Abstract
CXCR7 is an atypical chemokine receptor that signals through β-arrestin in response to agonists without detectable activation of heterotrimeric G-proteins. Its cognate chemokine ligand CXCL12 also binds CXCR4, a chemokine receptor of considerable clinical interest. Here we report that TC14012, a peptidomimetic inverse agonist of CXCR4, is an agonist on CXCR7. The potency of β-arrestin recruitment to CXCR7 by TC14012 is much higher than that of the previously reported CXCR4 antagonist AMD3100 and differs only by one log from that of the natural ligand CXCL12 (EC50 350 nm for TC14012, as compared with 30 nm for CXCL12 and 140 μm for AMD3100). Moreover, like CXCL12, TC14012 leads to Erk 1/2 activation in U373 glioma cells that express only CXCR7, but not CXCR4. Given that with TC14012 and AMD3100 two structurally unrelated CXCR4 antagonists turn out to be agonists on CXCR7, this likely reflects differences in the activation mechanism of the arrestin pathway by both receptors. To identify the receptor domain responsible for these opposed effects, we investigated CXCR4 and CXCR7 C terminus-swapping chimeras. Using quantitative bioluminescence resonance energy transfer, we find that the CXCR7 receptor core formed by the seven-transmembrane domains and the connecting loops determines the agonistic activity of both TC14012 and AMD3100. Moreover, we find that the CXCR7 chimera bearing the CXCR4 C-terminal constitutively associates with arrestin in the absence of ligands. Our data suggest that the CXCR4 and CXCR7 cores share ligand-binding surfaces for the binding of the synthetic ligands, indicating that CXCR4 inhibitors should be tested also on CXCR7.
Keywords: Cancer Therapy, Chemokines, G Protein-coupled Receptors (GPCR), Mutant, Protein Domains, Signal Transduction, Beta-Arrestin, CXCL12, CXCR4, CXCR7
Introduction
CXCR4 is a seven-transmembrane domain (7TMR)4 chemokine receptor of considerable clinical interest, involved in stem cell homing to bone marrow niches, cancer biology and metastasis, and HIV infection. Synthetic CXCR4 ligands are being developed, and the CXCR4 blocker AMD3100 has reached the clinic for the mobilization of hematopoietic stem cells from donor bone marrow. The natural CXCR4 ligand, the chemokine CXCL12 (also called SDF-1), has later been found to also bind the atypical chemokine receptor CXCR7, which in addition recognizes the chemokine CXCL11 (also called I-TAC) (1, 2). CXCR7 is atypical in that no classical heterotrimeric G-protein signaling pathways are observed and in that it does not induce chemotaxis of motile cells. However, CXCR7 sets off G-protein-independent signaling via the β-arrestin pathway, leading to activation of the Erk 1/2 kinases (3, 4). Functionally, CXCR7 has been implicated in cancer cell growth and transendothelial migration (2, 5, 6). Intriguingly, CXCR7 and CXCR4 can heteromerize, and this attenuates CXCR4 G-protein signaling (7, 8). Modulation of CXCR4 has been suggested as a major role of CXCR7 (9).
Given that CXCR4 and CXCR7 share a chemokine ligand, we have previously tested the effects of AMD3100 on CXCR7 and found that it induces arrestin recruitment at high doses on this receptor, contrary to its effect on CXCR4 (3). This finding prompted us to investigate another, structurally unrelated CXCR4 inhibitor. T140 is a horseshoe crab polyphemusin-derived peptidomimetic described as an inverse agonist of CXCR4 (10–12). Here we report that TC14012, a serum-stable derivative of T140 (13), recruits β-arrestin 2 to CXCR7. Using C terminus-swapping mutants of CXCR4 and CXCR7, we demonstrate that the CXCR7 core receptor, formed by the transmembrane helices and the connecting loops, but not the CXCR7 C terminus, is the determinant for the agonist activity of structurally unrelated synthetic CXCR4 blockers on the arrestin pathway mediated by CXCR7.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant CXCL12 was from PeproTech, AMD3100 was from Sigma, and [125I]-CXCL12 was from PerkinElmer Life Sciences. TC14012 was synthesized as described (13).
