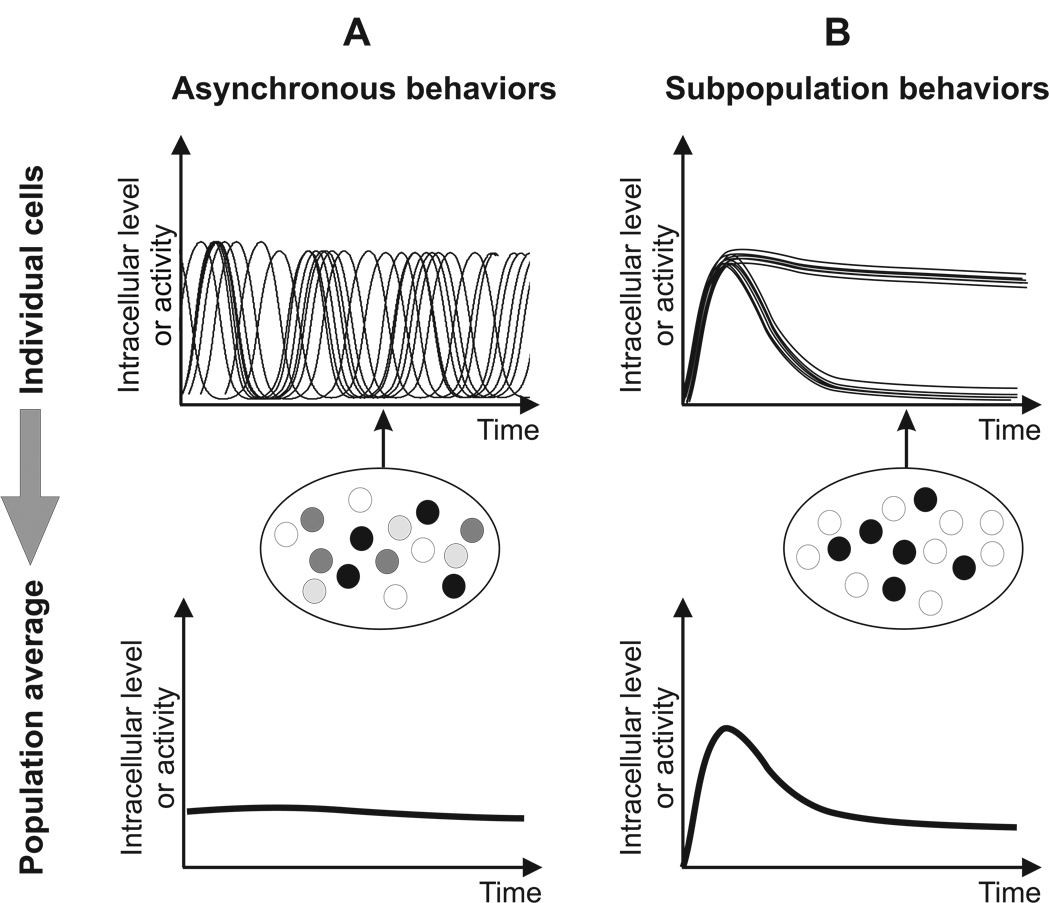
The consequences for systems models can be profound. The most striking examples come from studies in which oscillations in a molecular concentration have been analyzed (Fig. 1A). For example, in the NF-κB signaling pathway, it is now known that there are multiple periodic accumulations of NF-κB in the nucleus, and that the oscillation phase within individual cells becomes asynchronous shortly after activation by TNF-α [2, 25]. When averaged across the population of cells, these long-term asynchronous oscillations yield instead a strikingly damped biphasic profile that artifactually reaches a steady-state after only one cycle [21]. A similar discrepancy between single cell behavior and the population average has been uncovered for oscillations in ERK [14, 26] and p53 [1, 27].
Figure 1. Cellular behaviors that are not captured by population average measurements.
A. Individual cells may have dynamic fluctuations that are out of phase (upper panel). This variability disappears in a population average (lower panel). Snapshot measurements in single cells (depicted as grayscale circles in the middle panel) will also yield a misleading result by displaying a range of measured levels at a single time point (arrow). This could be incorrectly interpreted as variability in steady state levels within cells. B. Alternatively, the population may be composed of two different subsets of cells (upper panel). This heterogeneity will be smoothed out in a population average (lower panel), but in this case a snapshot measurement will detect the two populations (middle panel).
While oscillations are a particularly striking example of behavior masked by population assays, it is straightforward to see how other temporal fluctuations in molecular concentrations might also be missed. In general, temporal fluctuations of any kind may either be undetectable or grossly misrepresented by population averages. If such temporal fluctuations carry signaling information (such as affinity or concentration of the extracellular ligand) that can shape the response outcome (such as differentiation versus proliferation), then accurate measurements from single cells becomes essential.
Another flaw in population averages is that they fail to detect the presence of two or more subpopulations of cells exhibiting qualitatively different behaviors [4, 28]. For example, only a subpopulation of B. subtilis cells exhibits a transient competent state in nutrient-deprived conditions [4]. Similarly, only a subpopulation of fibroblast cells exhibits a sustained inflammatory response due to the probabilistic nature of autocrine/paracrine feedback [28]. Such dichotomies will not be detected in population averages (Fig. 1B).
In sum, under many biological conditions, population averages can give rise to inaccurate estimates for both the key variables and parameters in a network model. Clearly, the underlying mechanisms of any molecular network model are occurring at the single cell level, and thus measurement of the relevant system variables within single cells should be the goal.
Single cell kinetic measurements by live cell microscopy
The limitations of population average assays are naturally overcome by live cell microscopy, an approach that is now widely exploited in many cell biological applications. A microscope is an ideal system to collect data from a single cell, or from many cells in parallel, thereby providing direct information about cell heterogeneity. Recent advances in fluorescence light microscopy have made it possible to measure a number of cellular properties that are relevant for systems modeling [29–31]. These include the concentration levels of proteins [32], mRNAs [33], or small molecule second messengers [34, 35], the existence of protein-protein interactions [36, 37], the presence of protein modifications [38, 39], and the in vivo binding rates of proteins to immobile substrates [40].
In addition to overcoming the limitations of population averaging, these live cell approaches have a number of other advantages. Most approaches are quantitative [41], lending themselves quite naturally to computational or mathematical modeling.
A unique advantage of live cell approaches is that real-time measurements can be made at the temporal frequency necessary to adequately sample the dynamics of most biological processes. Such real-time, single-cell measurements eliminate artifacts that can arise from attempting to reconstruct time courses from snapshots of different cells taken at different times. Snapshot measurements cannot distinguish temporal from cell-to-cell variability.
Finally, a key advantage of live cell imaging is that by definition the living cell contains all of the complexity of real life, whereas in vitro studies inevitably simplify cellular complexity by extracting selected molecules in order to make measurements. For example, in vitro binding measurements often include only the two isolated molecules dissolved in buffer. Even in more complex whole cell extracts, some components may be lost or degraded during the isolation procedure. Furthermore, in all of these cases, the final test tube solution lacks the structural components and the spatial heterogeneity of a normal cell. These various deficiencies of in vitro assays can have serious consequences as demonstrated by a number of recent live cell studies, which have revealed that behaviors in the living cell can be very different than those inferred from the test tube. For example, the in vivo binding rates of transcription factors to chromatin have been found to be four orders of magnitude faster than the in vitro measurements [45].
Limited applications of live cell microscopy in current network modeling
Due to its many advantages, there has been increasing use of live cell imaging to gather data for modeling. Several studies have used FRAP analysis to quantify the assembly of multi-protein complexes involved in transcription or DNA repair [46, 47]. Some complex signaling networks in lower eukaryotes and bacteria have been analyzed by time lapse imaging [48]. Elowitz and coworkers used this approach to show that differentiation of B. subtilis is probabilistic and triggered by noise in the molecular network [4]. Time lapse analysis of a DNA repair pathway in E. coli revealed rather precise timing of oscillatory peaks, despite the fact that the number of peaks and their amplitudes vary in individual cells [3]. Fluorescent reporters have also been used to monitor the dynamics of an oscillating synthetic gene network in E. coli [49]. These latter studies on complex networks have demonstrated how time lapse imaging can be used to dissect the true behaviors of real and synthetic molecular networks that are otherwise undetectable in population assays.
