Abstract
OBJECTIVE
Whole-grain foods are touted for multiple health benefits, including enhancing insulin sensitivity and reducing type 2 diabetes risk. Recent genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs) associated with fasting glucose and insulin concentrations in individuals free of diabetes. We tested the hypothesis that whole-grain food intake and genetic variation interact to influence concentrations of fasting glucose and insulin.
RESEARCH DESIGN AND METHODS
Via meta-analysis of data from 14 cohorts comprising ∼48,000 participants of European descent, we studied interactions of whole-grain intake with loci previously associated in GWAS with fasting glucose (16 loci) and/or insulin (2 loci) concentrations. For tests of interaction, we considered a P value <0.0028 (0.05 of 18 tests) as statistically significant.
RESULTS
Greater whole-grain food intake was associated with lower fasting glucose and insulin concentrations independent of demographics, other dietary and lifestyle factors, and BMI (β [95% CI] per 1-serving-greater whole-grain intake: −0.009 mmol/l glucose [−0.013 to −0.005], P < 0.0001 and −0.011 pmol/l [ln] insulin [−0.015 to −0.007], P = 0.0003). No interactions met our multiple testing–adjusted statistical significance threshold. The strongest SNP interaction with whole-grain intake was rs780094 (GCKR) for fasting insulin (P = 0.006), where greater whole-grain intake was associated with a smaller reduction in fasting insulin concentrations in those with the insulin-raising allele.
CONCLUSIONS
Our results support the favorable association of whole-grain intake with fasting glucose and insulin and suggest a potential interaction between variation in GCKR and whole-grain intake in influencing fasting insulin concentrations.
Diet modification is among the premier targets for the prevention of many chronic diseases and has proven particularly effective for prevention and management of type 2 diabetes. For example, improvement in dietary quality, in conjunction with other lifestyle modifications like increased physical activity, was shown to be more effective than pharmacological treatment in prevention of diabetes in individuals at high risk (1). Further, lifestyle modification may mitigate the risk associated with the strongest known diabetes risk loci (2). While the existence of environmental influences on genetic risk (and vice versa, gene × environment interaction) is generally accepted, few examples have been empirically demonstrated and replicated using population-based or trial data (3).
Measures of carbohydrate source, quality, or quantity, like whole-grain intake, fiber intake, glycemic index, and glycemic load, are of particular interest in relation to glucose metabolism and diabetes risk (4). Carbohydrate quality and whole-grain intake have been tested in recent nested diabetes case-control studies of diet × gene interaction (5–7). Findings from these studies, while intriguing, need replication in studies of larger sample size and uniform design to more thoroughly elucidate the relationships among diet, genetic factors, and diabetes risk (8,9).
Polymorphic regions in the human genome associated with risk of diabetes (10,11) and related quantitative traits (12) have been identified and replicated in populations of European ancestry. Information on personal genetic risk is already being disseminated to individuals within the general population and touted for its potential contribution to personalized medicine (13–15), although the underlying clinical utility has yet to be demonstrated (16,17). Given the potential for individual genetic risk to be empirically quantified and rapidly communicated, it is of interest to both clinicians and the general public to discover if modifiable characteristics like diet can mitigate risk in individuals empirically defined as “high risk” on the basis of genotype.
The aims of the current cross-sectional investigation were accomplished through a multicohort collaboration (18,19) including ∼48,000 individuals of European descent originating from 14 cohort studies conducted in North America and northern and southern Europe. Our hypotheses were that 1) whole-grain food intake is inversely associated with fasting glucose and insulin concentrations and 2) single nucleotide polymorphisms (SNPs), previously identified as predictive of fasting glucose (16 SNPs) and fasting insulin (2 SNPs) concentrations (12), and whole-grain intake interact to influence these traits in individuals without diabetes.
RESEARCH DESIGN AND METHODS
Participants from each of the 14 cohorts (Table 1; supplemental Table S1 in the online appendix, available at http://care.diabetesjournals.org/cgi/content/full/dc10-11150/DC1) were excluded if diabetes was present at the time of glucose and insulin measurement (defined by self-reported diabetes, pharmacologic treatment for diabetes, or fasting glucose concentrations ≥7 mmol/l), if consent to genetic research was not provided, or diet and genotype information did not meet cohort-specific quality-control standards (supplemental Tables S2 and S3). Participants provided written informed consent, and protocols were approved by local institutional review boards.
Table 1.