Cell Culture and Transfections
Human embryonic kidney (HEK) 293E cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Wisent, Rocklin, CA), 100 units/ml penicillin/streptomycin, and 2 mm l-glutamine (Invitrogen). Transient transfections were performed in six-well dishes using the polyethylenimine (Polysciences, Warrington, PA) method.
Plasmids
CXCR4-YFP and CXCR7-YFP have been described previously (3). Plasmids encoding β-arrestin 2-Rluc (a generous gift of Michel Bouvier) have been described previously (15). To generate the C-terminal chimers, a unique BsiWI restriction site was inserted in CXCR4-YFP and CXCR7-YFP using the QuikChange Multi site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. The C-terminal domains were excised by BsiWI/NotI digestion and ligated into the respective opposite plasmid. The BsiWI site was then removed by site-directed mutagenesis, restoring the respective CXCR7 and CXCR4 sequences. The primers used for the site-directed mutagenesis were: CXCR4-mut916-BsiWI, 5′-CATCCTCTATGCTTTCCTCGTACGCAAATTTAAAACCTCTGCC-3′; CXCR7-mut955-BsiWI, 5′-CCCTGTCCTCTACAGCTTCATCGTACGCAACTACAGGTACGAGC-3′; CXCR4-X7Cter-WT, 5′-CATCCTCTATGCTTTCCTTAATCGCAACTACAGGTACGAGC-3′; CXCR7-X4Cter-WT, 5′-CCCTGTCCTCTACAGCTTCATCGGAGCCAAATTTAAAACCTCTGCCC-3′.
Radioligand Binding Assays
Cell membrane preparation and binding assays were performed as described previously (16) with minor modifications. Briefly, HEK293E cells expressing the respective receptor were washed once with PBS and subjected to one freeze-thaw cycle. Broken cells were then gently scraped in resuspension buffer (50 mm Hepes, pH 7.4, 1 mm CaCl2, and 5 mm MgCl2), centrifuged at 3500 × g for 15 min at 4 °C, and resuspended in binding buffer (50 mm Hepes, pH 7.4, 1 mm CaCl2 5 mm MgCl2, 140 mm NaCl, 0.5% BSA). For competition binding assays, broken cells (1 μg of protein) were incubated for 1 h at room temperature in binding buffer with 0.03 nm [125I]-SDF-1α as a tracer and increasing concentrations of competitor. Bound radioactivity was separated from free ligand by filtration, and receptor-bound radioactivity was quantified by γ-radiation counting.
BRET Measurements
β-Arrestin recruitment was measured by BRET essentially as described previously (17). HEK293T cells were cotransfected with 1 μg of receptor-eYFP construct with 0.05 μg of β-arrestin 2-Rluc. For [acceptor]/[donor] titrations, 0.05 μg of β-arrestin 2-Rluc was cotransfected with increasing amounts of the receptor-eYFP construct. All transfections were completed to 2 μg/well with empty vector. Following overnight culture, transiently transfected HEK293 cells were seeded in 96-well, white, clear bottom microplates (ViewPlate; PerkinElmer Life Sciences) coated with poly(d-lysine) and left in culture for 24 h. Cells were washed once with PBS, and the Rluc substrate coelenterazine h (NanoLight Technology, Pinetop, AZ) was added at a final concentration of 5 μm to BRET buffer (PBS, 0.5 mm MgCl2, 0.1% glucose). BRET readings were collected using a Mithras LB940 plate reader (Berthold Technologies, Bad Wildbad, Germany) and MicroWin2000 software. BRET measurement between Rluc and YFP was obtained by sequential integration of the signals in the 460–500 nm (Rluc) and 510–550 nm (YFP) windows. The BRET signal was calculated as the ratio of light emitted by acceptor (YFP) over the light emitted by donor (Rluc). The values were corrected to net BRET by subtracting the background BRET signal obtained in cells transfected with the Rluc construct alone. β-Arrestin recruitment was measured 30 min after ligand addition.
