Abstract
Using a pharmacological inhibitor of Hsp90 in cultured malarial parasite, we have previously implicated Plasmodium falciparum Hsp90 (PfHsp90) as a drug target against malaria. In this study, we have biochemically characterized PfHsp90 in terms of its ATPase activity and interaction with its inhibitor geldanamycin (GA) and evaluated its potential as a drug target in a preclinical mouse model of malaria. In addition, we have explored the potential of Hsp90 inhibitors as drugs for the treatment of Trypanosoma infection in animals. Our studies with full-length PfHsp90 showed it to have the highest ATPase activity of all known Hsp90s; its ATPase activity was 6 times higher than that of human Hsp90. Also, GA brought about more robust inhibition of PfHsp90 ATPase activity as compared with human Hsp90. Mass spectrometric analysis of PfHsp90 expressed in P. falciparum identified a site of acetylation that overlapped with Aha1 and p23 binding domain, suggesting its role in modulating Hsp90 multichaperone complex assembly. Indeed, treatment of P. falciparum cultures with a histone deacetylase inhibitor resulted in a partial dissociation of PfHsp90 complex. Furthermore, we found a well known, semisynthetic Hsp90 inhibitor, namely 17-(allylamino)-17-demethoxygeldanamycin, to be effective in attenuating parasite growth and prolonging survival in a mouse model of malaria. We also characterized GA binding to Hsp90 from another protozoan parasite, namely Trypanosoma evansi. We found 17-(allylamino)-17-demethoxygeldanamycin to potently inhibit T. evansi growth in a mouse model of trypanosomiasis. In all, our biochemical characterization, drug interaction, and animal studies supported Hsp90 as a drug target and its inhibitor as a potential drug against protozoan diseases.
Keywords: Heat Shock Protein, Parasitology, Post-translational Modification, Protein-Drug Interactions, Protozoan, Geldanamycin, Hsp90, Plasmodium falciparum, Preclinical Study, Trypanosoma evansi
Introduction
In vivo functions of molecular chaperones are not just limited to helping newly synthesized proteins to fold but also include regulation of gene expression and signal transduction events (1). This is well exemplified by the cellular activities supported by heat shock protein 90 (Hsp90).4 By modulating the functions of key protein kinases and nuclear receptors, Hsp90 is known to regulate cell cycle progression and signal transduction (2). The cellular substrates modulated by Hsp90 include AKT, p53, telomerase, heat shock factor, and other transcription factors involved in cell signaling events (3). This chaperone has therefore been implicated in supporting important cellular events including cell growth, signaling, and development (4, 5).
The ability of Hsp90 to affect important cellular transformations is well exploited by intracellular protozoan parasites like Trypanosoma, Leishmania, Toxoplasma, and Plasmodium (3, 6, 7). All these human pathogens have been shown to utilize Hsp90 in triggering important stage transitions during their life cycles. Our laboratory has previously implicated Hsp90 from the malarial parasite Plasmodium falciparum in regulating its asexual development in human erythrocytes. Using a specific inhibitor of Hsp90 function, namely geldanamycin (GA) (8), we have shown P. falciparum Hsp90 (PfHsp90) to play a critical role in regulating ring to trophozoite stage transition in the parasite (7).
Hsp90s from different organisms show a high degree of similarity in their primary and higher structural organization. Despite overall sequence conservation, there is about 40% difference between PfHsp90 and human Hsp90 (hHsp90). The most significant difference is observed in the linker region of PfHsp90, which uniquely contains an additional 30-amino acid-long stretch adjoining the ATP binding domain. Recent studies suggest that the linker domain of Hsp90 affects its ATPase activity as well as its overall regulation (9, 10).
In this study, we have evaluated the potential of PfHsp90 as an antimalarial drug target. In addition to cloning, expression, and purification of full-length Pfhsp90, we have systematically characterized its biochemical properties and drug binding abilities in comparison with hHsp90. We found that PfHsp90 binds and hydrolyzes ATP more efficiently as compared with hHsp90. GA was found to bind purified PfHsp90 with high affinity and brought about robust inhibition of PfHsp90 ATPase activity. Importantly, we found a GA derivative to be effective as an antimalarial in a mouse model of malaria. We found GA-coupled beads to specifically pull down PfHsp90 from the whole cell lysate of the parasite demonstrating that its in vivo effect is specifically through PfHsp90. Mass spectrometric analysis of PfHsp90 from the parasite lysate revealed it to be acetylated at sites required for p23 and Aha1 binding indicating that acetylation may play a role in regulating the PfHsp90 chaperone complex and thus GA binding affinity. Most significantly, we found a GA derivative, 17-(allylamino)-17-demethoxygeldanamycin (17AAG), to inhibit parasite growth in vitro and in a preclinical rodent model of malaria. We further extended our study to another protozoan parasite, Trypanosoma evansi, which causes surra in domestic animals. We were able to demonstrate specific binding of GA to purified T. evansi Hsp90 (TeHsp90) in whole cell lysate as well as in its purified form. Importantly, 17AAG was able to inhibit T. evansi growth and cure mice infected with T. evansi. In all, our studies support the potential of PfHsp90 and TeHsp90 as drug targets and also suggest the possibility of targeting Hsp90 of protozoan parasites for the treatment of a variety of human and animal infections.
EXPERIMENTAL PROCEDURES
Cloning and Purification of Recombinant PfHsp90
For cloning PfHsp90, cDNA was prepared from total RNA isolated from parasite cultures by RT-PCR. PfHsp90 was amplified using the following primers: CGGGATCCAAATGTCAACGGAAACATTCG (sense) and GGAATTCTTAGTCAACTTCTTCCATTTTA (antisense). The 2238-bp product was cloned into pGEM-T and subcloned into pRSETA. R98K PfHsp90 mutant was generated by PCR-based site-directed mutagenesis.
TeHsp90 was similarly cloned by preparing cDNA from total RNA isolated from T. evansi. TeHsp90 was amplified using the following primers: GGATCCATGACGGAGACATTCGCATTCC (sense) and GAATTCTCAGTCCACTTCCTC (antisense). The amplicon was cloned into pRSETA. For purification, the recombinant wild type and mutant His-tagged PfHsp90 and TeHsp90 were expressed in Escherichia coli Rosetta strain and purified using nickel-nitrilotriacetic acid affinity chromatography (Qiagen).
Kd Determination for GA and ATP Using Fluorescence Spectroscopy
To determine the binding affinity of ATP, 25 μg/ml PfHsp90 in 40 mm HEPES-KOH buffer, pH 7.4, 5 mm MgCl2, and 100 mm KCl was incubated with different concentrations of ATP (50–1000 μm). For estimating the binding affinity of GA, 25 μg/ml PfHsp90, R98K PfHsp90, hHsp90, or TeHsp90 in 50 mm Tris and 1 mm EDTA was incubated with different concentrations of GA (100 nm–10 μm). The final concentration of DMSO in the assay was 1%. Tryptophan fluorescence measurements were carried out by scanning the emission spectrum in the wavelength range of 300–400 nm. The wavelength of excitation spectrum used was 280 nm. The intensity at λmax 346 nm was selected for calculations. The intensity of protein quenched with GA was subtracted from the intensity of pure protein. This difference in intensity was then plotted against the concentration of GA on the x axis. The resultant hyperbolic curve was analyzed with GraphPad® Prism 5 statistical software using non-linear regression analysis with single site-specific binding. A similar procedure was used to determine the binding affinity for ATP.