Participant characteristics in 14 participating cohorts
| N* | Age (years) | Sex (% women) | Fasting glucose (mmol/l) | Fasting insulin (pmol/l)† | Whole-grain intake (servings/day)‡ | Energy intake (kcal/day) | BMI (kg/m2) | |
|---|---|---|---|---|---|---|---|---|
| Health, Aging, and Body Composition Study (Health ABC) (U.S.) | 1,249 | 74.8 ± 2.9 | 51.0% | 5.1 ± 0.6 | 44.7 (43.4–46.1) | 0.96 (0.99) | 1,807 ± 599 | 26.2 ± 4.0 |
| Cardiovascular Health Study (CHS) (U.S.)§¶ | 2,765 (2,753) | 72.3 ± 5.4 | 62.0% | 5.5 ± 0.5 | 84.8 (83.5–86.2) | 0.94 (1.13) | 1,807 ± 641 | 26.0 ± 4.3 |
| Framingham Heart Study (FHS) (U.S.)§¶ | 5,835 | 46.1 ± 12 | 54.7% | 5.2 ± 0.5 | 27.0 (26.7–27.3) | 0.92 (1.08) | 1,982 ± 662 | 26.7 ± 5.0 |
| Atherosclerosis Risk in Communities (ARIC) Study (U.S.)§¶ | 7,201 | 54.2 ± 5.7 | 53.7% | 5.5 ± 0.5 | 58.9 (58.0–59.9) | 1.01 (1.44) | 1,644 ± 603 | 26.7 ± 4.7 |
| Family Heart Study (FamHS) (U.S.)¶ | 2,094 (2,089) | 50.1 0.5 | 55.5% | 5.2 ± 0.5 | 58.3 (56.8–59.7) | 1.14 (1.64) | 1,733 ± 603 | 27.3 ± 5.1 |
| The Age, Gene/Environment Susceptibility-Reykjavik Study (AGES) (Iceland)§ | 2,819 | 76.4 ± 5.5 | 59.7% | 5.5 ± 0.5 | 55.1 (53.9–56.4) | 1.79 (1.07) | NA | 26.9 ± 4.3 |
| Fenland (U.K.)¶ | 720 | 45.0 ± 7.3 | 56.1% | 4.9 ± 0.5 | 38.5 (37.3–39.7) | 1.28 (1.15) | 1,949 ± 702 | 27.0 ± 4.9 |
| Malmö Diet and Cancer Study (Malmö) (Sweden) | 4,924 (4,765) | 57.5 ± 5.9 | 59.0% | 5.5 ± 0.5 | 37.4 (36.8–38.0) | 1.49 (2.02) | 2,324 ± 672 | 25.4 ± 3.8 |
| Uppsala Longitudinal Study of Adult Men (ULSAM) (Sweden)¶ | 942 (932) | 71.0 ± 0.6 | 0% | 5.4 ± 0.6 | 64.5 (62.3–66.8) | 2.04 (1.48) | 1,749 ± 462 | 26.0 ± 3.2 |
| Gene-Lifestyle interactions And Complex traits In Elevated disease Risk (GLACIER) (Sweden) | 14,913 (891) | 52.2 ± 8.7 | 59.9% | 5.4 ± 0.6 | 41.0 (39.3–42.8) | 1.66 (2.09) | 1,823 ± 549 | 25.8 ± 4.0 |
| Rotterdam Study (the Netherlands)§¶ | 2,304 | 65.4 ± 6.6 | 58.7% | 5.5 ± 0.5 | 63.4 (62.1–64.8) | 3.50 (3.00) | 1,991 ± 505 | 26.6 ± 3.8 |
| Invecchiare in Chianti (Aging in the Chianti Area; InCHIANTI) (Italy)¶ | 1,071 (1,044) | 67.7 ± 16 | 56.3% | 4.8 ± 0.6 | 56.3 (54.4–58.1) | 0.00 (2.22) | 2,014 ± 601 | 27.0 ± 4.1 |
| Gene-Diet Attica Investigation on Childhood Obesity (GENDAI) (Greece)¶ | 1,087 (1,064) | 11.2 ± 0.7 | 53.2% | 4.8 ± 0.5 | 40.0 (38.7–41.3) | 0.00 (0.50) | 1,891 ± 595 | 20.0 ± 3.4 |
| Greek Health Randomized Aging Study (GHRAS) (Greece)¶ | 865 (670) | 71.8 ± 7.5 | 71.2% | 5.8 ± 1.6 | 43.1 (41.3–45.0) | 0.00 (2.00) | 2,156 ± 693 | 29.7 ± 4.8 |
Data are means ± SD, median (interquartile range)
‡, or geometric mean (95% CI).
*Maximum available observations for interactions between whole-grain intake and SNPs in glucose analyses; values vary in some cohorts depending on availability of genotype information (in parentheses, where sample size for insulin analyses differs from glucose analyses).
†Insulin was analyzed on the natural log scale and back transformed to the geometric scale for presentation. Presented values are means (95% CI).
§Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium cohorts.
¶Meta-Analyses of Glucose and Insulin-Related Traits Consortium cohorts. NA, not available.
Characterization of whole-grain intake
Daily servings of whole-grain foods were estimated in each cohort as the sum of daily servings of whole-grain items included on food frequency questionnaires (FFQs) (11 cohorts), a lifestyle questionnaire (1 cohort), reported during multiple 24-h recalls (1 cohort), or recorded in 7-day dietary diaries (1 cohort). Breakfast cereals containing ≥25% whole grain or bran by weight were considered whole grain when brand name information and corresponding industry-provided ingredients were available (20). In cohorts where food reference portions were given alongside frequency options (i.e., semiquantitative FFQ), the reference portion was assigned as one serving. In cohorts where food items were quantified in daily grams, uniform weights were assigned as one serving on a food-by-food basis. (details in supplemental Table S2).
Genotyping, fasting glucose, and insulin quantification: assessment of other relevant variables
Cohort-specific methods for genotyping, fasting glucose and insulin quantification, and assessment of other participant characteristics, as well as allele frequencies at each locus are described in supplemental Tables S3, S4, and S5. The SNPs used in the present analysis were associated (P < 5 × 10−8) with fasting glucose and/or fasting insulin in a previous meta-analysis of genome-wide association studies with independent replication (12); 15 SNPs were associated with only fasting glucose, 1 SNP with only fasting insulin, and 1 SNP with both fasting glucose and insulin (listed in Table 3). Fasting glucose and insulin were quantified by enzymatic methods and radioimmunoassay, respectively.