Flow Cytometric Analysis
Receptor cell surface expression was confirmed by flow cytometry using anti-CXCR7-APC (clone 358426) and anti-CXCR4-APC (clone 12G5, both from R&D Systems). Cells were washed three times in ice-cold PBS, resuspended, and stained with antibody for 30 min at 4 °C. After a final wash, the cells were resuspended in 0.5% paraformaldehyde and analyzed using a FACSCalibur Flow Cytometer (BD Biosciences).
Data Analysis
Data from BRET assays were the mean of independent experiments, each of which was performed in triplicate. Curve fitting by nonlinear regression and statistical analysis was conducted using GraphPad Prism 4 software (GraphPad Software Inc., San Diego, CA). Statistical significance of the differences between more than two groups was calculated by one-way analysis of variance followed by Tukey's post test.
RESULTS
β-Arrestin Recruitment to CXCR7 by TC14012
We previously found that a small molecule antagonist of CXCR4, AMD3100, acted as an agonist on CXCR7 in that it induced recruitment of β-arrestin 2 to the receptor, albeit with low potency. Based on this finding, we tested whether this property was shared by different CXCR4 inhibitors. We thus tested the ability of TC14012, a serum-stable derivative of the peptidomimetic T140, to induce recruitment of β-arrestin 2 to CXCR7, using a previously reported BRET-based experimental system (17). As shown in Fig. 1A, TC14012 was found to be a potent and efficient agonist of β-arrestin recruitment to CXCR7, with an apparent EC50 of 350 nm. This is almost 3 logs more potent than AMD3100 (EC50 of 138 μm) and approximately one log less potent than the efficiency of the cognate CXCR7 chemokine ligand CXCL12 in this system (30 nm). The EC50 is in line with the IC50 of TC14012 observed in radioligand displacement assays using HEK293 cells stably expressing CXCR7 and radiolabeled CXCL12 (Ki of 157 nm ± 36, n = 3, data not shown). These experiments show that the previously reported capacity of AMD3100 to recruit β-arrestin to CXCR7 is shared by a second, structurally unrelated CXCR4 antagonist. To further confirm signaling downstream of arrestin (4), we addressed Erk phosphorylation by TC14012 via CXCR7 in untransfected U373 glioma cells that express endogenous CXCR7 but no CXCR4, unlike HEK293 cells that express trace amounts of both receptors. TC14012, like CXCL12, leads to sustained Erk 1/2 phosphorylation in these cells (supplemental methods and Fig. S1).
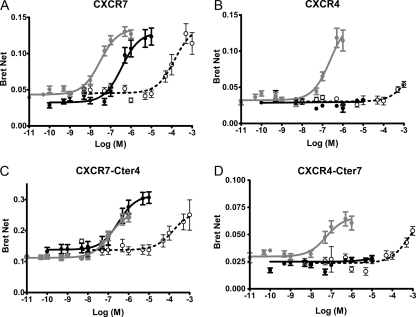
FIGURE 1.
Effect of natural and synthetic ligands on the β-arrestin recruitment to CXCR4, CXCR7, and receptor chimeras. HEK293 cells transiently coexpressing β-arrestin 2-RLuc as a BRET donor and respective receptors fused to the BRET acceptor YFP were stimulated with the indicated concentrations of CXCL12 (gray circles), TC14012 (black circles), or AMD3100 (open circles). Resulting BRET measurements are given as BRETnet. Data are mean values from 5–9 (CXCL12) or 3–7 (TC14012 and AMD3100) independent experiments, each performed in triplicate. Error bars indicate S.E. For statistical analysis, see Table 1. A, CXCR7; B, CXCR4; C, CXCR7-Cter4; D, CXCR4-Cter4.