ATPase Assay
The ATPase assay was carried out with 1.5 μm PfHsp90/hHsp90 in 40 mm HEPES-KOH, 100 mm KCl, and 5 mm MgCl2, pH 7.5 (buffer A) as described previously (11). The concentration of ATP was varied from 50 to 4000 μm in a final reaction volume of 10 μl. [γ32P*]ATP (specific activity, 0.55 Ci/mmol) was used as a tracer. To rule out any nonspecific contribution to ATPase activity, 30 μm GA was added to the control (12). The residual value of ATPase was subtracted from total activity to get the ATPase activity of Hsp90.
Determination of IC50 Value of Hsp90 ATPase Activity
The IC50(ATPase) assay was carried out in buffer A in a similar manner as for the determination of Hsp90 ATPase activity except that for the IC50 assay purified PfHsp90/hHsp90 was incubated with a constant concentration of ATP in the presence of different concentrations of GA. A control experiment with 30 μm GA was done. The percentage of residual ATPase activity was then plotted against the log of the inhibitor concentration, and the result was analyzed using GraphPad Prism 5.0.
In Vivo Acetylation of PfHsp90
For determination of in vivo acetylation, asynchronous P. falciparum-infected erythrocytes were incubated in RPMI 1640 medium with 10% serum with or without 500 nm trichostatin A (TSA) for 8 h at 37 °C. The cells were washed twice in PBS, and the parasites were released by saponin lysis. Saponin-released parasites were briefly sonicated in modified radioimmune precipitation assay buffer (50 mm Tris, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 10 mm sodium butyrate, 5 mm sodium acetate, 1 mm EDTA, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). For identification of sites of acetylation on PfHsp90, proteins from TSA-treated cells were separated by SDS-PAGE, and the band corresponding to PfHsp90 was excised and subjected to mass spectrometry analysis.
Gel Filtration Analysis
P. falciparum-infected erythrocytes were incubated in RPMI 1640 medium with 10% serum with or without 500 nm TSA for 8 h at 37 °C. Saponin-released parasites were briefly sonicated in 20 mm sodium phosphate, pH 7.4 containing protease inhibitors and passed through a Superdex 200 column (Amersham Biosciences) at a flow rate of 0.5 ml/min. Fractions were collected and analyzed by immunoblotting using anti-PfHsp90. Thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (220 kDa), and bovine serum albumin (66 kDa) were used as molecular size standards.
In-gel Digestion
The band corresponding to PfHsp90 or TeHsp90 was cut from the SDS-PAGE gel after colloidal Coomassie staining and further sliced into smaller gel plugs. After several washes with 100 mm ammonium bicarbonate (NH4HCO3) (Sigma-Aldrich) buffer in 50% acetonitrile, the gel plugs were subjected to a reduction step using 10 mm dithiothreitol (DTT) (Sigma-Aldrich) in 100 mm NH4HCO3 buffer (45 min at 56 °C). Alkylation was performed with a solution of 55 mm iodoacetamide (Sigma-Aldrich) in 100 mm NH4HCO3 (30 min at room temperature in the dark) followed by in-gel digestion with 20 μl of trypsin (10 ng/μl) (Promega) in 50 mm NH4HCO3 (overnight at 37 °C). The reaction was stopped by storing at −20 °C, and peptides were extracted in 5% formic acid. Samples were vacuum-dried and reconstituted in 5% formic acid.
Mass Spectrometry and Database Searching
The protein digest was analyzed by automated nanoflow LC-MS/MS. The sample was loaded onto a PepMap C18 reverse phase column connected to Tempo nano-HPLC system. The peptides were eluted from the analytical column by a linear gradient of 95% Solvent A (98% H2O, 2% acetonitrile, and 0.1% formic acid) to 10% Solvent A and Solvent B (2% H2O, 98% acetonitrile, and 0.1% formic acid). The spectra were acquired on a Q-STAR Elite mass spectrometer equipped with an Applied Biosystems NanoSpray II ion source. The data were acquired in a data-dependent mode with one MS spectrum followed by three MS/MS spectra. Data analysis was performed using Analyst QS 2.0 software. For identification of proteins, the processed data were searched against the NCBInr database using the Mascot protein identification tool (version 2.0) with precursor and fragment mass tolerance of 0.15 Da; cysteine carbamidomethylation as a fixed modification; and methionine oxidation, lysine acetylation, and glutamine and asparagine deamidation as variable modifications. The resulting MS/MS-based peptide identifications were manually verified using ProteinPilot 2.0 software.
Cytotoxicity Assay
P. falciparum (3D7 laboratory strain) growth was assessed by measuring the incorporation of [3H]hypoxanthine (13). The inhibitor was serially diluted into hypoxanthine-free complete RPMI 1640 medium ranging from 10 nm to 10 μm. 200 μl of synchronized rings were added to a microtiter plate that was subsequently incubated at 37 °C for 48 h. This was followed by 18-h incubation with 1 μCi of [3H]hypoxanthine (specific activity, 10–30 mCi/mmol). The radioactivity was counted in a scintillation counter, and the mean values for [3H]hypoxanthine incorporation were calculated. IC50(Growth) values were determined by plotting the percentage of incorporation of [3H]hypoxanthine against inhibitor concentrations in logarithmic scale.
Clinical isolates were acquired from Malaria Research Centre, Delhi, India. Clinical isolates used in this study were originally collected from a patient exhibiting complicated malaria (RK1) from Rourkela, India and a patient exhibiting uncomplicated malaria (DL1) from Delhi, India. Clinical isolates were maintained in continuous culture using A+ human erythrocytes in RPMI 1640 medium supplemented with 10% human AB+ serum with 5% hematocrit. The growth inhibitory assay was done as described for the 3D7 laboratory strain.
GA Pulldown Assay
GA-coupled beads were prepared as described previously by Pavithra et al. (18). P. falciparum-infected erythrocytes were freed using saponin treatment followed by lysis in TNESV buffer containing protease inhibitors (50 mm Tris, pH 7.5, 0.1% Nonidet P-40, 2 mm EDTA, 100 mm NaCl, and 1 mm sodium orthovanadate). T. evansi was purified from infected mice using DEAE-cellulose (14) and lysed in TNESV buffer. Precleared lysates from both parasites were incubated separately with GA-coupled beads or control beads, and samples were analyzed by SDS-PAGE or two-dimensional gel electrophoresis.
In Vivo Antimalarial and Antitrypanosomal Activity of GA Analog 17AAG
For determination of antimalarial activity, female Swiss mice (22–25 g) were infected intraperitoneally with Plasmodium berghei. After confirmation of infection by Giemsa-stained tail smears, mice were divided into two groups, each having four mice. 17AAG was dissolved in 20% Cremophor® EL (Sigma) in PBS (50 mg/kg of body weight) and injected intraperitoneally for 4 consecutive days. Vehicle-treated infected mice served as controls. Survival of mice was monitored for a period of 3 weeks. Every alternate day tail smears were taken, and the number of infected RBCs was counted and plotted against days postinfection. Percent survival was plotted as a function of time.
Similarly for antitrypanosomal activity, Swiss female mice were infected with 105 purified T. evansi and divided into two groups, each having five mice. These groups of mice were treated with either 30 mg/kg of body weight 17AAG or vehicle and were scored for survivability. Experimental studies with mice were conducted adhering to the institution's guidelines for animal husbandry at the Indian Institute of Science.