Despite the progress represented by these applications, integration of imaging methods into systems biology research has remained a rarity. Two recent reviews have recognized some of the potential of applying these approaches to network modeling [22, 50], but a comprehensive discussion of what live cell imaging can accomplish and how it might be explicitly applied in systems biology is currently unavailable. In the present review, we provide an introduction into the capabilities of live cell imaging relevant for systems biology, and also detail concrete examples of how imaging methods might be profitably applied to analyze a specific molecular network, namely NF-κB.
A live cell microscopy primer
Fluorescent fusion proteins and reporters
Most experiments in live-cell imaging today are performed using fluorescent fusion proteins [51–53]. There is an ever-expanding list of fluorescent proteins, including GFP (green) from the jellyfish and its many derivatives such as CFP (cyan) and YFP (yellow). In recent years, the palette has further diversified with the addition of proteins such as mCherry (red) derived from other species. Photo-activatable (from dark to fluorescent) and photo-convertible proteins (from green to red and back again) have also been developed [54]. The standard strategy to create a fluorescent reporter protein is to fuse the gene for one of the fluorescent proteins to either the N- or C-terminus of the protein of interest. In many cases, fusion proteins already exist. In fact genome-scale libraries of fluorescently tagged strains have been generated for yeast, bacteria, and human cells [42, 55, 56], although in some cases functionality of the fusion proteins may still need testing.
Ideally, the fluorescently tagged version of the protein replaces the endogenous protein, while retaining the endogenous promoter. This is best accomplished by gene replacement, which preserves the chromatin context of the gene. Alternatively, the chimeric gene driven by its normal promoter could be transfected into a null background. In either of these cases, absolute concentration levels of the fluorescently tagged protein should faithfully reflect endogenous levels. In some cases, however, it is sufficient to measure relative concentration levels, for example when comparing protein levels between different cellular compartments. In this case, the fusion protein can be introduced in a wild type background under a constitutive promoter, creating additional copies of the protein of interest.
When regulated gene expression must be monitored, luminescent reporters can be useful to achieve sensitive detection of low level expression [2]. For example, a promoter of interest can be placed upstream of a luciferase gene to drive its expression. However, the temporal resolution of time lapse luminescence imaging may be limited due to the necessary steps of substrate addition and photon counting.
Time-lapse
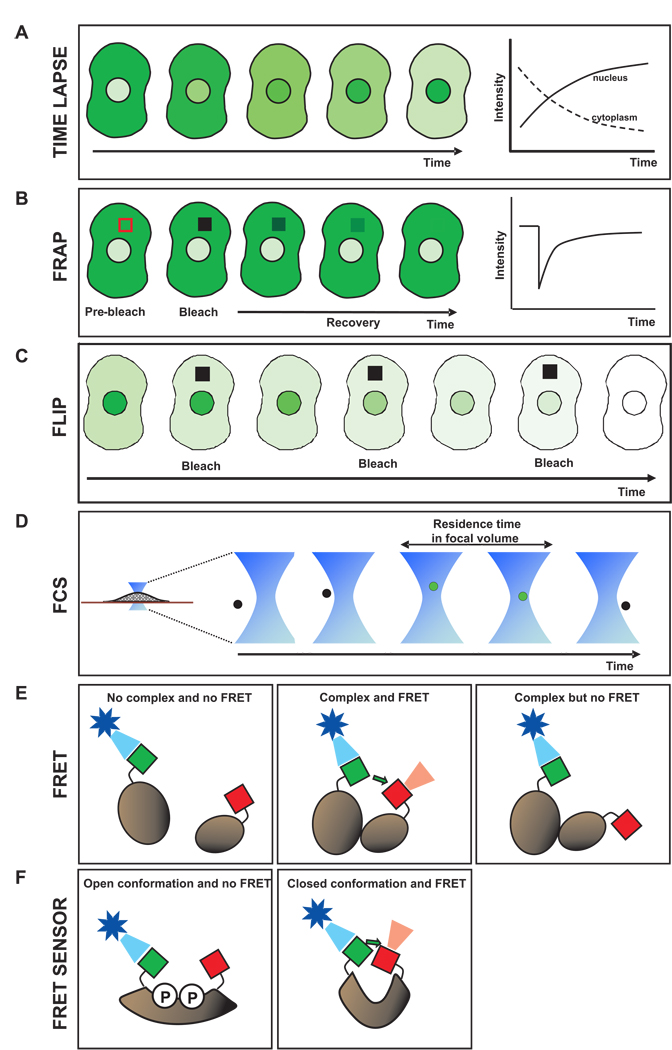
Time-lapse microscopy is used to assay how protein levels change over time within different cellular compartments [57]. For example, the protein of interest is tagged with GFP, and then fluorescence intensity is measured within single cells at a series of time points (Fig. 2A). If the protein concentration in a compartment changes due to turnover or flux into another compartment, it can be detected by quantifying the fluorescent intensity as a function of time.
Figure 2. Quantitative live cell imaging techniques.
A. Time lapse microscopy. Fluorescence intensity in distinct cellular compartments of the same cell can be monitored in real time. B. Fluorescence recovery after photobleaching. A region of interest, shown by the red box, is photobleached with a strong laser pulse and the recovery of intensity within the region is monitored in time. Slower recovery arises if the GFP fusion protein is bound longer to an immobile structure and/or diffusing at a slower rate. C. Fluorescence loss in photobleaching. Repetitive bleaching within a region of interest in one cellular compartment (cytoplasm in this example) is alternated with measurement of intensity in another cellular compartment (nucleus in this example). Fluorescence decays in the second compartment if there is a steady-state exchange between the two compartments. Faster loss in fluorescence generally corresponds to faster exchange. D. Fluorescence correlation spectroscopy. Photon counting in a diffraction-limited spot enables estimates of molecular residence times due to diffusion and/or binding. E. Fluorescence resonance energy transfer. Formation of a complex can be detected when donor (green) and acceptor (red) tags are brought in close proximity (second panel). Improper positioning of the tags can lead to no FRET even when a complex forms (third panel). F. Biosensors based on FRET for detection of small molecules. Binding of the small molecule(s) to the FRET sensor leads to a change in conformation and a corresponding change in FRET. In this example, phosphorylation due to the activity of a kinase would lead to a loss of FRET.
This strategy can be repeated to tag another protein in a complementary color and thereby monitor two different protein levels simultaneously within the same cell. Temporal profiles of such a correlated pair of molecules are far more informative for modeling of system dynamics.
Fluorescence recovery after photobleaching (FRAP)
FRAP is used to measure the diffusion and binding rates of fluorescently tagged proteins in live cells, typically on time scales from seconds to minutes [40, 58]. A region of interest in a cell is irreversibly photobleached by exposing only the region to a short pulse of high intensity light (Fig. 2B). Fluorescent molecules in the photobleached region enter into a permanent dark state, thereby creating a selected region of the cell that lacks (or has significantly reduced) fluorescence.
The bleached zone will gradually become brighter if unbleached fluorescent molecules enter from outside. The movement of molecules into and out of the bleached region occurs by diffusion (and possibly also directed transport). Thus the rate at which fluorescence recovers in the bleached region is a measure of how fast molecules are exchanging at that site in the cell.
Of particular interest to systems modeling is that FRAP reveals quantitative information about in vivo binding interactions. If the fluorescently tagged protein is bound to a relatively immobile scaffold such as DNA or cytoskeletal filaments, then the bleached molecules will remain longer within the bleach spot. These FRAP data can be mathematically analyzed with reaction-diffusion models to obtain estimates for the association and dissociation rates of the molecule with an immobile substrate and the fraction of bound molecules [40]. Such information can be directly translated into systems modeling and simulations.