Design and Expression of CXCR4-CXCR7 Chimeras
Although limited receptor selectivity of synthetic chemokine receptor ligands is not uncommon, we were intrigued by the finding that both shared ligands of CXCR4 and CXCR7 had antagonistic activity on CXCR4, whereas they agonistically induced β-arrestin recruitment to CXCR7. Our interpretation is that these divergent effects are not fortuitous but rather indicate differences between the two receptors in the activation mechanism of β-arrestin recruitment. Such differences might be due to differences in regulatory determinants for the recruitment of arrestin encoded by the receptor C termini. Alternatively, differences between the CXCR4 and CXCR7 cores could entail different ligand-induced receptor rearrangements that translate into inhibition of arrestin recruitment in one case but activation in the other.
To address this issue, we constructed chimeric CXCR4-CXCR7 receptors by exchanging the C-terminal domains of one receptor onto the other (named CXCR4-Cter7 and CXCR7-Cter4, supplemental Fig. S2A). All constructs were expressed at the cell surface and detected by respective monoclonal antibodies by flow cytometry (supplemental Fig. S2B).
β-Arrestin Recruitment to Chimeric Receptors Induced by Natural and Synthetic Ligands
Using the respective receptors/chimeras, we then tested β-arrestin recruitment induced by the different ligands in dose-response experiments (Fig. 1 and Table 1). Significantly higher BRET was observed with the CXCR7-Cter4 chimera in the absence of ligand, suggesting constitutive recruitment of arrestin by this chimera. Upon stimulation with TC14012, CXCR7 and the CXCR7-Cter4 chimera were able to recruit arrestin, whereas CXCR4 and CXCR4-Cter7 remained silent. A similar pattern was observed with AMD3100. These data show that it is the core of CXCR7, and not its C terminus, that is responsible for the CXCR7 response to the synthetic ligands.
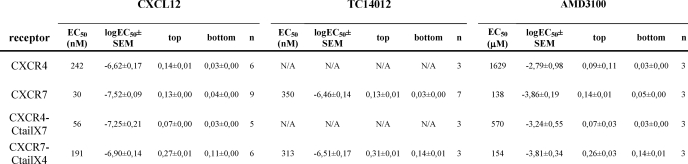
TABLE 1.
Curve-fitting parameters of arrestin recruitment to CXCR4, CXCR7, and the respective chimeras
The table summarizes the curve parameters of the activation of arrestin recruitment to the different receptors and chimera by CXCL12, TC14012, and AMD3100 shown in Fig. 1. The differences in EC50 upon stimulation with CXCL12 were statistically significant between CXCR4 and CXCR7 (p < 0.001); CXCR4 and CXCR4-Cter7 (p < 0.05); and CXCR7 and CXCR7-Cter4 (p < 0.01) (one-way analysis of variance with Tukey's post test). The difference of the curve bottom between CXCR7-Cter4 and all other receptors was also significant (p < 0.001). The difference of the curve bottom between CXCR7 and CXCR4 was not statistically significant. The very high EC50 values observed for AMD3100 (1.5 and 0.5 mm) on CXCR4 and CXCR4-Cter7 may reflect experimental artifacts at extreme doses of the compound (higher than those applied in our previous report (3)). Alternatively, they may represent weak agonist activity of AMD3100 also on CXCR4, in line with its previous description as a partial agonist (11, 19). NA, not applicable.