RESULTS
PfHsp90 Is a Hyperactive ATPase
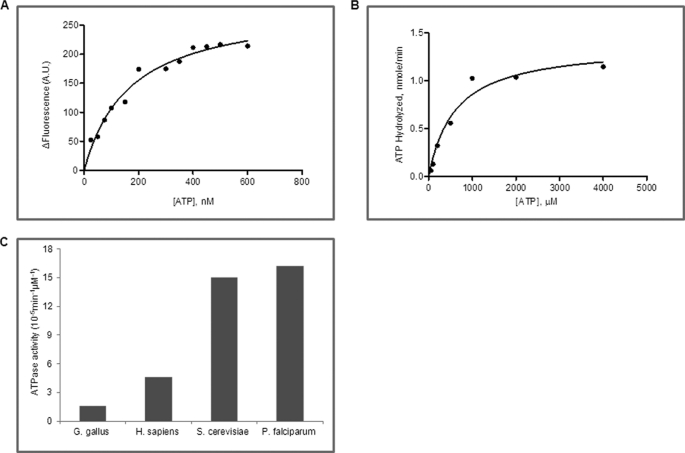
All Hsp90s are known to bind ATP and possess a weak ATPase activity. ATP-bound and -free forms of Hsp90 exhibit measurable differences in tryptophan fluorescence intensity, and this property has been exploited to quantify the binding affinity of Hsp90 for ATP. PfHsp90 was purified to homogeneity (supplemental Fig. S1) and incubated with varying concentrations of ATP, and the difference in tryptophan fluorescence was plotted against increasing concentrations of ATP. The dissociation constant was determined by analyzing the data using GraphPad Prism 5.0 as described under “Experimental Procedures.” The dissociation constant (Kd) of ATP with PfHsp90 was determined to be 168 ± 25 μm (Fig. 1A and Table 1). Using the same experimental approach, previous studies found ATP to bind human Hsp90 and yeast Hsp90 with dissociation constants of 240 ± 14 and 132 ± 47 μm, respectively (Table 1) (11, 12). This result suggests that PfHsp90 binds ATP with a 30% higher affinity as compared with its human host.
FIGURE 1.
PfHsp90 is a hyperactive ATPase. A, determination of binding affinity of ATP to PfHsp90 using tryptophan fluorescence. The change in fluorescence (ΔF) was plotted against ATP concentrations. B, rate of ATP hydrolysis was measured by the direct conversion of radiolabeled ATP to ADP. A Michaelis-Menten plot shows the fractional cleavage of γ-32P-labeled ATP plotted against ATP concentration. C, a comparison of ATP hydrolysis of Hsp90 from chicken (Gallus gallus), human (Homo sapiens), yeast, and Plasmodium. Plasmodium Hsp90 has higher ATPase activity as compared with human and chicken and is similar to yeast Hsp90. A.U., arbitrary units.
TABLE 1.
Biochemical properties of Hsp90 from different organisms
| Organism | ATP |
Hsp90 ATPase IC50 | GA Kd | |||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Kd | |||
| μm | min−1 | min−1 μm−1 | μm | nm GA | μm | |
| P. falciparum | 611 | 9.9 × 10−2 | 16.2 × 10−5 | 168 ± 25 | 207 | 1.05a |
| H. sapiens | 324b | 1.5 × 10−2b | 4.6 × 10−5b | 240 ± 14c | 702 | 4.4,a 0.480,d,e >1f |
| S. cerevisiae | 511b | 8.0 × 10−2b | 15.6 × 10−5b | 132 ± 47g | NDh | 1.2i,j |
| G. gallus | 1530b | 2.5 × 10−2b | 1.6 × 10−5b | ND | ND | ND |
a Data obtained by fluorescence spectroscopy.
b Data from the work done by Owen et al. (15).
c Data from the work done by McLaughlin et al. (11).
d Data from the work done by Onuoha et al. (39).
e Data obtained by fluorescence anisotropy using 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene geldanamycin.
f Data from the work done by Gooljarsingh et al. (40).
g Data from the work done by Prodromou et al. (12).
h ND, not determined.
i Data from the work done by Roe et al. (41).
j Data obtained by isothermal titration calorimetry.
To examine the ATPase activity of PfHsp90, we incubated the purified recombinant protein with different concentrations of [γ-32P*]ATP and monitored the direct conversion of [γ-32P*]ATP to 32Pi* by thin layer chromatography. As observed from Fig. 1B, the ATPase activity of PfHsp90 follows Michaelis-Menten kinetics. PfHsp90 hydrolyzed ATP with a Km of 611 μm and kcat of 9.9 × 10−2 min−1. Therefore, the catalytic efficiency (kcat/Km) for PfHsp90 is 16.2 × 10−5 min−1 μm−1. Interestingly, the ATPase activity of PfHsp90 was 6 times higher than that reported for its human host indicating that parasite Hsp90 is highly active (Fig. 1C and Table 1) (15).
GA Binds to Purified PfHsp90 with High Affinity
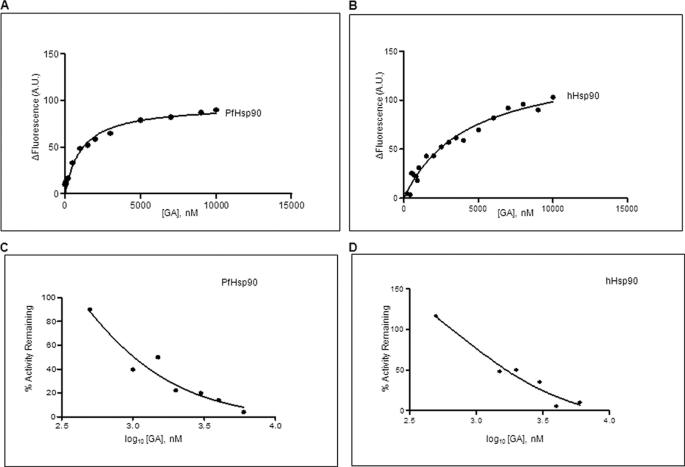
GA is known to bind to the amino-terminal domain of Hsp90, an ATP binding site, inhibiting Hsp90 function by blocking substrate maturation and subsequently leading to its degradation. Furthermore, despite overall similarity in the GA binding domains of Hsp90s from different organisms, there is a discrepancy in the binding ability of GA. A recent study in free living nematode showed that its Hsp90 is unable to bind GA, whereas Hsp90 of the obligate nematode parasite binds to GA (16, 17). The binding affinity of GA to Hsp90 from human host is well established, but its interaction with the malaria parasite Hsp90 has not been elucidated. Therefore, we decided to examine the binding ability of P. falciparum Hsp90 to GA and compare it with host Hsp90. Equal concentrations of purified proteins from parasite and human host were incubated with varying concentrations of GA, and the difference in tryptophan fluorescence was plotted against increasing concentrations of GA (Fig. 2, A and B). The dissociation constant was obtained by analyzing the resulting data using non-linear regression for single site-specific binding using GraphPad Prism 5.0. As shown in Table 1, the dissociation constants of GA for PfHsp90 and hHsp90 were found to be 1.05 and 4.4 μm, respectively. We also explored the slow binding nature of GA by determining binding affinity at longer time points of incubation with PfHsp90. However, because of the instability of purified PfHsp90 on prolonged incubation at ambient temperature, we were unable to determine accurate binding affinity under these conditions.
FIGURE 2.
GA potently inhibits PfHsp90 ATPase activity. A and B, determination of binding affinity of GA toward PfHsp90 and hHsp90 using tryptophan fluorescence. The change in fluorescence (ΔF) was plotted against GA concentrations for PfHsp90 (A) and hHsp90 (B). C and D, the IC50(ATPase) of PfHsp90 (F) and hHsp90 (G) ATPase activity was determined by plotting percent activity remaining versus GA concentration in logarithmic scale. A.U., arbitrary units.