Table 3.
Meta-analyzed interactions between daily whole-grain intake and genotype for select SNPs for fasting glucose and fasting insulin in 14 cohorts*
| SNP | Nearest gene | Glucose- or insulin-raising allele/other allele | Number of cohorts | n | Regression coefficient for interaction between daily servings of whole grains × SNP for fasting glucose (mmol/l) |
I2 (95% uncertainty interval) (%) | ||
|---|---|---|---|---|---|---|---|---|
| β | SE | P | ||||||
| Glucose-related SNP | ||||||||
| rs340874 | PROX1 | C/T | 13 | 43,527 | −0.0011 | 0.0030 | 0.71 | 0 (0–57) |
| rs780094 | GCKR | C/T | 14 | 48,303 | 0.0040 | 0.0027 | 0.13 | 0 (0–55) |
| rs560887 | G6PC2 | C/T | 13 | 43,488 | −0.0001 | 0.0032 | 0.98 | 0 (0–57) |
| rs11708067 | ADCY5 | A/G | 13 | 43,555 | 0.0039 | 0.0036 | 0.28 | 24 (0–61) |
| rs11920090 | SLC2A2 | T/A | 13 | 43,451 | 0.0006 | 0.0043 | 0.89 | 0 (0–57) |
| rs2191349 | DGKB/TMEM195 | T/G | 13 | 43,561 | −0.0044 | 0.0029 | 0.13 | 0 (0–57) |
| rs4607517 | GCK | A/G | 14 | 48,323 | 0.0002 | 0.0035 | 0.95 | 0 (0–55) |
| rs11558471 | SLC30A8 | A/G | 10 | 40,776 | −0.0007 | 0.0034 | 0.84 | 0 (0–62) |
| rs7034200 | GLIS3 | A/C | 13 | 43,362 | 0.0015 | 0.0029 | 0.60 | 0 (0–57) |
| rs10885122 | ADRA2A | G/T | 13 | 43,391 | 0.0082 | 0.0044 | 0.06 | 0 (0–57) |
| rs4506565 | TCF7L2 | T/A | 12 | 45,911 | 0.0004 | 0.0030 | 0.88 | 51 (6–75) |
| rs11605924 | CRY2 | A/C | 13 | 43,567 | −0.0016 | 0.0029 | 0.58 | 0 (0–57) |
| rs7944584 | MADD | A/T | 13 | 43,361 | 0.0049 | 0.0033 | 0.14 | 0 (0–57) |
| rs174550 | FADS1 | T/C | 14 | 48,162 | −0.0027 | 0.0028 | 0.34 | 32 (0–64) |
| rs10830963 | MTNR1B | G/C | 13 | 43,433 | 0.0028 | 0.0035 | 0.42 | 32 (0–65) |
| rs11071657 | C2CD4B | A/G | 13 | 42,500 | 0.0035 | 0.0031 | 0.26 | 0 (0–57) |
| Insulin-related SNP | Regression coefficient for interaction between daily servings of whole grains × SNP for fasting insulin [(ln)pmol/l] | |||||||
| rs780094 | GCKR | C/T | 14 | 33,784 | 0.0091 | 0.003 | 0.006 | 1 (0–36) |
| rs35767 | IGF1 | G/A | 13 | 29,078 | 0.0022 | 0.005 | 0.69 | 0 (0–57) |
*Regression coefficient for interaction between daily servings of whole grains × SNP for fasting glucose (mmol/l) and fasting insulin [(ln)pmol/l], adjusted for age, sex, energy intake (not in the Age, Gene/Environment Susceptibility-Reykjavik Study), and field center (Health, Aging, and Body Composition Study; the Cardiovascular Health Study; the Atherosclerosis Risk in Communities Study; and the Invecchiare in Chianti [Aging in the Chianti Area] Study) and population structure by principal components in the Framingham Heart Study and the Family Heart Study.
Statistical analysis
Glucose was analyzed without transformation and insulin was natural log transformed before analysis. β-Coefficients from regression analyses are presented for (ln)insulin. For descriptive purposes, cohort mean insulin concentrations were back transformed and presented as geometric means with 95% CIs.
Cohort-specific analyses
Each cohort provided β-coefficients and SEs for the following linear regression models: 1) association between daily servings of whole-grain foods and fasting glucose or fasting insulin concentrations, 2) interactions between daily servings of whole-grain foods and 16 SNPs for fasting glucose concentrations, and 3) interactions between daily servings of whole-grain foods and 2 SNPs for fasting insulin concentrations. To evaluate associations of whole-grain intake with fasting glucose and insulin concentrations, we used the following four linear regression models (listed in Table 2 and defined in supplemental Table S6; linear mixed-effects models were used to account for familial correlation among participants in the Framingham Heart Study and the Family Heart Study): model 1, age (years, continuous), sex, energy intake (kcal/day, continuous) plus field center (in the Health, Aging, and Body Composition Study; the Cardiovascular Health Study; the Atherosclerosis Risk in Communities Study; the Family Heart Study, and the Invecchiare in Chianti [Aging in the Chianti Area] Study) and population substructure (by principal components in Framingham Heart Study and Family Health Study); model 2, model 1 plus lifestyle characteristics; model 3, model 2 plus select dietary factors; and model 4, model 3 plus BMI. For the interaction analyses, we used model 1 covariates. In accordance with an additive model where the SNPs were uniformly modeled for the glucose- or insulin-raising allele, the interaction regression coefficients represent the difference in the magnitude of the whole-grain association (per one daily serving) with glucose (mmol/l) or (ln) insulin (pmol/l) per copy of the glucose- or insulin-raising allele.