Fluorescence loss in photobleaching (FLIP)
FLIP can be used to detect protein shuttling between cellular compartments in live cells [59]. This may occur even though the apparent protein concentrations in the two compartments are not changing. For example, a protein localized predominantly in the nucleus could in fact be shuttling between the cytoplasm and nucleus if the nuclear import and export rates are not zero but balanced. Thus FLIP is useful because it can reveal intracellular transport that may be invisible to many other techniques. Identifying all the relevant shuttling processes not only informs model building, but it can also evaluate untested hypotheses about flow of molecules between any two cellular compartments [25].
FLIP reveals the steady state exchange of a protein between two separate compartments by repeatedly photobleaching a region in one compartment. Each round of photobleaching produces permanently inactivated (dark) fluorescent molecules that were present within the first compartment at that time. If there is no exchange with a second compartment, then the first compartment will become progressively darker while the second compartment will be unaffected. However if exchange is occurring between the compartments, then the second compartment will also become progressively darker over time (Fig. 2C).
Fluorescence correlation spectroscopy (FCS)
FCS is an alternate approach to measure diffusion and binding rates in live cells [60]. Compared to FRAP, it is better suited to faster processes such as transient binding events of less than 1 second in duration. The technique does not involve strong laser pulses or selecting the shape/size of a region for bleaching.
To perform FCS, the fluorescent intensity is measured within a diffraction-limited focal volume (smallest possible optical spot) at some intracellular location of interest (Fig. 2D). The intensity value of the spot is collected typically at microsecond or millisecond intervals, yielding a time series with fluctuations due to fluorescent molecules leaving and entering the volume. A slowly diffusing or tightly bound protein, having a longer residence time in the spot, will exhibit less frequent fluctuations than a rapidly diffusing or transiently bound protein. The autocorrelation function of the time series of intensity is then fit with reaction-diffusion models. This analysis provides estimates for kinetic parameters such as association and dissociation rates and the residence time of binding to any immobile scaffolds [61].
FCS can also be used to detect oligomerization of a protein. This could be useful, for example, to measure dimerization of a membrane receptor as a function of time in live cells. A dimer will yield fluctuating intensity that is on average twice as bright as for a monomer [62].
The formation of a complex between two different proteins can also be detected by performing a variant of FCS known as fluorescence cross correlation spectroscopy (FCCS) [60]. This method can resolve when and where two differentially labeled (e.g. red and green) molecules interact with each other. These protein:protein interactions are detected by FCCS because when a complex is formed, the GFP and RFP tags of the proteins in the complex will be present in the focal volume at the same time points. Thus the cross-correlation function between the green and red intensities reflects this relationship and can be used for model fitting similarly as for one color FCS.
Fluorescence resonance energy transfer (FRET)
FRET, like FCCS, is most often used in live cells to determine whether two proteins are binding to each other, or are at least close enough to be in the same complex [36, 37]. Specifically, occurrence of FRET between two fluorescently labeled proteins shows that they are within 5 nm of each other.
To perform FRET, the two proteins of interest are labeled with fluorescent tags that can interact, termed the donor and the acceptor. Good donor / acceptor pairs are CFP and YFP, or GFP and mCherry. When these fluorescent tags are close enough, their fluorescence properties change. The changes in fluorescence can be detected in live cells by light microscopy (Fig. 2E).
The simplest change in fluorescence is that excitation of the donor leads to an increase in fluorescence of the acceptor. Other changes can also be utilized to assay for FRET, such as an increase in donor fluorescence when the acceptor is photobleached [63]. Whatever diagnostic is used, it is important to recognize that a lack of FRET signal does not rule out protein – protein interactions, since FRET can fail for a variety of trivial reasons, such as placement of the donor and acceptor tags at locations which may not be in close enough proximity (Fig. 2E, third panel).
There is another technique that provides an alternative to FRET and FCCS, namely bimolecular fluorescence complementation (BiFC) [64]. Here the two proteins of interest are each labeled with fragments of a fluorescent protein which, upon complex formation, are joined to yield a signal-emitting fluorophore.
In sum, there are several methods (FRET, BiFC, and FCCS) for detecting protein:protein interactions in live cells that allow systems biologists to determine the spatiotemporal profiles for interactions of interest.
FRET sensors
FRET sensors use the principles of FRET to detect post-translational modification of proteins or the level of second messenger molecules. These small molecules are key players in many signaling contexts, where it would be valuable for systems modeling to quantify the messenger signaling and correlate it with other components of the pathway.
In all cases, the conformation of a FRET sensor is altered upon arrival of the target molecule(s), which induces a change in the FRET signal (Fig. 2F). Thus the change signifies the presence of the molecule and the magnitude of change reflects the concentration of the molecule. Second messenger sensors have been developed for Ca++, IP3, and cGMP [34, 65], and also for protein phosphorylation, glycosylation, ubiquitination, methylation, and acetylation [38, 39, 66]. The repertoire of these probes will expand and make many biochemical assays feasible in living cells.
Potential applications of live cell microscopy in network modeling
The canonical NF-κB network
We now consider a simple model system to illustrate how imaging approaches could provide important insights into the model parameter estimation and validation process (Fig. 3). NF-κB is a classical example of an inducible transcription factor regulated by various feedback loops [67]. It has also been the subject of several different network modeling studies [21, 25, 68–70].
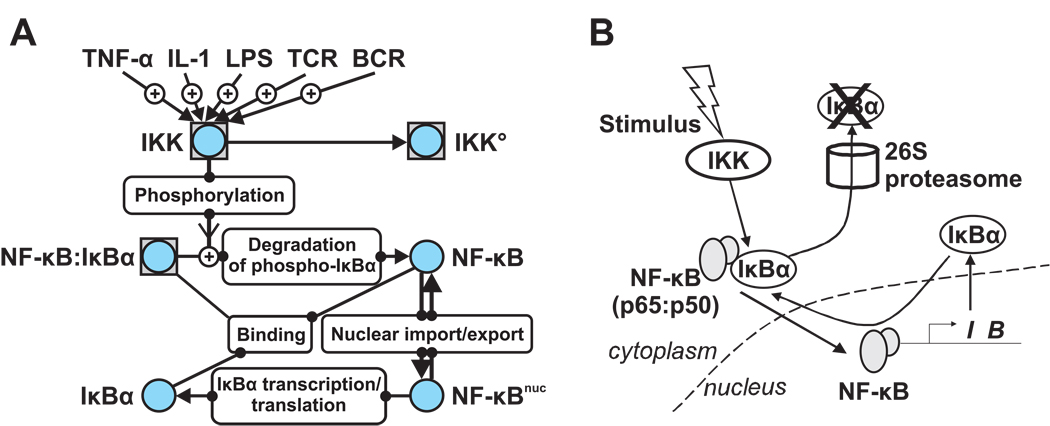
Figure 3. NF-κB as an example molecular network.
A simplified network for the canonical NF-κB pathway consisting of active IKK, IκBα, and NF-κB are represented in a navigational map (A) and in a schematic diagram (B). Many upsteam signals (factors like Tumor Necrosis Factor-α, Interleukin-1, and Lipopolysaccharide, or cell surface receptor signaling from T cell/B cell receptors) induce the kinase activity of IKK. Phosphorylation of two specific serines in IκBα by IKK initiates the degradation of IκBα by the proteasome. Free NF-κB then translocates to the nucleus and activates IκBα gene transcription. Newly synthesized IκBα binds to NF-κB in the nucleus or in the cytoplasm. Numerous molecular mechanisms down-regulate the post-stimulus kinase activity of IKK. IKKO indicates an inactive form without catalytic activity. NF-κBnuc represents the nuclear species. NF-κB itself exists most predominantly as a heterodimer of p65 (transcriptionally active) and p50 subunits. Symbols were used in the navigational map (A) according to the WIRE guideline: Briefly, a circle indicates a protein molecule. Boxed circles are protein complexes. A ‘+’ sign in an arrow represents a positive (activating) effect.