To examine any differences in amino acid sequence that could explain the slight increase in the binding affinity of GA to PfHsp90, we compared the primary structure of PfHsp90 and its human counterpart, hHsp90. As shown in supplemental Fig. S2, the residues required for GA binding were conserved in PfHsp90 except for a homologous substitution of Lys-112 in hHsp90 to Arg-98 in PfHsp90. To explore the possible involvement of this homologous substitution on GA binding, we mutated Arg-98 to Lys. The R98K mutant did not show any significant difference in the binding affinity of GA suggesting that this homologous substitution has minimal or no effect on drug-protein interactions.
PfHsp90 Is Hypersensitive to GA-mediated Inhibition of Its ATPase Activity
GA exerts its inhibitory effect by inhibiting the ATPase cycle of Hsp90 that is mandatory for its function. We next examined the extent of GA-mediated inhibition of Hsp90 from malaria parasites and human host. For this, we monitored conversion of [γ-32P]ATP to 32Pi* by incubating equal concentrations of either PfHsp90 or hHsp90 with increasing concentrations of GA. The percentage of remaining Hsp90 ATPase activity was calculated and plotted against log10[GA] (Fig. 2, C and D). The IC50(ATPase) of GA for PfHsp90 (207 nm) was found to be about 3 times lower than that for hHsp90 (702 nm) indicating that PfHsp90 is more sensitive to GA-mediated inhibition.
GA Specifically Pulls Down PfHsp90 from P. falciparum Lysate
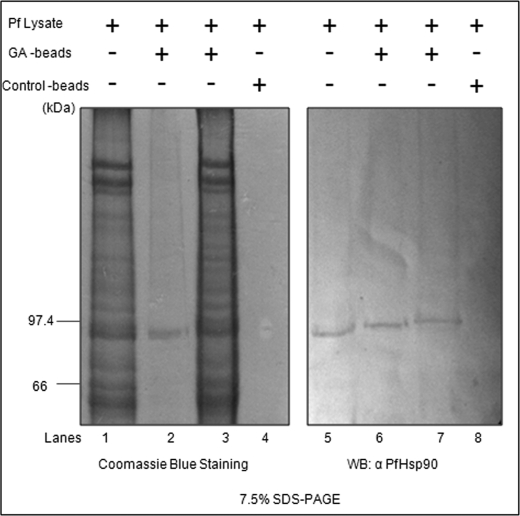
Previously, we have shown that GA inhibits P. falciparum growth in erythrocytes and binds to PfHsp90. To establish that the inhibitory effect of GA is mediated by its interaction with PfHsp90 only, we carried out an in vivo inhibitor pulldown assay. For this, GA was coupled to N-hydroxysuccinimide-Sepharose beads as described previously by Pavithra et al. (18). Saponin-freed parasites were lysed in TNESV buffer containing protease inhibitors. Precleared P. falciparum lysate was incubated overnight with GA-coupled beads at 4 °C. Lysate incubated with uncoupled beads served as a control. The bound proteins were analyzed by SDS-PAGE. As shown in Fig. 3, control beads did not show any signal, whereas GA-coupled beads showed a band corresponding to an 86-kDa protein, which was confirmed as PfHsp90 after immunoblotting using antibody specific to PfHsp90.
FIGURE 3.
GA specifically binds to PfHsp90. Parasite lysate was incubated with GA-coupled beads or control beads. Bound (lanes 2 and 6) and unbound proteins (lanes 3 and 7) were analyzed by SDS-PAGE followed by Coomassie staining (left side) or immunoblotting using anti-PfHsp90 (right side). WB, Western blot.
PfHsp90 Acetylation and Its Influence on Multichaperone Complex
Previous studies in cancer cells have shown that the sensitivity of cancer cells to Hsp90 inhibitors is influenced by acetylation of Hsp90 as well as the expression levels of its co-chaperones p23 and Aha1 (19–21). The acetylation status of PfHsp90 has not been examined previously. We decided to address two questions. 1) Does PfHsp90 undergo acetylation during asexual multiplication of the parasite in human erythrocytes, and 2) if yes, will acetylation status of PfHsp90 affect its multichaperone complex stability? Parasite culture was treated with histone deacetylase inhibitor TSA for 8 h to enrich acetylated PfHsp90 and lysed as described under “Experimental Procedures.” To confirm that TSA treatment increases the acetylation level of PfHsp90, we carried out immunoprecipitation of TSA-treated sample and control sample using anti-PfHsp90 antibody followed by immunoblotting using either αPfHsp90 antibody or anti-acetyllysine antibody. We found an increase in the acetylation level of PfHsp90 in the sample treated with TSA. This suggested that inhibition of histone deacetylase using TSA increased the acetylation of PfHsp90 (supplemental Fig. S3). To determine the identity of the lysine residue that undergoes acetylation, the lysate of TSA-treated parasites was resolved by SDS-PAGE, and the band corresponding to PfHsp90 was subjected to in-gel trypsinization. The resulting peptides were analyzed on a nano-LC/quadrupole TOF instrument to examine potential post-translational modifications. Peak list data obtained from MS were searched against the NCBI protein database using the Mascot search algorithm, and PfHsp90 was found to be the first hit with a score of 3201 and 46% coverage (supplemental Fig. S4).
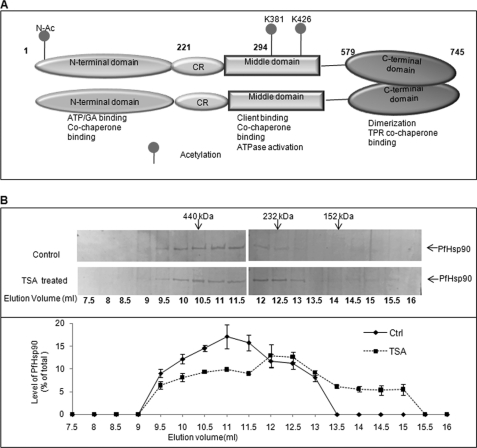
Furthermore, analysis of the MS/MS spectra using ProteinPilot 2.0 identified three sites of acetylation on PfHsp90. In addition to amino-terminal acetylation at a serine residue, PfHsp90 was also acetylated at Lys-381 and Lys-426 (in its middle domain) (supplemental Fig. S5, A–C) (Fig. 4A). Although human Hsp90 is also reported to be acetylated (22), the sites of acetylation do not coincide with those for PfHsp90 found in this study. Interestingly, the interaction site of Hsp90 with co-chaperones p23 and Aha1, as reported in yeast, was found to coincide with Lys-426 of PfHsp90 (supplemental Fig. S6, A and B) (23, 24). An overlap in the site of co-chaperone binding and acetylation suggested a possible involvement of acetylation in regulating co-chaperone binding and thus the formation of Hsp90 chaperone complex.
FIGURE 4.
Acetylation disrupts PfHsp90 complex. PfHsp90 acetylation status was determined by treating parasite-infected erythrocytes using histone deacetylase inhibitor TSA. A, domain distribution of acetylation sites in PfHsp90. CR, charged linker region; TPR, tetratricopeptide repeat. B, P. falciparum-infected erythrocytes were treated/untreated with histone deacetylase inhibitors. Parasite lysate was prepared as described under “Experimental Procedures” and analyzed using a Superdex 200 gel filtration column. 500-μl fractions were collected and immunoblotted for PfHsp90 (top panel). The bottom panel shows quantitation of the profile. Error bars have been represented to indicate standard error from three independent experiments.
We have previously shown that PfHsp90 complex fractionates around 440 kDa (7, 18). To examine the effect of acetylation on PfHsp90 multichaperone complex, gel filtration analysis was performed for TSA-treated parasites as described under “Experimental Procedures.” The fractions were collected, acetone-precipitated, and analyzed by SDS-PAGE followed by immunoblotting with an antibody specific to PfHsp90. As shown previously, the control sample fractionated around 440 kDa. However, for sample that was treated with TSA, the elution profile of the complex shifted toward the lower mass range with peak intensity around 232 kDa (Fig. 4B). This suggested that acetylation played an important role in regulating the composition of PfHsp90 chaperone complex.