Table 2.
Meta-analyzed association between daily whole-grain intake and fasting glucose and fasting insulin in 14 cohorts
| n | Regression coefficient (β [95% CI] representing expected change in fasting glucose [mmol/l] per one-daily-serving–greater whole-grain intake) | P | n | Regression coefficient (β [95% CI] representing expected change in fasting insulin [{ln}pmol/l] per one-daily-serving–greater whole-grain intake) | P | |
|---|---|---|---|---|---|---|
| Model 1: age, sex, energy intake, field center, or population structure* | 48,723 | −0.019 (−0.022 to −0.015) | <0.0001 | 34,201 | −0.021 (−0.025 to −0.017) | <0.0001 |
| Model 2: model 1 + education level, physical activity, alcohol intake, and smoking status† | 48,207 | −0.013 (−0.017 to −0.010) | <0.0001 | 34,108 | −0.022 (−0.026 to −0.017) | <0.0001 |
| Model 3: model 2 + red or processed meat, fish, vegetables, fruit, coffee, nuts, and seeds‡ | 46,985 | −0.012 (−0.016 to −0.008) | <0.0001 | 33,993 | −0.016 (−0.021 to −0.011) | <0.0001 |
| Model 4: model 3 + BMI§ | 46,928 | −0.009 (−0.013 to −0.005) | <0.0001 | 33,937 | −0.011 (−0.015 to −0.007) | 0.0003 |
*Energy intake was not estimated in the Age, Gene/Environment Susceptibility-Reykjavik Study cohort. Field center was included as a covariate in the Health, Aging, and Body Composition Study; the Cardiovascular Health Study, the Atherosclerosis Risk in Communities Study, the Family Heart Study, and the Invecchiare in Chianti (Aging in the Chianti Area) Study. Principal components were used to adjust for population structure in the Framingham Heart Study and the Family Heart Study.
†Education level and physical activity were defined uniquely by cohort. Smoking status was characterized as current, former, or never in 12 cohorts and as current or not current in 3 cohorts (Framingham Heart Study; Age, Gene/Environment Susceptibility-Reykjavik Study; Uppsala Longitudinal Study of Adult Men). Education level, smoking status, and alcohol intake were not adjusted in the Gene-Diet Attica Investigation on Childhood Obesity cohort (fifth and sixth graders).
‡Most cohorts included each of dietary covariates listed in the table as servings per day or grams per day; exceptions are noted in the online supplement.
§BMI was modeled as a continuous variable in all cohorts (kg/m2).
Meta-analyses
We used an inverse variance–weighted meta-analysis with fixed effects to estimate summary effects (METAL software [http://www.sph.umich.edu/csg/abecasis/metal/index.html] for whole-grain × SNP interaction tests; and Stata 11.0, Stata Corporation, College Station, TX, for whole-grain outcome associations) and assessed heterogeneity by the I2 index (21). Bonferroni correction was used to determine the level of statistical significance; with 16 tests for glucose and 2 for insulin, we used α = 0.05/18 = 0.0028.
The sample sizes for the whole-grain associations with fasting glucose ranged from 48,723 to 46,928 in models 1 and 4, respectively, and for fasting insulin, samples ranged from 34,201 to 33,937 in models 1 and 4, respectively. The sample sizes for the whole-grain × SNP interaction analyses for fasting glucose ranged from 40,776 for rs11558471 to 48,323 for rs4607517 and rs174550, with samples sizes for the other 13 SNPs between those values. The sample sizes for the whole-grain × SNP interaction analyses for fasting insulin was 29,078 for rs35767 and 33,784 for rs780094. Post hoc estimates of study power are provided in supplemental Fig. S1.
RESULTS
Table 1 summarizes the basic demographic characteristics of the 14 contributing cohorts. The mean self-reported daily whole-grain intake was lowest in Mediterranean regions and highest in northern European regions. Variation did not appear to correspond to measurement method (FFQ vs. 24-h recalls versus dietary records) (supplemental Fig. S2).
Associations of whole-grain intake with fasting glucose and insulin concentrations
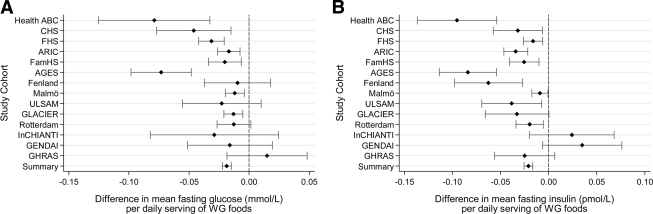
With adjustment for sex, age, and energy intake, greater whole-grain intake was associated with lower fasting glucose and insulin concentrations. For each one-daily-serving–greater intake of whole-grain foods, fasting glucose concentrations were 0.019 units lower (β [95% CI]: −0.019 mmol/l [−0.022 to −0.015], P < 0.0001) (Fig. 1A; Table 2) and fasting insulin concentrations were 0.021 units lower (β [95% CI]: −0.021 [ln] pmol/l [−0.025 to −0.017], P < 0.0001) (Fig. 1B; Table 2). Results from models 2–4 were similar (Table 2), showing only slight attenuation in the regression estimates (Table 2; see also supplemental Figs. S3 and S4 and supplemental Table S7).