The use of quantitative BRET permitted additional observations concerning the respective responses to CXCL12. The responses of CXCR7 and the CXCR4-Cter7 chimera were significantly more potent (EC50 of 30 and 58 nm, respectively) than those of CXCR4 and the CXCR7-Cter4 chimera (EC50 of 242 and 191 nm, respectively) (CXCR4 versus CXCR7, p < 0.01; CXCR4 versus CXCR4-Cter7 and CXCR7 versus CXCR7-Cter4, p < 0.05; see also Table 1). This suggests that unlike the ability to respond to TC14012, the potency of the response to CXCL12 was determined by the respective C termini, possibly reflecting their effectiveness in translating ligand-induced conformational changes into arrestin recruitment. We cannot formally exclude the possibility that this was due to the C-terminal YFP BRET-fusion that might affect the regulatory function of this domain. However, our observation that the potency of arrestin recruitment to CXCR7 was identical in an alternative BRET system with unfused CXCR7 and a dual brilliance Rluc-arrestin-YFP fusion (18) (data not shown) speaks against a role of the fusion for potency and supports the idea that the receptor C terminus indeed determines the potency of the arrestin response.
Spontaneous Arrestin Recruitment to CXCR7-Cter4 in Absence of Ligand
To further evaluate the constitutive BRET signal yielded by the CXCR7-Cter4 chimera, we performed BRET acceptor/donor titrations in the absence and presence of 100 nm CXCL12. In the absence of chemokine, CXCR7-YFP titrations over β-arrestin 2-Rluc yielded a straight line representing increasing nonspecific bystander BRET (19), in line with the absence of baseline arrestin recruitment (supplemental Fig. S3). However, the CXCR7-Cter4 chimera yielded a saturable hyperbolic curve, in line with specific BRET resulting from spontaneous arrestin recruitment by this chimera. In curve-fitting analysis, the preferred model for the curve yielded by CXCR7-Cter4 in the absence of ligand was consistently hyperbolic (p < 0.001, n = 4), unlike for CXCR7, where the preferred model in the absence of ligand was a straight line. In the presence of 100 nm CXCL12, [acceptor]/[donor] titrations of both receptors yielded specific BRET as hyperbolic curves. The BRET50 is a measure of the propensity with which an interaction takes place (19). Remarkably, in simultaneous curve fittings, the BRET50 of the CXCR7-Cter4 mutant is significantly smaller in the presence of the chemokine than in its absence (p < 0.001 in 3 out of 4 experiments, and p < 0.01 in one 1 out of 4 experiments). This indicates that the constitutive activity of the CXCR7-Cter4 chimera is further activated by the presence of CXCL12.
DISCUSSION
The main finding of the present report is that the polyphemusin derivative TC14012, a CXCR4 inverse agonist (11, 12), also binds CXCR7 but acts here as an agonist of the arrestin pathway. Although this is similar to the previously reported agonist activity on CXCR7 of the structurally unrelated CXCR4 inhibitor AMD3100, TC14012 is a much more potent agonist on CXCR7 (EC50 of 350 nm for TC14012 versus 140 μm for AMD3100) and only one log weaker than the natural chemokine agonist CXCL12 (35 nm). Given that AMD3100 and TC14012 are structurally unrelated and that both receptors also share a natural ligand, we envision that the cross-reactivity of both synthetic ligands results from structural similarities of the ligand-binding surfaces of CXCR4 and CXCR7.