Furthermore, to examine whether TSA treatment can sensitize the parasite for GA, we performed in vitro drug interaction by treating Plasmodium-infected erythrocytes with GA and TSA in combination and individually as described in the supplemental experimental procedures. Fractional inhibitory concentrations of the two drugs at various ratios were determined. We found an additive and synergistic interaction between GA and TSA in inhibiting Plasmodium growth (supplemental Table 1 and supplemental Fig. S7).
GA Analog 17AAG Inhibits P. falciparum Growth in Vitro
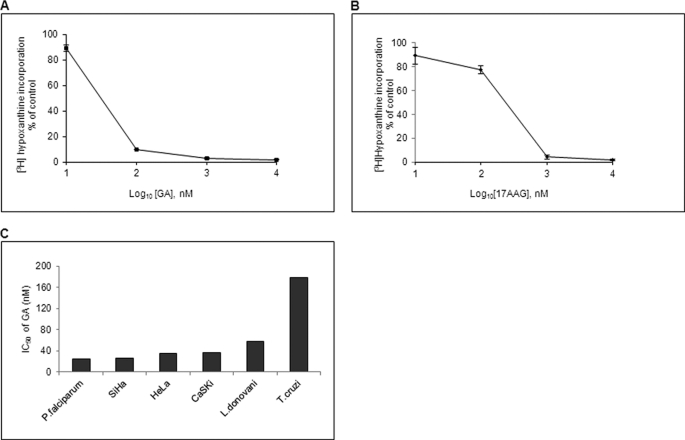
17AAG, a soluble analog of GA, has been shown to be effective in cancer patients because of its lesser hepatotoxicity, and it is believed to be more promising than GA despite its lower potency in vitro. Although the inhibitory effect of GA in parasite development has been shown before, the effect of 17AAG has not been explored. Therefore, we decided to quantitatively analyze the growth inhibition conferred by 17AAG in addition to GA. We used a [3H]hypoxanthine incorporation assay to determine the cytotoxic effect of GA and 17AAG. As shown in Fig. 5, A and B, with an increase in concentration of GA or 17AAG, there was a corresponding decrease in the incorporation of [3H]hypoxanthine in P. falciparum (supplemental Fig. S8, A and B). The concentrations of GA and 17AAG required to cause 50% inhibition in [3H]hypoxanthine incorporation (IC50(Growth)) were found to be 25 and 160 nm, respectively, after 48 h. Even though 17AAG showed less potency as compared with GA, its IC50 value in P. falciparum was comparable with cancerous cells indicating its potential as an antimalarial. A comparison of IC50(Growth) values of GA for P. falciparum determined in this study with those reported for cancer cells (SiHa, HeLa, and CaSig) and other protozoan parasites such as Leishmania donovani and Trypanosoma cruzi (3, 25, 26) indicated that P. falciparum is more sensitive toward GA (Fig. 5C).
FIGURE 5.
Hsp90 inhibitors potently inhibit parasite growth. A and B, IC50 values for GA and 17AAG in the P. falciparum 3D7 strain were determined using a [3H]hypoxanthine incorporation assay. IC50 values were determined by plotting the percentage of incorporation of [3H]hypoxanthine against different concentrations of drug in logarithmic scale. A, percentage of [3H]hypoxanthine incorporation upon GA treatment for 48 h. B, percentage of [3H]hypoxanthine incorporation upon 17AAG treatment for 48 h. C, comparison of the IC50 values among different organisms. Error bars have been represented to indicate standard error from three independent experiments.
We further determined the efficacy of GA and 17AAG in inhibiting the growth of parasites collected from malaria patients. GA and 17AAG were indeed able to inhibit growth of malaria parasites isolated from patients exhibiting complicated (RK1) as well as uncomplicated (DL1) malaria. GA exhibited IC50(Growth) values of 20 nm for RK1 and 5 nm for DL1 after 48 h. 17AAG exhibited IC50(Growth) values of 150 nm for RK1 and 50 nm for DL1 after 48 h of treatment. The above results indicated that both GA and 17AAG effectively inhibit the growth of the clinical parasites in addition to 3D7 laboratory culture.
In Vivo Efficacy of PfHsp90 Inhibitors in Rodent Model of Malaria
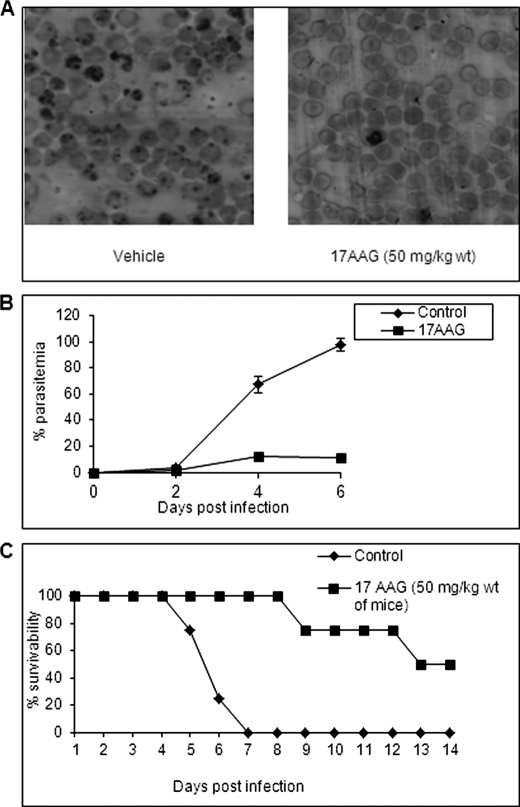
We next determined the ability of 17AAG to inhibit parasite growth in P. berghei-infected mice. For this, Peter's 4-day suppressive test against P. berghei infection in mice was performed as described under “Experimental Procedures.” Fig. 6A is a representative Giemsa-stained tail smear of 17AAG-untreated and -treated mice. As evident from the smear, 17AAG was able to inhibit parasite growth. Fig. 6B shows the percentage of parasitemia observed in the control and drug-treated experimental mice until the 6th day following infection when the majority of control mice had succumbed to infection. It is evident that in the control group the parasitemia rose steadily peaking at 80–90% until the death of the animal, whereas in drug-treated mice, the parasitemia was significantly attenuated resulting in about 2-fold prolonged survival of the drug-treated mice (Fig. 6C). An overall survival rate of 40–50% in 17AAG-treated as compared with 0% in vehicle-treated animals was observed 21 days postinfection. The “p” value for this experiment was found to be 0.00692 (p < 0.01) by log rank test. In an independent experiment, we examined the efficacy of 17AAG in a group of 10 mice. Here too we found a significant protection in 17AAG-treated mice (33% survival as compared with none in control mice at the end of the 11th day postinfection). The p value for this experiment was again found to be below 0.001 (supplemental Fig. S9). The low p value suggests that the difference as observed in the survivability curve is not merely chance but is an outcome of drug treatment. Although much further refinement is necessary in optimizing the efficacy of the GA derivative in an animal model, the results shown here provide a proof of principle for the efficacy of an Hsp90 inhibitor as an antimalarial in the preclinical rodent model of malaria.
FIGURE 6.
Efficacy of 17AAG, a GA derivative, in rodent model of malaria. A, representative Giemsa-stained tail smears of 17AAG-untreated (left) and -treated (right) P. berghei-infected mice after 6 days postinfection. B, plot of percentage of parasitemia against the number of days postinfection. (C) Percentage viability of 17AAG treated/untreated mice plotted against time. Error bars have been represented to indicate standard error from three independent experiments.