NF-κB activity is controlled in part by the level of nuclear translocation. In resting cells, the predominant dimer p65:p50 exists mostly as a cytoplasmic complex bound to its inhibitor IκB proteins. Inhibition is released after an inflammatory signal stimulates the IκB kinase complex (IKK) to phosphorylate IκB proteins, which leads to their degradation by the proteasome. NF-κB is then released from cytoplasmic latency and translocates into the nucleus to activate numerous target genes, including several genes that encode feedback proteins [67]. One such target is the Nfkbia gene which codes for IκBα. The re-synthesis of IκBα provides strong negative feedback, causing the inactivation of NF-κB and its re-localization to the cytoplasm. Such a negative feedback loop can lead to oscillations. These have been detected experimentally by real time monitoring of the nuclear level of NF-κB [2, 25].
We have constructed a simplified NF-κB network that captures the essence of this pathway (Fig. 3) and now use this example to illustrate how live cell microscopy techniques might be applied to gather data about a molecular network from single cells.
Measuring protein concentration levels in the nucleus or cytoplasm
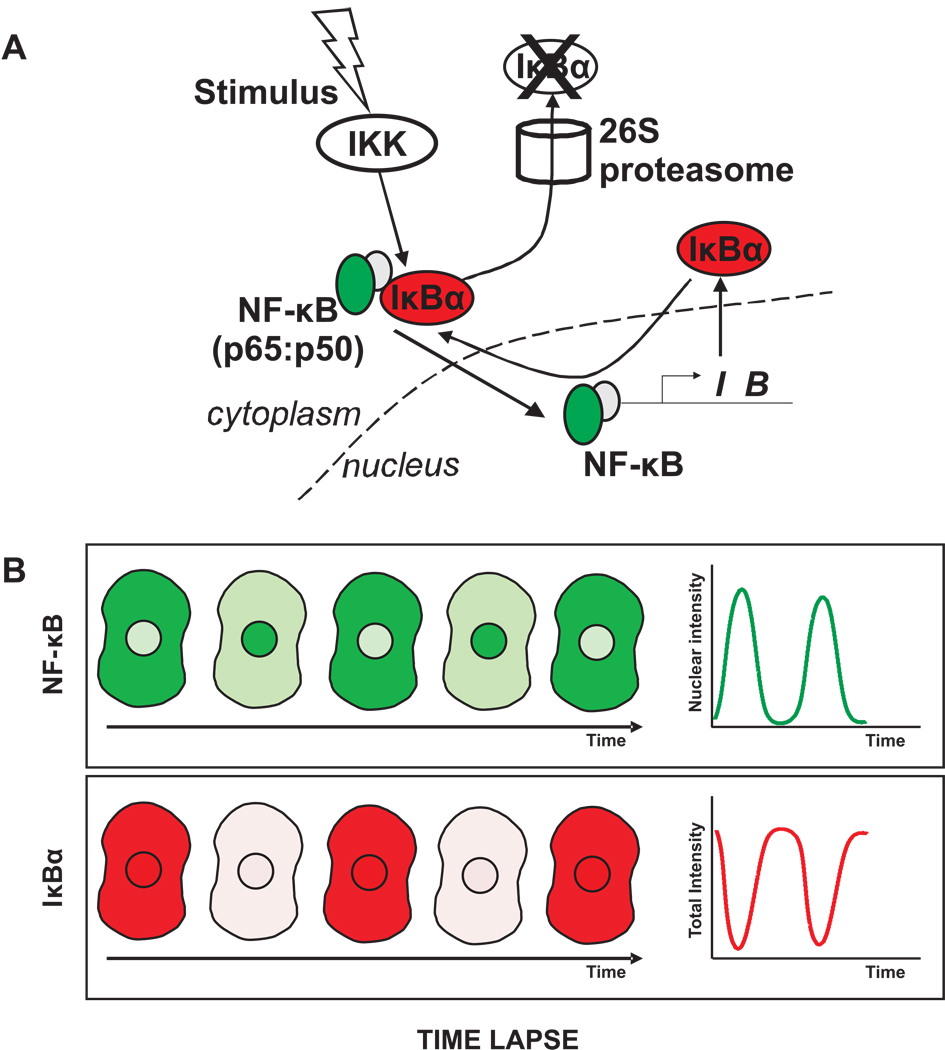
Time-lapse microscopy of a GFP-tagged NF-κB has already been used to measure its concentration levels in the nucleus and cytoplasm [25]. These experiments could be extended by measuring concentration levels of a second protein tagged with a complementary color. An attractive possibility would be to tag different IκB isoforms1 responsible for sequestering NF-κB predominantly in the cytoplasm. In a two-color time-lapse experiment, the NF-κB oscillation profiles could be correlated with the IκB isoform levels in each cell (Fig. 4). Given the range of NF-κB oscillation profiles observed in live cells, a double-label analysis would provide a rich dataset for dissecting the coupling between NF-κB and IκB levels.
Figure 4. Measuring protein levels by time lapse imaging.
Time lapse imaging could be used to follow changes in concentration and localization of NF-κB and IκBα within individual cells. A. NF-κB could be tagged with GFP (green) and IκBα with mCherry (red). B. Following stimulation of the pathway, fluorescence images in red and green could be acquired at a series of time points. Due to the IKK-induced degradation and re-synthesis of IκBα proteins, the levels of IκBα are expected to decrease and increase, while the nuclear level of NF-κB is expected to concomitantly increase and decrease. These changes could be quantified by measuring NF-κB fluorescence intensities in the nucleus and IκBα intensities in the whole cell.
Measuring nuclear / cytoplasmic exchange rates by FRAP or photo-convertible proteins
NF-κB, IκB proteins, and subunits of the IKK complex can all enter the nucleus under various conditions [25, 67, 71–74]. The rates of import can be assayed in live cells using FRAP. In the simplest scenario, the entire nuclear fluorescence is photobleached, and then the rate of increase of fluorescence in the nucleus is measured corresponding to the import rate of the tagged molecule. A limitation of this approach is that it can only be applied a few times in the same cell before the total cellular fluorescence is depleted.
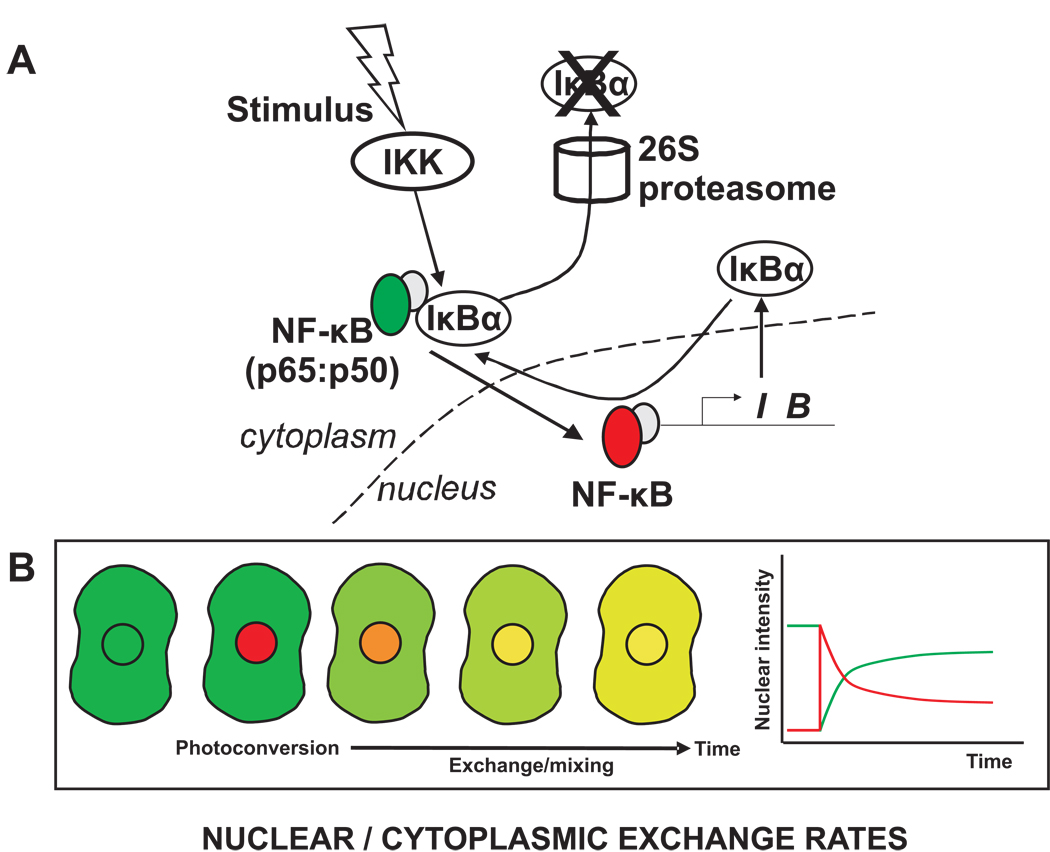
This drawback might be overcome by using photo-convertible proteins which cycle between red and green states [54, 75, 76]. Thus the nuclear fluorescence could be converted to red and then the rate of green nuclear import measured, along with the rate of red nuclear export (Fig. 5). For the next measurement, all of the molecules could be converted back to green, and after that only those molecules in the nucleus could be converted to red, and then the measurement shown in Figure 5 could be repeated.