Lack of selectivity for one given chemokine receptor of synthetic ligands has hampered the development of drug candidates targeting chemokine receptors, and our results suggest that newly developed CXCR4 inhibitors should also be routinely tested on CXCR7. However, previous work with different T140 analogues (20) and the recent findings that the small molecule FC131 does not bind to CXCR7 (21) and does not induce arrestin recruitment to CXCR7 5 indicate that CXCR4 inhibitors do not inherently also bind CXCR7 and that receptor selectivity can be achieved. To our knowledge, synthetic chemokine receptor ligands that exert opposite effects on two different receptors are still unreported. Of note, receptor promiscuity being a hallmark of natural chemokine receptor ligands, such inverse action on different receptors also exists among natural chemokine receptor ligands, but the structural basis for these opposite effects remains yet unknown. Although nonselectivity of synthetic 7TMR ligands is generally seen as a drawback in drug development, simultaneous agonism on one receptor and antagonism on a second one might actually be of advantage in specific settings. This emerging concept especially applies to 7TMRs that functionally and/or physically interact and that share endogenous ligands. For example, simultaneous activation of the μ-opioid receptor and inhibition of the δ-opioid receptor are desired properties that have been shown to positively alter the side-effect profile (tolerance and dependence) of analgesics such as morphine, which activates both the μ-opioid receptor and the δ-opioid receptor (22). In this context, the documented physical and functional interactions between CXCR4 and CXCR7 (5, 7, 8) should be kept in mind. CXCR4 inhibition interferes with cancer biology at multiple steps including cancer cell growth and dissemination. Moreover, synthetic ligands of CXCR7, CCX451, CCX754, and CCX771 also reduce tumor growth and transendothelial migration (2, 6). Interestingly, rather than being a CXCR7 inhibitor, at least CCX771 turned out to be a potent activator of arrestin recruitment to CXCR7 (6). Taken together, these data suggest that simultaneous inhibition of CXCR4 and activation of CXCR7 might indeed be of interest in the context of cancer treatment.
The results obtained with CXCR4-CXCR7 C terminus-swapping chimera identified the receptor core as the determinant for the agonistic activity of TC14012 and AMD3100 on CXCR7. This finding was unexpected insofar as that for both CXCR4 and CXCR7, the C terminus is a crucial arrestin recruitment determinant. Phosphorylation of C-terminal serine residues promotes β-arrestin 2 recruitment to CXCR4 (14), and deletion of 43 CXCR7 C-terminal residues results in loss of arrestin recruitment (6). Our finding that the potency of arrestin recruitment in response to CXCL12 depends on the receptor C terminus is in line with a regulatory role of this domain. Although independence of arrestin recruitment from receptor phosphorylation (but still dependence on the receptor C terminus) has been described for some 7TMRs, among which is the chemokine scavenger receptor D6, which constitutively recruits β-arrestin (23), we find that CXCR7, which has also been suggested to be a chemokine scavenger receptor (24, 25), does not constitutively recruit β-arrestin. In this context, our finding that the CXCR7-Cter4 chimera associates with β-arrestin in the absence of ligand is intriguing and might reflect the overall greater proclivity of CXCR7 to recruit arrestin.
The opposite effects of TC14012 and AMD3100 on CXCR4 and CXCR7 thus reflect differences between their respective activation mechanisms of the arrestin pathway that are located in the receptor core. Despite growing information about interactions that contribute to binding of chemokines to their receptors (mostly involving the structured chemokine core and the receptor N terminus), the chemokine receptor determinants for activation still remain elusive. Similar to other studied chemokine-receptor couples, CXCR4 activation requires the flexible N terminus of CXCL12, and in particular, the lysine and proline residues in positions 1 and 2 (26). However, to date, and because of the lacking identification of receptor residues that are directly involved in interactions with the chemokine N terminus, only speculative models of chemokine receptor activation have been forwarded. This remains true despite the recent publication of the CXCR4 crystal structure in the presence of small antagonists (27) (PDB codes 3ODU, 3OE0, 3OE9, and 3OE6). With reference to earlier models (26), a recent report puts forward the hypothesis that insertion of the CXCL12 N terminus into the cavity formed by the CXCR4 transmembrane helices was required for activation, similar to binding pockets for small agonists of other class A 7TMRs (28). This is supported by data that show that AMD3100 prevents interaction of the CXCL12 N terminus with this cavity but not other receptor-chemokine interactions (28).