Hsp90 Inhibitor as a Cure for Trypanosomiasis
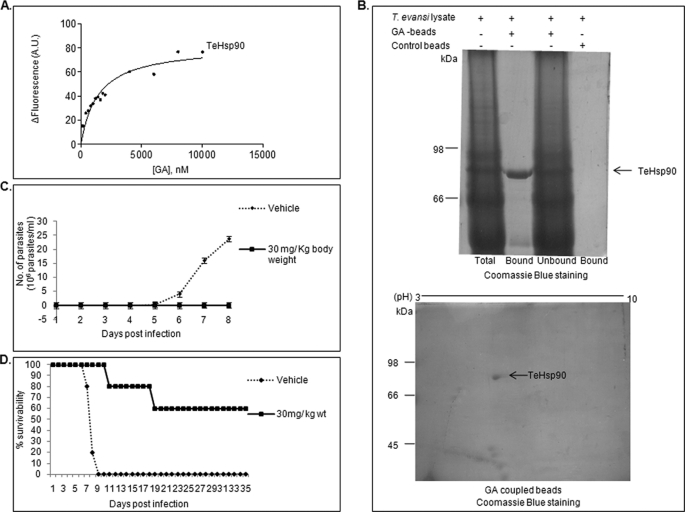
The ability of Hsp90 inhibitor to inhibit malaria parasite infection in mice raises the possibility of treating other protozoan infections by targeting Hsp90. To investigate this possibility, we chose extracellular protozoan parasite T. evansi, which causes surra in domestic animals like camels, horses, cattle, and buffaloes. For this, we cloned Hsp90 from T. evansi and overexpressed and purified it in bacteria as described under “Experimental Procedures” (supplemental Fig. S10). We are the first to provide the sequence of Hsp90 from T. evansi in the absence of any genomic sequence. Multiple sequence alignment of Hsp90 from various protozoan parasites is shown in Fig. 7. As can be seen, the residues required for GA binding are conserved across protozoan parasites. We examined the ability of Hsp90 from T. evansi to bind GA. Using tryptophan fluorescence spectroscopy, we found that purified TeHp90 binds to GA with a binding affinity of 1.4 μm (Fig. 8A).
FIGURE 7.
Multiple sequence alignment of Hsp90s from various protozoan parasites. Hsp90 sequences for P. falciparum (PfHsp90; GenBankTM accession number CAA82765.1), Plasmodium vivax (PvHsp90; GenBank accession number EDL43724.1), Eimeria tenella (EtHsp90; GenBank accession number AAB97088.1), Toxoplasma gondii (TgHsp90; GenBank accession number AAP44977.1), Cryptosporidium parvum Iowa II (CpHsp90; GenBank accession number EAK89246.1), Babesia bovis (BbHsp90; RefSeq accession number XP_001611554.1), L. donovani infantum (LdHsp90; Protein Information Resource accession number S57415), Leishmania major (LmHsp90; GenBank accession number CAJ05959.1), T. brucei (TbHsp90; Protein Information Resource accession number A44983), T. cruzi strain CL Brener (TcHsp90; RefSeq accession number XP_814892.1), Entamoeba histolytica (EhHsp90; RefSeq accession number XP_653132.1), and Trichomonas vaginalis (Tvhsp90; RefSeq accession number XP_001583398.1) were obtained from NCBI. The Tehsp90 sequence was obtained from the sequence of the cloned gene obtained in this study. All the residues required for GA binding are conserved across protozoan parasites. The residues that interact with GA are indicated (●) and marked (♦, hydrogen bond; ■, van der Waals; ♢, with H2O).
FIGURE 8.
Hsp90 inhibitor binds to TeHsp90 and cures infection in mice. A, determination of binding affinity of GA toward TeHsp90 by tryptophan fluorescence. The change in fluorescence (ΔF) was plotted against GA concentration for TeHsp90. B, GA-immobilized beads specifically pull down TeHsp90 from T. evansi lysate. The GA pulldown fraction was analyzed by SDS-PAGE (upper panel) and two-dimensional electrophoresis (lower panel). C and D, in vivo efficacy of Hsp90 inhibitors on the survivability of T. evansi-infected mice. C, plot of number of parasites against the number of days postinfection. D, percentage of viability of 17AAG-treated/untreated mice plotted against time. A.U., arbitrary units. Error bars have been represented to indicate standard error from three independent experiments.
Furthermore, to examine whether GA specifically binds to TeHsp90 in T. evansi whole cell lysate, we performed a GA bead pulldown assay. T. evansi was purified from infected mouse blood using DEAE-cellulose (14). Purified T. evansi was lysed in TNESV buffer containing protease inhibitors. Parasite lysate was incubated overnight with GA-coupled beads at 4 °C. Uncoupled beads served as control. As can be seen from Fig. 8B (upper panel), GA-coupled beads specifically pulled down a band corresponding to an 83-kDa protein. Furthermore, the identity of this band was confirmed by mass spectrometry, which gave the first hit for Hsp90 from Trypanosoma brucei, a closely related species of T. evansi, because the T. evansi genome is not sequenced (supplemental Fig. S11). We also performed two-dimensional gel electrophoresis for the GA-bound fraction, which gave a specific spot of 83 kDa with a pI of 5.1, which matches the pI calculated from its sequence (Fig. 8B, lower panel). The above results confirmed that GA specifically binds to TeHsp90.
We further examined the ability of 17AAG to inhibit T. evansi infection at the preclinical level. For this, Swiss female mice were infected with 105 purified T. evansi cells and treated with 17AAG for 4 consecutive days. 17AAG was administrated intraperitoneally at a concentration of 30 mg/kg of body weight. T. evansi-infected mice that were injected intraperitoneally with vehicle (20% Cremophor in PBS) served as an untreated control. Mice were examined daily by wet tail blood examination under a microscope, and the number of parasites was counted using a hematocytometer. The average number of parasites in each group was plotted against days postinfection. Fig. 8C shows the number of parasites in untreated and 17AAG-treated mouse groups. It is evident that in mice treated with vehicle the number of parasites increased rapidly to 108 parasites/ml and resulted in death of all mice by the 9th day following infection, whereas in the drug-treated mice, no parasites were detected resulting in curing of disease in those mice. An overall survival rate of 60% was observed for more than 90 days in 17AAG-treated mice (Fig. 8D). All the mice that survived showed no signs of infection and are still healthy. The p value for this experiment was found to be 0.002 (p < 0.01) by log rank test. Hsp90 inhibitor was thus effective in inhibiting the growth of two parasites, P. berghei and T. evansi, in the in vivo mice model. The results shown here suggest that Hsp90 inhibitor can be used in the treatment of a wide range of diseases caused by protozoan parasites both in humans and animals.
DISCUSSION
Hsp90s from different organisms share a significant homology at the level of their primary structures. All Hsp90 family members show a domain organization consisting of 1) an amino-terminal nucleotide binding region, 2) a charged linker region, 3) a middle domain, and 4) a carboxyl-terminal dimerization domain (12). All the Hsp90 family members are known to form homodimers through their carboxyl-terminal domains. There is also similarity in Hsp90s from different species at the level of their higher order structure. The functional unit of Hsp90 in the cell consists of a multichaperone complex containing Hsp70 and other co-chaperones (3). We have previously shown the presence of a similar Hsp90 multichaperone complex in the malaria parasite (7, 18).