Figure 1.
Associations between daily whole-grain intake (A) and fasting glucose (B) and fasting insulin in 14 cohorts. A: Regression coefficient (β [95% CI]) representing expected change in fasting glucose (mmol/l) per one-daily-serving–greater whole-grain intake. B: Regression coefficient (β [95% CI]) representing expected change in fasting insulin [(ln)pmol/l] per one-daily-serving–greater whole-grain intake. Data are adjusted for model one covariates: age, sex, energy intake, field center, or population structure (Note: energy intake was not estimated in the AGES cohort; field center was included as a covariate in Health ABC, CHS, ARIC, FamHS, and InCHIANTI; population structure by principal components in FHS and FamHS).
Interactions of whole-grain intake and SNPs with respect to fasting glucose and insulin concentrations. The strongest identified interaction was between whole-grain intake and rs780094 (in GCKR) in association with fasting insulin concentrations (βinteraction ± SE: 0.009 [ln] pmol/l ± 0.003, P = 0.006). Translated, this interaction regression coefficient indicates that greater whole-grain intake had a weaker insulin-lowering effect in the presence of the insulin-raising C allele. For example, in individuals carrying one copy of the insulin-raising C allele, the lower insulin concentration observed in association with greater whole-grain intake would be reduced by 0.009 units (that is, 0.010 units lower insulin in association with one daily whole-grain serving instead of 0.019 units lower). Correspondingly, in individuals carrying two copies of the insulin-raising C allele, the lower insulin concentration observed in association with greater whole-grain intake would be reduced by 0.018 units (that is, 0.001 units lower insulin in association with one daily whole-grain serving instead of 0.019 units lower). After correction for multiple hypothesis testing, none of the interactions between whole-grain intake and the preselected SNPs (including rs780094) met our a priori cut point for significance (P < 0.0028) (Table 3 and supplemental Figs. S5 and S6).
CONCLUSIONS
Understanding how a potentially modifiable dietary characteristic like whole-grain food intake influences genetic effects on metabolic homeostasis may help elucidate the therapeutic potential of personalized medicine. We have performed a meta-analysis evaluating interactions between whole-grain food intake, an easily modifiable dietary characteristic with known associations with fasting glucose, insulin and diabetes risk, and loci previously identified as significantly and reproducibly associated with concentrations of fasting glucose and insulin (12). This is, to our knowledge, the largest and most comprehensive study of gene × lifestyle interactions conducted to date. In over 48,000 European individuals, we observed robust associations of whole-grain intake with fasting glucose and fasting insulin concentrations, firmly supporting observations previously made in other, smaller studies (22–25). The most promising interaction we identified was between whole grains and variation in GCKR (rs780094) in association with fasting insulin, where the inverse association between whole-grain intake and fasting insulin concentrations was weakened in the presence of the insulin-raising allele. However, for the majority of loci studied, the inverse association of whole-grain intake with fasting glucose or fasting insulin was present regardless of allelic variation at these loci.
Current findings in the context of gene × environment interaction investigations
The polymorphic locus rs780094 lies near a splice site in intron 18 of the GCKR gene whose product is a regulatory protein that inhibits glucokinase, a key regulatory step in glucose metabolism that is influenced by dietary composition (26). The locus was originally identified in the Diabetes Genetics Initiative GWAS for triglyceride levels (27). Later, the triglyceride-raising T allele was associated with lower fasting glucose and insulin concentrations (28) and confirmed in a meta-analysis of several GWAS (12). Fine mapping of the region for association with triglyceride levels pinpointed a Pro446Leu missense variant in GCKR (28) that is less responsive to regulation by concentrations of fructose-6-phospate, resulting in increased liver glucokinase activity, enhanced glycolysis, and elevated liver malonyl-CoA. The consequence of this metabolic shift manifests in lower fasting glucose and elevated triglyceride concentrations (29). The mechanism by which whole-grain food intake improves insulin resistance may involve glucokinase, and our results suggest that allelic variation at GCKR could diminish the beneficial effects of whole-grain foods on insulin homeostasis, possibly via the strong effect of GCKR variant on both triglyceride and glucose levels.
No other studied interaction met our Bonferroni-corrected cut point for statistical significance. Aside from the possibility that there really is no interaction between whole grains and these loci, the null results could still reflect insufficient statistical power or misclassification in the quantification of whole-grain intake. It is also possible that latent interactions might be observable in acute diet intake settings, that is, after a whole-grain–enriched meal where postmeal measures of insulin sensitivity are obtained.