Following this model, our results indicate that CXCR7 activation does not require the CXCL12 N terminus interaction in the same way as does CXCR4 because activation by the chemokine is not blocked by the small molecule ligands. Rather, CXCR7 activation determinants actually overlap with the AMD3100-CXCR7 and TC14012-CXCR7 interaction determinants because both ligands promote CXCR7 activation. Intriguingly, with CXCR4, the interactions of both compounds have been mapped to the transmembrane domain/extracellular loop intersection rather than to the depth of the transmembrane crevice (11, 29, 30). At least for AMD3100, a similar binding mode to both CXCR4 and CXCR7 can be inferred by the conservation of CXCR4 key residues Asp-171 and Asp-262 for AMD3100 interaction in CXCR7. It is thus tempting to speculate that CXCR7 activation determinants are rather close to the surface, whereas those of CXCR4 are located deeper in the crevice of the receptor. More work about the respective activation mechanisms of CXCR4 and CXCR7 by CXCL12 will be needed to test this hypothesis.
Supplementary Material
Acknowledgments
We are indebted to Geneviève Saint-Onge, Marie-Ève Pelletier, and Guillaume Sylvian-Drolet for expert technical assistance.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, scholarships from the “Fondation de l'Hôpital Sainte-Justine” and the “Groupe de Recherche Universitaire sur le Médicament” (to S. G.), a grant from the “Fonds de la recherche en santé du Québec” (FRSQ) (to C. M.), and CIHR fellowships (to P. E. B. and Y. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and Figs. S1–S3.
S. Gravel and N. Heveker, unpublished data.
- 7TMR
- seven-transmembrane domain receptor
- BRET
- bioluminescence resonance energy transfer
- Rluc
- renilla luciferase.
REFERENCES
- 1.Balabanian K., Lagane B., Infantino S., Chow K. Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. (2005) J. Biol. Chem. 280, 35760–35766 [DOI] [PubMed] [Google Scholar]
- 2.Burns J. M., Summers B. C., Wang Y., Melikian A., Berahovich R., Miao Z., Penfold M. E., Sunshine M. J., Littman D. R., Kuo C. J., Wei K., McMaster B. E., Wright K., Howard M. C., Schall T. J. (2006) J. Exp. Med. 203, 2201–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalatskaya I., Berchiche Y. A., Gravel S., Limberg B. J., Rosenbaum J. S., Heveker N. (2009) Mol. Pharmacol. 75, 1240–1247 [DOI] [PubMed] [Google Scholar]
- 4.Rajagopal S., Kim J., Ahn S., Craig S., Lam C. M., Gerard N. P., Gerard C., Lefkowitz R. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann T. N., Grabovsky V., Pasvolsky R., Shulman Z., Buss E. C., Spiegel A., Nagler A., Lapidot T., Thelen M., Alon R. (2008) J. Leukoc Biol. 84, 1130–1140 [DOI] [PubMed] [Google Scholar]
- 6.Zabel B. A., Wang Y., Lewén S., Berahovich R. D., Penfold M. E., Zhang P., Powers J., Summers B. C., Miao Z., Zhao B., Jalili A., Janowska-Wieczorek A., Jaen J. C., Schall T. J. (2009) J. Immunol. 183, 3204–3211 [DOI] [PubMed] [Google Scholar]
- 7.Sierro F., Biben C., Martínez-Muñoz L., Mellado M., Ransohoff R. M., Li M., Woehl B., Leung H., Groom J., Batten M., Harvey R. P., Martínez-A. C., Mackay C. R., Mackay F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14759–14764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levoye A., Balabanian K., Baleux F., Bachelerie F., Lagane B. (2009) Blood 113, 6085–6093 [DOI] [PubMed] [Google Scholar]
- 9.Thelen M., Thelen S. (2008) J. Neuroimmunol. 198, 9–13 [DOI] [PubMed] [Google Scholar]