Structural conservation of Hsp90 is also reflected in its function. As a molecular chaperone, Hsp90 is unique in its ability to modulate functions of native proteins. Hsp90 is known to regulate activities of key protein kinases and nuclear receptors involved in controlling cell cycle progression and signal transduction pathways. The list of substrates of Hsp90 includes transcription factors like p53 and cell cycle-regulating kinases like AKT (3). Such a regulatory role of Hsp90 has been demonstrated in lower eukaryotes like yeast as well as in higher organisms like mammals. It is not surprising that Hsp90 function has been shown to be essential in yeast, flies, worm, and plants (17, 27–30) as well as in the malaria parasite (7).
The pivotal role played by Hsp90 in growth and development in eukaryotes has been exploited in implicating it as a therapeutic target in various infectious as well as non-infectious diseases. Its proposed role in the treatment of cancer may soon become a reality as Hsp90-specific drugs have been tested in phase III clinical trials in humans (31–33). Although there are many studies implicating Hsp90 as a target for the treatment of human diseases, apart from cancer therapy, details regarding its structure-activity relationship have not been addressed systematically.
We have previously shown that inhibition of Hsp90 function prevents growth of the malaria parasite in human erythrocytes in vitro (7). However, this previous study mainly focused on the growth inhibition exhibited by GA on P. falciparum in vitro. Aspects related to biochemical properties of PfHsp90 and its interaction with GA were not examined in detail. It was also not clear whether inhibition of PfHsp90 would be effective in inhibiting parasite growth in vivo in an animal model of malaria. As Hsp90 chaperone function is dependent on its ATPase activity, which requires cooperative interaction between different domains, it would be desirable to biochemically characterize full-length Hsp90 of P. falciparum to gain insights into its potential as an antimalarial target. With this view in mind, here we have for the first time carried out systematic biochemical characterization of full-length PfHsp90, studied its interaction in purified form with GA, and also examined its efficacy in a mouse model of malaria.
Hsp90 from all known systems is an ATP-dependent chaperone and is able to bind and hydrolyze ATP. We addressed whether Hsp90 from P. falciparum can also bind and hydrolyze ATP. We found PfHsp90 to bind ATP with about 30% higher affinity and to have 6 times higher ATPase activity as compared with its human host. It is well known that despite conservation in the overall structural organization from prokaryotes to eukaryotes as demonstrated by the crystal structures of HtpG from E. coli (34) and Hsp90 from Saccharomyces cerevisiae (23), there is a vast difference in their abilities to bind and hydrolyze ATP. It is noteworthy that the ATPase activity of PfHsp90 was highest among any reported value for Hsp90. Higher catalytic efficiency implies a higher rate of substrate cycling through the Hsp90 chaperone cycle. This may be essential in the case of P. falciparum, which experiences stress throughout its life cycle. Therefore, PfHsp90 may have evolved to combat high stress conditions requiring higher client turnover. The ATPase activity of Hsp90 in vivo is known to be regulated by a cohort of co-chaperones. Although Hsp90 ATPase activity was studied using purified protein, the in vivo ATPase may in fact reach higher values because the homologs of Hsp90 co-chaperones known to increase the ATPase activity are well conserved in P. falciparum. Such a hyperactive ATPase of PfHsp90 may also contribute to the hypersensitivity of malarial parasite to Hsp90 inhibitors.
GA, a structural analog of ATP, binds to Hsp90 with a far higher affinity than ATP and thus inhibits its chaperone cycle leading to proteosomal degradation of client proteins. Despite conservation in the nucleotide binding domain of Hsp90 from different organisms, sensitivity to GA is known to vary. For example, Hsp90s from free living nematodes do not bind to GA even though the residues involved in GA binding are conserved indicating that properties beyond primary structure may modulate GA sensitivity. Previous studies in cancer cells have addressed parameters influencing GA binding to Hsp90. These studies show co-chaperone interaction, increased ATPase activity, and post-translational modifications to influence GA binding. A virtual prototyping study has shown that Hsp90 with higher ATPase activity is more sensitive to GA-mediated inhibition (35).
We found GA to bind PfHsp90 with an affinity in the lower μm range. Most importantly, GA shows more potent inhibition of PfHsp90 ATPase activity as compared with hHsp90. Furthermore, higher ATPase activity will shift the equilibrium toward the ADP-bound form of Hsp90, which is more sensitive to GA (36). In agreement with its affinity to Hsp90, GA-coupled beads specifically pulled down a single Coomassie-stainable band from P. falciparum cell lysate that was confirmed to be PfHsp90 by immunoblotting, thus establishing GA specificity.
The binding affinity of GA to Hsp90 in vivo is known to be sensitive to its acetylation status and the composition of multichaperone complex. Although we too found PfHsp90 from P. falciparum lysate to be acetylated, the sites of modifications do not coincide with those reported for hHsp90. One of the acetylation sites on Pfhsp90, namely Lys-426, overlaps with Aha1 and p23 binding domains reported in yeast Hsp90. It is likely that acetylation at Lys-426 may regulate co-chaperone binding and thereby PfHsp90 ATPase activity and GA binding affinity. Indeed, co-treatment with TSA and GA showed a synergistic and additive effect in inhibiting parasite growth in vitro.
17AAG, a clinically tested approved ansamycin, inhibits the 3D7 laboratory strain of the parasite as well as parasites isolated from a malaria patient. Additionally, GA and 17AAG could inhibit the growth of chloroquine-resistant parasites collected from malaria patients in India. Our results suggest that P. falciparum is hypersensitive to GA compared with other organisms for which GA sensitivities have been determined (Fig. 2C). Furthermore, yeast expressing PfHsp90 in place of its endogenous Hsp82 exhibited hypersensitivity to GA (37). Interestingly, the IC50(Growth) values exhibited by parasites were comparable with that of cancerous cells. 17AAG is in a phase III clinical trial as an anticancer drug. Because of its potent inhibition of P. falciparum growth, it also has potential as an antimalarial drug.
Finally, the inhibitory effects of 17AAG on the growth of the malaria parasite in human erythrocytes in vitro also showed promise in vivo in a preclinical model of rodent malaria. Intraperitoneal administration of 17AAG to P. berghei-infected mice inhibited parasite growth. Most of the protozoans are known to depend on Hsp90 for their development. To examine whether GA and its analog could be used against a wide variety of protozoan infections, we extended our study to another protozoan parasite, T. evansi, which causes surra in animals. Although GA has been shown to interfere with the development of Leishmania, Toxoplasma, Eimeria, and Trypanosoma (3, 6, 24, 38), its efficacy at the preclinical level had not been addressed previously. Our study in T. evansi showed that GA specifically binds to TeHsp90, and inhibition of TeHsp90 using 17AAG in mice infected with T. evansi was able to cure trypanosomiasis. Moreover, sequence identity between Hsp90 from T. evansi and its closely related species T. brucei from which T. evansi is thought to have evolved suggests that 17AAG could also be used against T. brucei infection in humans. Our study showed 17AAG to be effective in treating malaria as well as trypanosomiasis. All together, our studies on the effect of GA on P. falciparum not only provide insights into Hsp90 biology in this important parasite but also raise the possibility of chaperone-targeted therapy for the treatment of a variety of protozoan infections.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of human Hsp90 protein from Prof. David O. Toft (Mayo Clinic, Rochester, MN). We thank Pragyan Acharya and Manish Grover (Department of Biochemistry, Indian Institute of Science, Bangalore, India) for giving valuable feedback on the manuscript.
This work was supported in part by the Department of Biotechnology, New Delhi, India; Malaria Signalling Consortium; Roche (University of Bern) and the Indo-Swiss Joint Research Programme of DST.

The on-line version of this article (available at http://www.jbc.org) contains supplemental experimental procedures, Table 1, and Figs. S1–S11.