Figure 5. Measuring nucleocytoplasmic exchange rates by photo-convertible proteins.
A. A photo-convertible protein such as Dronpa could be used to investigate exchange rates of NF-κB between the cytoplasm and nucleus. Dronpa initially exhibits green fluorescence, but activation with the appropriate wavelength converts Dronpa into a red fluorescent protein. B. If photo-conversion is restricted to the nucleus then NF-κB present in the nucleus at that moment will be labeled red and cytoplasmic NF-κB will remain green. If NF-κB exchange occurs between these two compartments, then green fluorescence will enter the nucleus and red fluorescence will enter the cytoplasm. Shown is the green/red overlay image, but measurements in the separate green and red channels would yield the changing intensity profiles shown in the plot.
Measuring protein-protein interactions by FRET or FCS
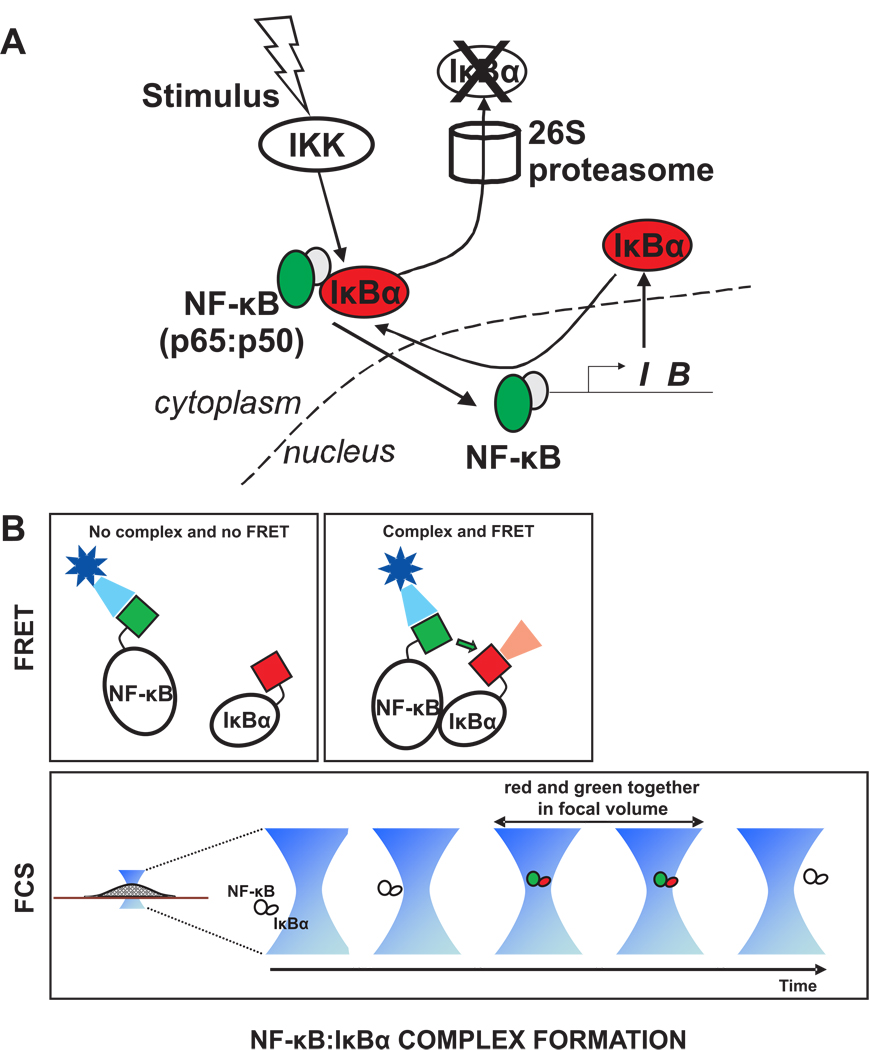
It is often presumed that the binding between NF-κB and IκB proteins is of high affinity and that when both are present a stable protein complex is formed. However, the binding reaction is actually reversible and represented as such in many mathematical models. Thus the concentration of the NF-κB:IκB complex may vary in space and time. These sorts of changing protein – protein interactions can be determined in live cells by either FRET or cross-correlation FCS (Fig. 6). Either of these measurements could be performed both at different locations within the cell and at a series of time points to provide spatiotemporal information about complex formation.
Figure 6. Measuring protein-protein interactions by FRET or FCS A.
Complex formation between NF-κB and IκBα could be detected by labeling NF-κB with GFP (green) and IκBα with mCherry (red). B. FRET could be used to determine if the two molecules are in close proximity, assuming the GFP and mCherry tags (green and red boxes) are appropriately situated on the NF-κB and IκBα molecules. Alternatively, cross-correlation FCS could be used to detect the complex, since red and green tags would be found simultaneously in the focal volume when a complex is present there. Note that for performing cross correlation FCS, the fluorescent tags should be situated to avoid FRET, otherwise the green signal will be reduced due to energy transfer to red.
Measuring phosphorylation status by a FRET sensor
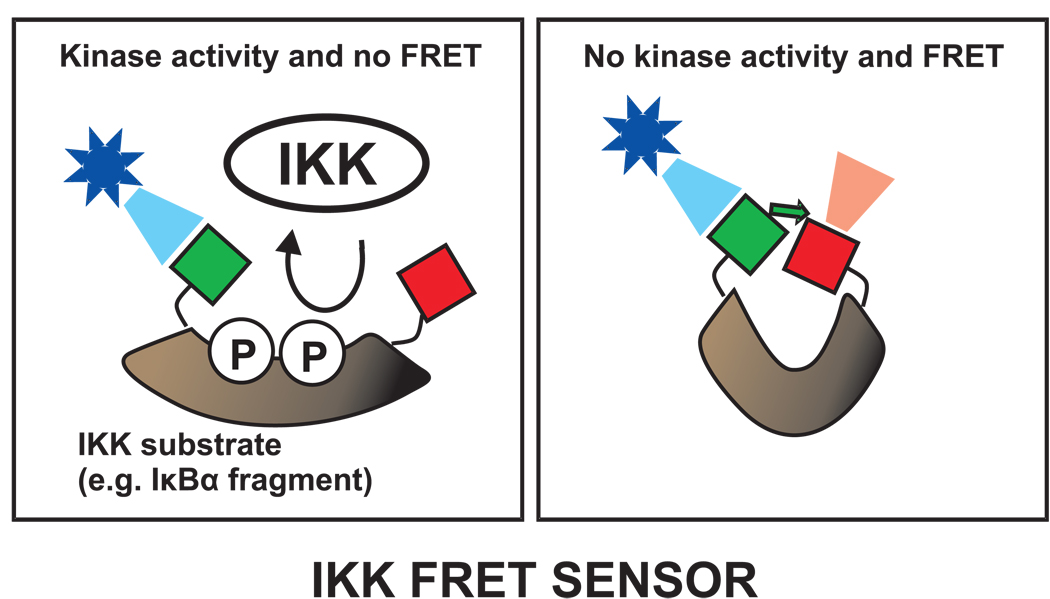
Upon stimulation, IKK phosphorylates the IκB proteins leading to their degradation, and ultimately to nuclear import of NF-κB. The direct measurement of IκB phosphorylation status would be particularly informative for modeling, providing accurate information about the stimulus input for the network. This phosphorylation could be measured in vivo by a FRET sensor (Fig. 7). Phosphorylation sensors contain a specific substrate sequence recognized by a particular kinase, in this case IKK. When this region is phosphorylated, the sensor molecule unfolds leading to a loss of FRET between, for example, GFP and mCherry molecules on either side of the phosphorylation sites.