Previous studies have evaluated interactions between diabetogenic loci and whole-grain intake or other proxies of carbohydrate intake or overall dietary quality. Three nested case-control studies previously investigated interactions of whole-grain intake (6), glycemic index/glycemic load (5), or a Western dietary pattern (7) with TCF7L2 SNPs (rs7903146 (6) and rs12255372 (5,6) or a genetic risk score that included a TCF7L2 marker among 10 risk loci (7). All three studies reported significant interactions (P < 0.05) between the TCF7L2 variants and the respective dietary factors on diabetes incidence. Unlike these studies, we found no evidence of interaction between whole-grain food intake and the rs4506565 variant (another TCF7L2 marker highly correlated [r2 0.68–0.917] with rs7903146 in Europeans) with respect to either fasting glucose or fasting insulin concentrations. We cannot exclude interactions between whole-grain intake and TCF7L2 variants on diabetes risk, as the mechanisms of interaction may differ in persons with established diabetes. On the other hand, these previous studies were relatively small and did not apply conservative corrections for multiple testing, raising the possibility of false-positive findings.
Strengths and limitations of the present work
The strengths of our study include its large sample size, clearly defined a priori hypotheses, control for multiple testing, comparable whole-grain definitions across cohorts, and inclusion of well-characterized cohorts with diverse underlying dietary patterns (i.e., unique correlation structure of foods), which reduces the potential for confounding by other foods correlated with whole-grain intake. However, studies such as ours also have some inherent limitations. For example, measurement error in epidemiological studies can seriously impact the ability to detect small gene × environment interaction effects (30). The study-specific interaction regression coefficients covered a wide range (i.e., we observed small regression coefficients and large within-study variances), suggesting that some random errors may have reduced study power. Thus, even though our study is large in relative terms, it may still lack power to detect small interaction effects. On the other hand, if too small to be detected by our analysis, such small interactions might have relatively limited population or clinical relevance. The role of measurement error in dietary assessment has been long debated (31), and it is possible that the influence of genetic factors on these outcomes may vary according to whole-grain intake in more well-controlled clinical settings. Furthermore, even though sequential adjustment for putative confounding factors had little impact on the effect sizes across models, we cannot exclude the possibility that residual confounding explains some of our findings. It may also be that because we used an overly conservative method for adjusting for multiple testing, some of our findings may be falsely negative.
Genome-wide scans typically rank the most significant effects highest. The statistical significance of a genotype-phenotype association is diminished in the presence of interaction (32). Thus, loci that interact with other loci or with environmental factors may be less likely to rank highly in conventional GWASs compared with those that have strong main effects that are not modified by other exposures. Thus, by examining only the top main effects from GWAS in the present study, we may have overlooked numerous valid gene × whole-grain interaction effects elsewhere in the genome. Furthermore, because it is unknown whether the SNPs studied here are the causal variants, it is possible that stronger effects attributable to rarer SNPs could underlie some of the examined loci. It is worth noting that for some SNPs, we observed a high degree of heterogeneity in interaction effects across cohorts, suggesting the possibility of multidimensional interactions, which could not be examined in the present study.
Results of this large, comprehensive investigation of gene-diet interaction, suggest that the association of whole-grain intake with fasting insulin may be modified by GCKR rs780094. While intriguing, the test of interaction did not meet our conservative Bonferroni-corrected cut point for statistical significance and requires confirmation in other studies. Our results do show that whole-grain food intake is similarly and inversely associated with fasting insulin and glucose irrespective of genetic variation at the other loci studied. Our work coincides with the dawn of a new age in genetic and nutritional research. Investigations such as ours contribute to a better understanding of how diet therapy may (or may not) be individualized to a person's genetic background. However, to fully realize this potential, studies will require more precisely measured exposures (such as nutritional biomarkers of whole-grain intake) and should include experimental settings where diet is manipulated in people of contrasting genetic risk profiles.
Supplementary Material
Acknowledgments
No potential conflicts of interest relevant to this article were reported.
APPENDIX