- Hsp
- heat shock protein
- GA
- geldanamycin
- 17AAG
- 17-(allylamino)-17-demethoxygeldanamycin
- IC50(Growth)
- concentration of GA required to inhibit growth by 50%
- IC50(ATPase)
- concentration of inhibitor required to inhibit ATPase activity by 50%
- PfHsp90
- P. falciparum Hsp90
- hHsp90
- human Hsp90
- TeHsp90
- T. evansi Hsp90
- TSA
- trichostatin A.
REFERENCES
- 1.Walter S., Buchner J. (2002) Angew. Chem. Int. Ed. Engl. 41, 1098–1113 [DOI] [PubMed] [Google Scholar]
- 2.Pratt W. B., Toft D. O. (2003) Exp. Biol. Med. 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 3.Wegele H., Müller L., Buchner J. (2004) Rev. Physiol. Biochem. Pharmacol. 151, 1–44 [DOI] [PubMed] [Google Scholar]
- 4.Chiosis G., Vilenchik M., Kim J., Solit D. (2004) Drug. Discov. Today 9, 881–888 [DOI] [PubMed] [Google Scholar]
- 5.Bagatell R., Whitesell L. (2004) Mol. Cancer Ther. 3, 1021–1030 [PubMed] [Google Scholar]
- 6.Echeverria P. C., Matrajt M., Harb O. S., Zappia M. P., Costas M. A., Roos D. S., Dubremetz J. F., Angel S. O. (2005) J. Mol. Biol. 350, 723–734 [DOI] [PubMed] [Google Scholar]
- 7.Banumathy G., Singh V., Pavithra S. R., Tatu U. (2003) J. Biol. Chem. 278, 18336–18345 [DOI] [PubMed] [Google Scholar]
- 8.Whitesell L., Mimnaugh E. G., De Costa B., Myers C. E., Neckers L. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8324–8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hainzl O., Lapina M. C., Buchner J., Richter K. (2009) J. Biol. Chem. 284, 22559–22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsutsumi S., Mollapour M., Graf C., Lee C. T., Scroggins B. T., Xu W., Haslerova L., Hessling M., Konstantinova A. A., Trepel J. B., Panaretou B., Buchner J., Mayer M. P., Prodromou C., Neckers L. (2009) Nat. Struct. Mol. Biol. 16, 1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaughlin S. H., Ventouras L. A., Lobbezoo B., Jackson S. E. (2004) J. Mol. Biol. 344, 813–826 [DOI] [PubMed] [Google Scholar]
- 12.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1997) Cell 90, 65–75 [DOI] [PubMed] [Google Scholar]
- 13.Chulay J. D., Haynes J. D., Diggs C. L. (1983) Exp. Parasitol. 55, 138–146 [DOI] [PubMed] [Google Scholar]
- 14.Roy N., Nageshan R. K., Pallavi R., Chakravarthy H., Chandran S., Kumar R., Gupta A. K., Singh R. K., Yadav S. C., Tatu U. (2010) PLoS One 5, e9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen B. A., Sullivan W. P., Felts S. J., Toft D. O. (2002) J. Biol. Chem. 277, 7086–7091 [DOI] [PubMed] [Google Scholar]
- 16.David C. L., Smith H. E., Raynes D. A., Pulcini E. J., Whitesell L. (2003) Cell Stress Chaperones 8, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devaney E., O'neill K., Harnett W., Whitesell L., Kinnaird J. H. (2005) Int. J. Parasitol. 35, 627–636 [DOI] [PubMed] [Google Scholar]
- 18.Pavithra S. R., Banumathy G., Joy O., Singh V., Tatu U. (2004) J. Biol. Chem. 5, 46692–46699 [DOI] [PubMed] [Google Scholar]
- 19.Rao R., Fiskus W., Yang Y., Lee P., Joshi R., Fernandez P., Mandawat A., Atadja P., Bradner J. E., Bhalla K. (2008) Blood 112, 1886–1893 [DOI] [PubMed] [Google Scholar]
- 20.Forafonov F., Toogun O. A., Grad I., Suslova E., Freeman B. C., Picard D. (2008) Mol. Cell. Biol. 28, 3446–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes J. L., Sharp S. Y., Hobbs S., Workman P. (2008) Cancer Res. 68, 1188–1197 [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Rao R., Shen J., Tang Y., Fiskus W., Nechtman J., Atadja P., Bhalla K. (2008) Cancer Res. 68, 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer P., Prodromou C., Liao C., Hu B., Roe S. M., Vaughan C. K., Vlasic I., Panaretou B., Piper P. W., Pearl L. H. (2004) EMBO J. 23, 1402–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwock J., Pham N. A., Cao M. P., Hedley D. W. (2008) Cancer Chemother. Pharmacol. 61, 669–681 [DOI] [PubMed] [Google Scholar]
- 26.Graefe S. E., Wiesgigl M., Gaworski I., Macdonald A., Clos J. (2002) Eukaryot. Cell 1, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. (1989) Mol. Cell. Biol. 9, 3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cutforth T., Rubin G. M. (1994) Cell 77, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 29.Rutherford S. L., Lindquist S. (1998) Nature 396, 336–342 [DOI] [PubMed] [Google Scholar]
- 30.Queitsch C., Sangster T. A., Lindquist S. (2002) Nature 417, 618–624 [DOI] [PubMed] [Google Scholar]
- 31.Heath E. I., Gaskins M., Pitot H. C., Pili R., Tan W., Marschke R., Liu G., Hillman D., Sarkar F., Sheng S., Erlichman C., Ivy P. (2005) Clin. Prostate Cancer 4, 138–141 [DOI] [PubMed] [Google Scholar]
- 32.Banerji U., O'Donnell A., Scurr M., Pacey S., Stapleton S., Asad Y., Simmons L., Maloney A., Raynaud F., Campbell M., Walton M., Lakhani S., Kaye S., Workman P., Judson I. (2005) J. Clin. Oncol. 23, 4152–4161 [DOI] [PubMed] [Google Scholar]
- 33.Usmani S. Z., Bona R., Li Z. (2009) Curr. Mol. Med. 9, 654–664 [DOI] [PubMed] [Google Scholar]
- 34.Krukenberg K. A., Förster F., Rice L. M., Sali A., Agard D. A. (2008) Structure 16, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vali S., Pallavi R., Kapoor S., Tatu U. (2010) Syst. Synth. Biol. 4, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan W., Stensgard B., Caucutt G., Bartha B., McMahon N., Alnemri E. S., Litwack G., Toft D. (1997) J. Biol. Chem. 272, 8007–8012 [DOI] [PubMed] [Google Scholar]
- 37.Wider D., Péli-Gulli M. P., Briand P. A., Tatu U., Picard D. (2009) Mol. Biochem. Parasitol. 164, 147–152 [DOI] [PubMed] [Google Scholar]
- 38.Péroval M., Péry P., Labbé M. (2006) Int. J. Parasitol. 36, 1205–1215 [DOI] [PubMed] [Google Scholar]
- 39.Onuoha S. C., Mukund S. R., Coulstock E. T., Sengerovà B., Shaw J., McLaughlin S. H., Jackson S. E. (2007) J. Mol. Biol. 372, 287–297 [DOI] [PubMed] [Google Scholar]
- 40.Gooljarsingh L. T., Fernandes C., Yan K., Zhang H., Grooms M., Johanson K., Sinnamon R. H., Kirkpatrick R. B., Kerrigan J., Lewis T., Arnone M., King A. J., Lai Z., Copeland R. A., Tummino P. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7625–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe S. M., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1999) J. Med. Chem. 42, 260–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.