Figure 7. Measuring phosphorylation events by a FRET sensor.
A FRET sensor could be used to detect phosphorylation of IκBα. The sensor would be a specially designed molecule (brown) that would be recognized by the IKK kinase. For example, the IκBα fragment containing the two specific serines (phosphorylated by IKK) would be a candidate. Typically, the phosphorylation sensors unfold after phosphorylation leading to loss of FRET between the donor and acceptor tags, shown here as green and red squares. Changes in FRET therefore reflect IKK activity.
Measuring DNA binding of a transcription factor
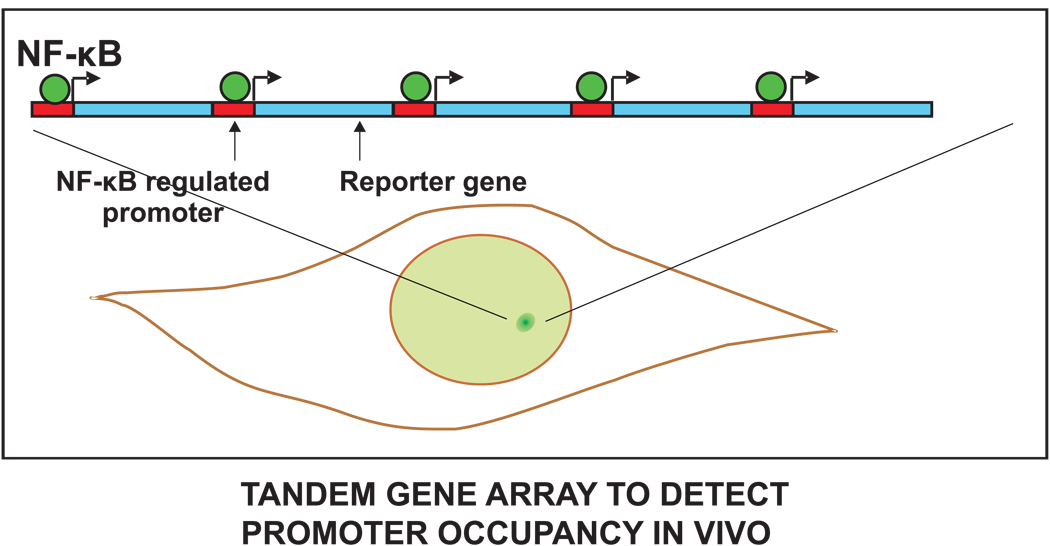
In the NF-κB network, NF-κB binds to the IκBα promoter and drives its expression. Transcription factor binding at a promoter can be measured in live cells using tandem arrays of promoter elements [77]. These arrays typically contain the same promoter sequence repeated several hundred times. In many cases, functional transcription units are created by including a reporter gene after each promoter. The arrays produce a locally high concentration of transcription factor binding sites that can be visualized as a discrete bright spot within live cell nuclei, usually by moderately over-expressing a fluorescently tagged version of the transcription factor. The fluorescence intensity of the spot corresponds to the amount of bound transcription factor fusion protein.
An array of a few thousand κB sites was created in fibroblasts [78], where it was used to measure binding residence times of NF-κB at the promoter using FRAP. The same system would be an effective one to measure the relative level of NF-κB at a promoter and correlate it with the level of NF-κB in the nucleus or with the levels of other factors in the regulatory network (Fig. 8). In addition to avoiding misleading effects of population averages, an advantage of this live-cell approach over traditional biochemical assays of promoter binding is that it more accurately accounts for chromatin accessibility in vivo, which is known to be an important component of gene regulation.
Figure 8. Measuring DNA binding of a transcription factor.
The levels of NF-κB binding to a target promoter could be measured by tagging NF-κB with GFP (green) and observing its binding to a tandem gene array. Such an array already exists for NF-κB and promoters containing κB consensus motifs or HIV LTR, and similar design principles could be used to construct an array cell line for any transcription factor. An array is composed of a series of NF-κB target sites (red rectangle) each followed by a reporter gene (blue). The array is stably integrated at a chromosomal locus. Live cells containing the GFP-tagged NF-κB (green circles) exhibit a bright green spot in the nucleus due to clustering of NF-κB sites at the tandem array. The brightness of this spot is proportional to the amount of NF-κB binding to the target site.
A drawback of the tandem array approach is that it is a time-consuming process to construct a cell line with an array. A simpler alternative is to use assays for measuring live cell binding such as FRAP or FCS to estimate the fraction of transcription factors that are bound to DNA throughout the nucleus. This can be done by performing a FRAP or FCS experiment at a random site in the nucleus. One uncertainty here is that it is not clear how much of the DNA binding measured by FRAP or FCS is due to specific interactions with promoter target sites and how much is due to non-specific interactions at other DNA sites throughout the genome. For this reason the tandem arrays have a distinct advantage as they unequivocally reflect promoter site binding, thereby providing the network model with important direct estimates for the level of NF-κB activity at a promoter.
Measuring mRNA production
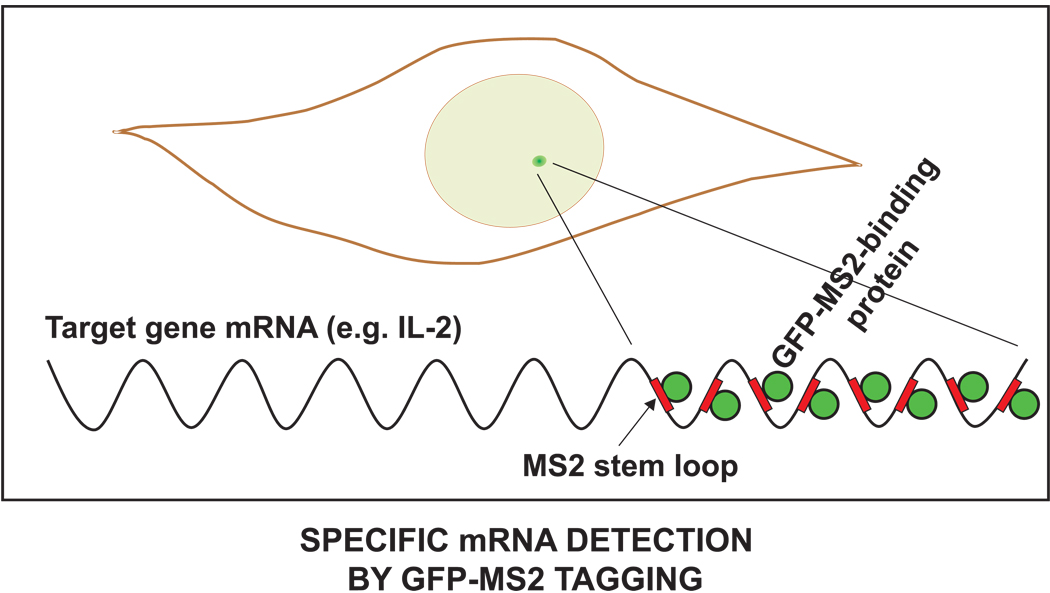
Once bound at the IκBα promoter, NF-κB then activates the transcription of this key gene which encodes the negative feedback protein. The rate of transcription for a target gene can be visualized and quantified in live cells [30, 79]. Although there are several related strategies for doing this, the most widely used involves the MS-2 tag which is an RNA motif that forms a looped structure. A series of these motifs can be added to the beginning or end of a gene transcript. These loops are recognized by the MS-2 binding protein, which can be fused to GFP and introduced into cells. As a result, the RNA transcripts from the promoter of interest are specifically labeled by GFP (Fig. 9). With this system in place, the levels of NF-κB in the nucleus or even at a promoter could be correlated with the levels of transcript produced. One limitation of this system is that it may not accurately report degradation of the mRNA since binding of the GFP-tagged MS-2 binding protein may protect the mRNA from degradation.