Full author list: the CHARGE Whole Grain Foods Study Group, in addition to the first 11 authors: Melissa Garcia, MPH12; Jennifer S. Anderson, MD, PhD13; Jack L. Follis, MS14; Luc Djousse, MD, DrPH15; Kenneth Mukamal, MD16; Constantina Papoutsakis, PhD3; Dariush Mozaffarian, MD, DrPH17; M. Carola Zillikens, MD18; Stefania Bandinelli, MD19; Amanda J. Bennett, PhD20; Ingrid B. Borecki, PhD9; Mary F. Feitosa, PhD9; Luigi Ferrucci, MD, PhD11; Nita G. Forouhi, MD10; Christopher J. Groves, PhD21; Goran Hallmans, PhD22; Tamara Harris, MD12; Albert Hofman, PhD7; Denise K. Houston, PhD13; Frank B. Hu, PhD23; Ingegerd Johansson, PhD24; Stephen B. Kritchevsky, PhD13; Claudia Langenberg, MD, PhD10; Lenore Launer, PhD12; Yongmei Liu, PhD13; Ruth J. Loos, PhD10; Michael Nalls, PhD25; Marju Orho-Melander, PhD8; Frida Renstrom, PhD26; Kenneth Rice, PhD4; Ulf Riserus, PhD27; Olov Rolandsson, PhD28; Jerome I. Rotter, MD29; Georgia Saylor, BS13; Eric J.G. Sijbrands, MD30; Per Sjogren, PhD26; Albert Smith, PhD31; Laufey Steingrímsdóttir, PhD32; André G. Uitterlinden, PhD18; Nicholas J. Wareham, PhD10; Inga Prokopenko, PhD20; James S. Pankow, PhD33; Cornelia M. van Duijn, PhD7; Jose C. Florez, MD, PhD34; Jacqueline C.M. Witteman, PhD7; the MAGIC Investigators (complete author list can be found in the online appendix); Josée Dupuis, PhD35; George V. Dedoussis, PhD3; Jose M. Ordovas, PhD36; Erik Ingelsson, PhD37; L. Adrienne Cupples, PhD6; David S. Siscovick, MD4; Paul W. Franks, PhD38; James B. Meigs, MD.39
The 12Laboratory of Epidemiology, Demography, and Biometry, National Institute on Aging, Bethesda, Maryland; the 13Sticht Center on Aging, Wake Forest University, Winston-Salem, North Carolina; the 14Division of Biostatistics, The University of Texas Health Sciences Center-Houston, Houston, Texas; the 15Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts, and Massachusetts Veterans Epidemiology and Research Information Center and Geriatric Research, Education, and Clinical Center, Boston Veterans Affairs Healthcare System, Boston, Massachusetts; the 16Division of General Medicine & Primary Care, Beth Israel Deaconess Medical Center, Boston, Massachusetts; the 17Division of Cardiovascular Medicine, Brigham and Women's Hospital and Harvard Medical School, and the Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts; the 18Department of Internal Medicine, Erasmus MC, Rotterdam, 3015GE, the Netherlands, and Netherlands Genomics Initiative (NGI)-sponsored Netherlands Consortium for Healthy Aging (NCHA); the 19Geriatric Unit, Azienda Sanitaria Firenze (ASF), Florence, Italy; the 20Oxford Centre for Diabetes, Endocrinology and Metabolism, Churchill Hospital, Oxford, U.K.; the 21Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, U.K., and the Oxford Centre for Diabetes, Endocrinology and Metabolism, University of Oxford, Oxford, U.K.; the 22Department of Public Health and Clinical Medicine, Nutritional Research, Umeå University, Umeå Sweden; 23Channing Laboratory, Department of Medicine, Brigham and Women's Hospital/Harvard Medical School, Boston, Massachusetts; the 24Department of Odontology, Umeå University, Umeå, Sweden; 25Laboratory of Neurogenetics, National Institute on Aging, Bethesda, Maryland; the 26Genetic Epidemiology & Clinical Research Group, Department of Public Health & Clinical Medicine, Section for Medicine, Umeå University Hospital, Umeå Sweden; the 27Department of Public Health and Caring Sciences, Clinical Nutrition and Metabolism, Uppsala University, Uppsala, Sweden; the 28Department of Public Health and Clinical Medicine, Family Medicine, Umeå University, Umeå, Sweden; the 29Division of Medical Genetics, Cedars-Sinai Medical Center, Los Angeles, California; the 30Department of Internal Medicine, Erasmus Medical Center Rotterdam, the Netherlands, and The Netherlands Genomics Initiative-sponsored Netherlands Consortium for Healthy Aging (NGI-NCHA), Leiden, the Netherlands; the 31Icelandic Heart Association, Kopavogur, Iceland; 32Icelandic Heart Association, Kopavogur, Iceland, and the Unit for Nutrition Research, Landspitali University Hospital and Faculty for Food Science and Nutrition, University of Iceland; the 33Department of Epidemiology, University of Minnesota, Minneapolis, Minnesota; the 34Center for Human Genetic Research, Massachusetts General Hospital, Boston, Massachusetts, and Program in Medical and Population Genetics, Broad Institute, Cambridge, Massachusetts, and Harvard Medical School, Boston, Massachusetts; the 35Department of Biostatistics, Boston University School of Public Health, Boston University, Boston, Massachusetts, and National Heart, Lung, and Blood Institute's Framingham Heart Study, Framingham, Massachusetts; 36Nutrition and Genomics Laboratory, Jean Mayer USDA Human Nutrition Research Center on Aging, Tufts University, Boston, Massachusetts; the 37Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden; the 38Genetic Epidemiology & Clinical Research Group, Department of Public Health & Clinical Medicine, Section for Medicine, Umeå University Hospital, Umeå Sweden, and the Department of Clinical Sciences, Lund University, Malmö, Sweden; and the 39General Medicine Division, Clinical Epidemiology Unit, and Diabetes Research Unit, Massachusetts General Hospital, Boston, Massachusetts.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM: Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D: TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corella D, Peloso G, Arnett DK, Demissie S, Cupples LA, Tucker K, Lai CQ, Parnell LD, Coltell O, Lee YC, Ordovas JM: APOA2, dietary fat, and body mass index: replication of a gene-diet interaction in 3 independent populations. Arch Intern Med 2009;169:1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bantle JP, Wylie-Rosett J, Albright AL, Apovian CM, Clark NG, Franz MJ, Hoogwerf BJ, Lichtenstein AH, Mayer-Davis E, Mooradian AD, Wheeler ML: Nutrition recommendations and interventions for diabetes: a position statement of the American Diabetes Association. Diabetes Care 31(Suppl.)2008;1:S61–S78 [DOI] [PubMed] [Google Scholar]