- 10.Tamamura H., Xu Y., Hattori T., Zhang X., Arakaki R., Kanbara K., Omagari A., Otaka A., Ibuka T., Yamamoto N., Nakashima H., Fujii N. (1998) Biochem. Biophys. Res. Commun. 253, 877–882 [DOI] [PubMed] [Google Scholar]
- 11.Trent J. O., Wang Z. X., Murray J. L., Shao W., Tamamura H., Fujii N., Peiper S. C. (2003) J. Biol. Chem. 278, 47136–47144 [DOI] [PubMed] [Google Scholar]
- 12.Zhang W. B., Navenot J. M., Haribabu B., Tamamura H., Hiramatu K., Omagari A., Pei G., Manfredi J. P., Fujii N., Broach J. R., Peiper S. C. (2002) J. Biol. Chem. 277, 24515–24521 [DOI] [PubMed] [Google Scholar]
- 13.Tamamura H., Omagari A., Hiramatsu K., Gotoh K., Kanamoto T., Xu Y., Kodama E., Matsuoka M., Hattori T., Yamamoto N., Nakashima H., Otaka A., Fujii N. (2001) Bioorg Med. Chem. Lett. 11, 1897–1902 [DOI] [PubMed] [Google Scholar]
- 14.Busillo J. M., Armando S., Sengupta R., Meucci O., Bouvier M., Benovic J. L. (2010) J. Biol. Chem. 285, 7805–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perroy J., Adam L., Qanbar R., Chénier S., Bouvier M. (2003) EMBO J. 22, 3816–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulais P. E., Dulude D., Cabana J., Heveker N., Escher E., Lavigne P., Leduc R. (2009) Biochem. Pharmacol. 78, 1382–1390 [DOI] [PubMed] [Google Scholar]
- 17.Hamdan F. F., Audet M., Garneau P., Pelletier J., Bouvier M. (2005) J. Biomol. Screen. 10, 463–475 [DOI] [PubMed] [Google Scholar]
- 18.Charest P. G., Terrillon S., Bouvier M. (2005) EMBO Rep. 6, 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercier J. F., Salahpour A., Angers S., Breit A., Bouvier M. (2002) J. Biol. Chem. 277, 44925–44931 [DOI] [PubMed] [Google Scholar]
- 20.Oishi S., Masuda R., Evans B., Ueda S., Goto Y., Ohno H., Hirasawa A., Tsujimoto G., Wang Z., Peiper S. C., Naito T., Kodama E., Matsuoka M., Fujii N. (2008) Chembiochem 9, 1154–1158 [DOI] [PubMed] [Google Scholar]
- 21.Narumi T., Hayashi R., Tomita K., Kobayashi K., Tanahara N., Ohno H., Naito T., Kodama E., Matsuoka M., Oishi S., Fujii N. (2010) Org Biomol Chem. 8, 616–621 [DOI] [PubMed] [Google Scholar]
- 22.Schiller P. W. (2010) Life Sci. 86, 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galliera E., Jala V. R., Trent J. O., Bonecchi R., Signorelli P., Lefkowitz R. J., Mantovani A., Locati M., Haribabu B. (2004) J. Biol. Chem. 279, 25590–25597 [DOI] [PubMed] [Google Scholar]
- 24.Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q., Raz E. (2008) Cell 132, 463–473 [DOI] [PubMed] [Google Scholar]
- 25.Naumann U., Cameroni E., Pruenster M., Mahabaleshwar H., Raz E., Zerwes H. G., Rot A., Thelen M. (2010) PLoS One 5, e9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crump M. P., Gong J. H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., Virelizier J. L., Baggiolini M., Sykes B. D., Clark-Lewis I. (1997) EMBO J. 16, 6996–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu B., Chien E. Y. T., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherzov V., Stevens R. C. (2010) Science, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kofuku Y., Yoshiura C., Ueda T., Terasawa H., Hirai T., Tominaga S., Hirose M., Maeda Y., Takahashi H., Terashima Y., Matsushima K., Shimada I. (2009) J. Biol. Chem. 284, 35240–35250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labrosse B., Brelot A., Heveker N., Sol N., Schols D., De Clercq E., Alizon M. (1998) J. Virol. 72, 6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerlach L. O., Skerlj R. T., Bridger G. J., Schwartz T. W. (2001) J. Biol. Chem. 276, 14153–14160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.