Figure 9. Measuring mRNA production.
Transcription driven by NF-κB can be detected by using the MS-2 RNA tagging system. A single copy gene is modified such that the transcript includes a series of MS-2 binding sites (red rectangles). The cells also express a GFP-tagged MS-2 binding protein (green circles) at low levels. This binds to the mRNA of the modified gene giving rise to a bright green spot in the nucleus where the newly transcribed mRNA is located. If sufficient MS-2 binding sites are present in the transcript, multiple GFP-MS-2 molecules will bind enabling a single mRNA molecule to be detected. The brightness of the GFP-MS-2 spot corresponds to the amount of mRNA produced.
Limitations of live cell microscopy
The preceding examples illustrate a number of powerful approaches for performing in vivo biochemical measurements. Although the live cell imaging methods have many advantages over their in vitro counterparts, there are still limitations to live cell imaging that must be appreciated.
In order to image a living cell, its natural state must be slightly perturbed. For example, cells can be damaged by extended exposure to the fluorescence excitation light that is required for all of the imaging approaches discussed here. An additional concern arises in those approaches that exploit intentional photobleaching, such as FRAP or FLIP, since these subject cells to a brief pulse of intense excitation light beyond the lower intensities used for normal imaging. Damage in any of these live cell experiments may be subtle and thus escape attention, and so controls must always be performed to confirm that cellular processes have not been adversely affected.
A second perturbation involves tagging proteins with a fluorescent marker such as GFP. Although a surprisingly large number of proteins seem to tolerate the addition of a GFP-like protein at their N- or C-terminus, control experiments are always necessary to show that a new fusion protein functions normally. Even when this is confirmed, overexpression of the fusion protein could still disrupt some cellular functions. For systems modeling, it is important to specifically check whether the molecular network under study has been perturbed by overexpressing one of its components.
Another concern arises specifically in the case of transcription factor binding at promoter target sites. Currently this binding can only be analyzed in live cells with the use of tandem gene arrays. Although some genes naturally arise in tandem copies, the copy number is generally low and the vast majority of genes exist in single copies. Thus, there is always the risk that an artificial tandem array may behave differently from a natural gene.
In addition to the preceding perturbations, there are also some technical limitations in live cell imaging. Currently, two to three different fluorescent markers can be visualized simultaneously, although new labeling methods and spectral imaging approaches may make it more routine in the future to do more than three markers at the same time.
Finally, although the ability to quantify binding rates in live cells is an especially important tool for systems biology approaches, work still remains to validate the current approaches and demonstrate that their estimates are in fact accurate [61, 80, 81].
Practical guidelines for implementation
Live cell experiments are non-trivial, and so as a guide for the novice, we define the workflow below and identify where some of the complications arise. In general, development of these techniques for every cellular variable in a systems model would be impractical, but application of selected techniques to a few key components in a model is feasible and should reap considerable insights.
Construction of the biological tools for live cell microscopy is relatively straightforward for FRAP, FLIP, FCS, and time-lapse imaging, since these can all be accomplished with the introduction of a fluorescent fusion protein into cells. It is more time consuming to generate the biological tools for FRET, mRNA detection, or promoter-specific binding. For FRET it is often necessary to test multiple fusion protein constructs, including both N- and C-terminal fusions with different spacer lengths before the right combination is found that permits FRET. These complexities also hold for developing a new FRET sensor, but here additional considerations arise to ensure that the sensor is specific and does not perturb the molecular concentrations it seeks to detect. For measuring mRNA levels, a cell line must be created that expresses the GFP-tagged MS-2 and also incorporates a modified gene containing the stem loops that the MS-2 protein binds to. Detection of promoter specific binding also requires construction of a cell line, in this case one that incorporates a tandem gene array. These are usually produced by applying some form of selection pressure to amplify a transgene that has been stably incorporated into the genome.
Once the biological tools are developed, then appropriate instrumentation is required for imaging. A well equipped light microscope is needed for FRAP, FLIP, FRET, and time-lapse experiments, as well as strong lasers for FRAP and FLIP where bleaching is necessary. Long-term time-lapse also requires an environmental chamber to keep the specimen in physiological culture conditions, as well as automated focus control to maintain stable focus. The latter control is especially important for quantitative measurements since even slight shifts of focus will change the measured intensity. FCS typically requires a microscope specifically designed for this purpose, namely one with a highly sensitive detector that can count photons from a diffraction-limited spot, although new approaches that can extract similar information from standard confocal images have recently been developed [82].
Data collection on all of these instruments is generally fast. For virtually all of the techniques, dozens of cells can be examined within a few hours. This generally provides sufficient sampling of the population variability. Long-term time lapse imaging is an exception. Dependent on the process under study, this can require several hours or days per cell. Some long-term time lapse applications have benefited from automated high throughput approaches [42–44].
Analysis of imaging data for many techniques requires some sophistication. Time-lapse data analysis benefits from automated data processing procedures (e.g. cell tracking) that often needs to be customized. This might involve automated identification of signal from certain cellular compartments or from a nuclear spot corresponding to either MS2-tagged mRNA or promoter-specific binding. Although macros to perform FRAP and FLIP are now available on most confocal microscopes, quantitative analysis of these data can become complicated particularly when extracting the kinetic estimates relevant for systems modeling. Software is available on many confocals to perform primary analysis of FRET data. Analysis of FCS data is non-trivial and must always be done by fitting mathematical models. Even when the microscope software performs the first-pass analysis, it is beneficial to have a deeper understanding of the mathematics underlying the procedures.
Conclusion
We have argued here that live cell imaging methods can and should find much broader application in systems biology research. It is likely that these approaches will ultimately set the standard for how best to measure the dynamics of molecular networks by providing information that no other approach can. At the moment, the major limitation in achieving this goal is the time to construct the biological tools and the expertise to acquire and analyze the image data. These limitations vary with the technique. It is anticipated that the various limitations will dwindle over time with increased application of live cell imaging approaches combined with the training of interdisciplinary scientists that are fluent in both systems biology and microscopy.
Footnotes
This experiment requires some care when examining those IκB isoforms whose genes are directly regulated by NF-κB [63, Tian B et al. 2005. JBC], since in these cases the IκB fusion gene should contain its endogenous promoter thereby retaining the natural regulatory circuit.
Contributor Information
Myong-Hee Sung, Email: sungm@mail.nih.gov.
James G McNally, Email: mcnallyj@exchange.nih.gov.