- 5.Cornelis MC, Qi L, Kraft P, Hu FB: TCF7L2, dietary carbohydrate, and risk of type 2 diabetes in US women. Am J Clin Nutr 2009;89:1256–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher E, Boeing H, Fritsche A, Doering F, Joost HG, Schulze MB: Whole-grain consumption and transcription factor-7-like 2 (TCF7L2) rs7903146: gene-diet interaction in modulating type 2 diabetes risk. Br J Nutr 2009;101:478–481 [DOI] [PubMed] [Google Scholar]
- 7.Qi L, Cornelis MC, Zhang C, van Dam RM, Hu FB: Genetic predisposition, Western dietary pattern, and the risk of type 2 diabetes in men. Am J Clin Nutr 2009;89:1453–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi L, Liang J: Interactions between genetic factors that predict diabetes and dietary factors that ultimately impact on risk of diabetes. Curr Opin Lipidol 2009;21:31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolio TA, Bailey-Wilson JE, Collins FS: Genes, environment and the value of prospective cohort studies. Nat Rev Genet 2006;7:812–820 [DOI] [PubMed] [Google Scholar]
- 10.Frayling TM: Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet 2007;8:657–662 [DOI] [PubMed] [Google Scholar]
- 11.Grant RW, Moore AF, Florez JC: Genetic architecture of type 2 diabetes: recent progress and clinical implications. Diabetes Care 2009;32:1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I: New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson H: Genetic testing for everyone. Nature 2008;453:570–571 [DOI] [PubMed] [Google Scholar]
- 14.Hunter DJ, Khoury MJ, Drazen JM: Letting the genome out of the bottle–will we get our wish? N Engl J Med 2008;358:105–107 [DOI] [PubMed] [Google Scholar]
- 15.Positively disruptive. Nat Genet 2008;40:119. [DOI] [PubMed] [Google Scholar]
- 16.Burke W, Psaty BM: Personalized medicine in the era of genomics. JAMA 2007;298:1682–1684 [DOI] [PubMed] [Google Scholar]
- 17.Gulcher J, Stefansson K: Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2009;360:1360; author reply 360:1361 [PubMed] [Google Scholar]
- 18.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orru M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR: Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Psaty BM, O'Donnel CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JL, Uitterlinden AG, Harris TB, Witteman JCM, Boerwinkle E: Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from five cohorts. Circ Cardiovasc Genet 2009;2:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs DR, Jr, Meyer KA, Kushi LH, Folsom AR: Whole-grain intake may reduce the risk of ischemic heart disease death in postmenopausal women: the Iowa Women's Health Study. Am J Clin Nutr 1998;68:248–257 [DOI] [PubMed] [Google Scholar]
- 21.Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, Rao DC, Sprafka JM, Williams R: NHLBI Family Heart Study: objectives and design. Am J Epidemiol 1996;143:1219–1228 [DOI] [PubMed] [Google Scholar]
- 22.McKeown NM, Meigs JB, Liu S, Wilson PW, Jacques PF: Whole-grain intake is favorably associated with metabolic risk factors for type 2 diabetes and cardiovascular disease in the Framingham Offspring Study. Am J Clin Nutr 2002;76:390–398 [DOI] [PubMed] [Google Scholar]
- 23.Pereira MA, Jacobs DR, Jr, Pins JJ, Raatz SK, Gross MD, Slavin JL, Seaquist ER: Effect of whole grains on insulin sensitivity in overweight hyperinsulinemic adults. Am J Clin Nutr 2002;75:848–855 [DOI] [PubMed] [Google Scholar]
- 24.Murtaugh MA, Jacobs DR, Jr, Jacob B, Steffen LM, Marquart L: Epidemiological support for the protection of whole grains against diabetes. Proc Nutr Soc 2003;62:143–149 [DOI] [PubMed] [Google Scholar]
- 25.de Munter JS, Hu FB, Spiegelman D, Franz M, van Dam RM: Whole grain, bran, and germ intake and risk of type 2 diabetes: a prospective cohort study and systematic review. PLoS Med 2007;4:e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiota M, Postic C, Fujimoto Y, Jetton TL, Dixon K, Pan D, Grimsby J, Grippo JF, Magnuson MA, Cherrington AD: Glucokinase gene locus transgenic mice are resistant to the development of obesity-induced type 2 diabetes. Diabetes 2001;50:622–629 [DOI] [PubMed] [Google Scholar]
- 27.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 28.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, Tewhey R, Rieder MJ, Hall J, Abecasis G, Tai ES, Welch C, Arnett DK, Lyssenko V, Lindholm E, Saxena R, de Bakker PI, Burtt N, Voight BF, Hirschhorn JN, Tucker KL, Hedner T, Tuomi T, Isomaa B, Eriksson KF, Taskinen MR, Wahlstrand B, Hughes TE, Parnell LD, Lai CQ, Berglund G, Peltonen L, Vartiainen E, Jousilahti P, Havulinna AS, Salomaa V, Nilsson P, Groop L, Altshuler D, Ordovas JM, Kathiresan S: Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 2008;57:3112–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL: The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet 2009;18:4081–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong MY, Day NE, Luan JA, Wareham NJ: Estimation of magnitude in gene-environment interactions in the presence of measurement error. Stat Med 2004;23:987–998 [DOI] [PubMed] [Google Scholar]
- 31.Kristal AR, Peters U, Potter JD: Is it time to abandon the food frequency questionnaire? Cancer Epidemiol Biomarkers Prev 2005;14:2826–2828 [DOI] [PubMed] [Google Scholar]
- 32.Murcray CE, Lewinger JP, Gauderman WJ: Gene-environment interaction in genome-wide association studies. Am J Epidemiol 2009;169:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.