References
- 1.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 2.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 3.Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 5.Mellman I, Misteli T. Computational cell biology. J Cell Biol. 2003;161:463–464. doi: 10.1083/jcb.200303202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogilner A, Wollman R, Marshall WF. Quantitative modeling in cell biology: what is it good for? Dev Cell. 2006;11:279–287. doi: 10.1016/j.devcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 8.Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sveiczer A, Csikasz-Nagy A, Gyorffy B, Tyson JJ, Novak B. Modeling the fission yeast cell cycle: quantized cycle times in wee1- cdc25Delta mutant cells. Proc Natl Acad Sci U S A. 2000;97:7865–7870. doi: 10.1073/pnas.97.14.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyson JJ, Novak B. Regulation of the eukaryotic cell cycle: molecular antagonism, hysteresis, and irreversible transitions. J Theor Biol. 2001;210:249–263. doi: 10.1006/jtbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 11.von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 12.Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:E10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozbudak EM, Lewis J. Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 2008;4:e15. doi: 10.1371/journal.pgen.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 15.DeWitt AE, Dong JY, Wiley HS, Lauffenburger DA. Quantitative analysis of the EGF receptor autocrine system reveals cryptic regulation of cell response by ligand capture. J Cell Sci. 2001;114:2301–2313. doi: 10.1242/jcs.114.12.2301. [DOI] [PubMed] [Google Scholar]
- 16.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, et al. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada S, Shiono S, Joo A, Yoshimura A. Control mechanism of JAK/STAT signal transduction pathway. FEBS Lett. 2003;534:190–196. doi: 10.1016/s0014-5793(02)03842-5. [DOI] [PubMed] [Google Scholar]
- 18.Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100068. 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 20.Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 22.Ankers JM, Spiller DG, White MR, Harper CV. Spatio-temporal protein dynamics in single living cells. Curr Opin Biotechnol. 2008;19:375–380. doi: 10.1016/j.copbio.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci U S A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung MH, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL, et al. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS One. 2009;4:e7163. doi: 10.1371/journal.pone.0007163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, et al. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol Syst Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, et al. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Sci Signal. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day RN, Schaufele F. Imaging molecular interactions in living cells. Mol Endocrinol. 2005;19:1675–1686. doi: 10.1210/me.2005-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shav-Tal Y, Singer RH, Darzacq X. Imaging gene expression in single living cells. Nat Rev Mol Cell Biol. 2004;5:855–861. doi: 10.1038/nrm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 32.Mavrakis M, Pourquie O, Lecuit T. Lighting up developmental mechanisms: how fluorescence imaging heralded a new era. Development. 137:373–387. doi: 10.1242/dev.031690. [DOI] [PubMed] [Google Scholar]
- 33.Bao G, Rhee WJ, Tsourkas A. Fluorescent probes for live-cell RNA detection. Annu Rev Biomed Eng. 2009;11:25–47. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalink K. Spying on cGMP with FRET. Nat Methods. 2006;3:11–12. doi: 10.1038/nmeth0106-11. [DOI] [PubMed] [Google Scholar]
- 35.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 37.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura H, Hayashi-Takanaka Y, Yamagata K. Visualization of DNA methylation and histone modifications in living cells. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.02.004. in press. [DOI] [PubMed] [Google Scholar]
- 39.Ni Q, Titov DV, Zhang J. Analyzing protein kinase dynamics in living cells with FRET reporters. Methods. 2006;40:279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Eils R, Athale C. Computational imaging in cell biology. J Cell Biol. 2003;161:477–481. doi: 10.1083/jcb.200302097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Alaluf I, et al. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat Methods. 2006;3:525–531. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- 43.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 44.Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 45.Voss TC, Hager GL. Visualizing chromatin dynamics in intact cells. Biochim Biophys Acta. 2008;1783:2044–2051. doi: 10.1016/j.bbamcr.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinant C, Luijsterburg MS, Hofer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, et al. Assembly of multiprotein complexes that control genome function. J Cell Biol. 2009;185:21–26. doi: 10.1083/jcb.200811080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 48.Locke JC, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol. 2009;7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 50.Mullassery D, Horton CA, Wood CD, White MR. Single live-cell imaging for systems biology. Essays Biochem. 2008;45:121–133. doi: 10.1042/BSE0450121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Day RN, Schaufele F. Fluorescent protein tools for studying protein dynamics in living cells: a review. J Biomed Opt. 2008;13:031202. doi: 10.1117/1.2939093. [DOI] [PubMed] [Google Scholar]
- 52.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 53.Wiedenmann J, Oswald F, Nienhaus GU. Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges. IUBMB Life. 2009;61:1029–1042. doi: 10.1002/iub.256. [DOI] [PubMed] [Google Scholar]
- 54.Nienhaus GU, Nienhaus K, Holzle A, Ivanchenko S, Renzi F, Oswald F, et al. Photoconvertible fluorescent protein EosFP: biophysical properties and cell biology applications. Photochem Photobiol. 2006;82:351–358. doi: 10.1562/2005-05-19-RA-533. [DOI] [PubMed] [Google Scholar]
- 55.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 56.Howson R, Huh WK, Ghaemmaghami S, Falvo JV, Bower K, Belle A, et al. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp Funct Genomics. 2005;6:2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muzzey D, van Oudenaarden A. Quantitative time-lapse fluorescence microscopy in single cells. Annu Rev Cell Dev Biol. 2009;25:301–327. doi: 10.1146/annurev.cellbio.042308.113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verkman AS. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem Sci. 2002;27:27–33. doi: 10.1016/s0968-0004(01)02003-5. [DOI] [PubMed] [Google Scholar]
- 59.Koster M, Frahm T, Hauser H. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr Opin Biotechnol. 2005;16:28–34. doi: 10.1016/j.copbio.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Bacia K, Schwille P. A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods. 2003;29:74–85. doi: 10.1016/s1046-2023(02)00291-8. [DOI] [PubMed] [Google Scholar]
- 61.Michelman-Ribeiro A, Mazza D, Rosales T, Stasevich TJ, Boukari H, Rishi V, et al. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys J. 2009;97:337–346. doi: 10.1016/j.bpj.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levin MK, Carson JH. Fluorescence correlation spectroscopy and quantitative cell biology. Differentiation. 2004;72:1–10. doi: 10.1111/j.1432-0436.2004.07201002.x. [DOI] [PubMed] [Google Scholar]
- 63.Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, et al. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J Microsc. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 64.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsien RY. Indicators based on fluorescence resonance energy transfer (FRET) CSH Protoc. 2009;2009 doi: 10.1101/pdb.top57. pdb top57. [DOI] [PubMed] [Google Scholar]
- 66.Aye-Han NN, Ni Q, Zhang J. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr Opin Chem Biol. 2009;13:392–397. doi: 10.1016/j.cbpa.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 68.Joo J, Plimpton S, Martin S, Swiler L, Faulon JL. Sensitivity analysis of a computational model of the IKK NF-kappaB IkappaBalpha A20 signal transduction network. Ann N Y Acad Sci. 2007;1115:221–239. doi: 10.1196/annals.1407.014. [DOI] [PubMed] [Google Scholar]
- 69.O'Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sung MH, Simon R. In silico simulation of inhibitor drug effects on nuclear factor-kappaB pathway dynamics. Mol Pharmacol. 2004;66:70–75. doi: 10.1124/mol.66.1.70. [DOI] [PubMed] [Google Scholar]
- 71.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida K, Ozaki T, Furuya K, Nakanishi M, Kikuchi H, Yamamoto H, et al. ATM-dependent nuclear accumulation of IKK-alpha plays an important role in the regulation of p73-mediated apoptosis in response to cisplatin. Oncogene. 2008;27:1183–1188. doi: 10.1038/sj.onc.1210722. [DOI] [PubMed] [Google Scholar]
- 73.Lee SH, Hannink M. Characterization of the nuclear import and export functions of Ikappa B(epsilon) J Biol Chem. 2002;277:23358–23366. doi: 10.1074/jbc.M111559200. [DOI] [PubMed] [Google Scholar]
- 74.Tam WF, Lee LH, Davis L, Sen R. Cytoplasmic sequestration of rel proteins by IkappaBalpha requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20:2269–2284. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci U S A. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rafalska-Metcalf IU, Janicki SM. Show and tell: visualizing gene expression in living cells. J Cell Sci. 2007;120:2301–2307. doi: 10.1242/jcs.008664. [DOI] [PubMed] [Google Scholar]
- 78.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. Embo J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silverman AP, Kool ET. Quenched probes for highly specific detection of cellular RNAs. Trends Biotechnol. 2005;23:225–230. doi: 10.1016/j.tibtech.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 80.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.03.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]