Abstract
In the natural environment, bacterial cells have to adjust their metabolism to alterations in the availability of food sources. The order and timing of gene expression are crucial in these situations to produce an appropriate response. We used the galactose regulation in Escherichia coli as a model system for understanding how cells integrate information about food availability and cAMP levels to adjust the timing and intensity of gene expression. We simulated the feast-famine cycle of bacterial growth by diluting stationary phase cells in fresh medium containing galactose as the sole carbon source. We followed the activities of six promoters of the galactose system as cells grew on and ran out of galactose. We found that the cell responds to a decreasing external galactose level by increasing the internal galactose level, which is achieved by limiting galactose metabolism and increasing the expression of transporters. We show that the cell alters gene expression based primarily on the current state of the cell and not on monitoring the level of extracellular galactose in real time. Some decisions have longer term effects; therefore, the current state does subtly encode the history of food availability. In summary, our measurements of timing of gene expression in the galactose system suggest that the system has evolved to respond to environments where future galactose levels are unpredictable rather than regular feast and famine cycles.
Keywords: Bacterial Metabolism, Bacterial Transcription, Evolution, Galactose, Gene Expression
Introduction
Transport and metabolism of several sugars are controlled via two feedback loops connected by a common regulator that senses the intracellular concentration of the small molecule (1, 2). The simplest systems (e.g. the lactose utilization system in Escherichia coli) consist of two operons, a regulator gene and a regulated operon containing at least two cistrons, one encoding a transporter and the other encoding an enzyme that modifies/degrades the small molecule (3). In such systems, the genes encoding the sugar transporter (e.g. lacY) and the enzyme for sugar degradation (e.g. lacZ) are regulated simultaneously. However, many sugar utilization systems reached higher levels of complexity, e.g. having multiple transporters, regulators, or several enzymes of a metabolic pathway. For example, the gal system of E. coli contains genes involved in the transport (galP and mglBAC) and amphibolic utilization (galETKM) of the sugar d-galactose. Genes of the gal regulon belong to different operons (4). This setup allows differential regulation of functions when needed. Regulation of the gal system is governed by two similar regulators, GalR and GalS, which are regulated in different ways (5–7). Previous studies suggested that GalS plays only a minor role in steady-state conditions but becomes important transiently when a high level of extracellular galactose is quickly decreased (8). Besides sensing the intracellular sugar level, galactose utilization is also regulated by the cAMP-cAMP receptor protein (CRP)2 complex. cAMP is a signal of carbon shortage and is sensed by CRP. cAMP is synthesized intracellularly by adenylate cyclase (CyaA), which is activated when phosphorylation of Enzyme IIAGlc of the phosphotransferase system is increased (9, 10). However, CyaA activity can also be regulated by a so far unknown mechanism in the presence of certain non-phosphotransferase system sugars. This mechanism does not depend on the phosphotransferase system enzymes (11, 12). Galactose availability also strongly influences the intracellular cAMP level; cells grown in a galactose-limited chemostat show substantially higher cAMP levels than cells grown in a high-galactose batch culture (13). Transcription of the galactose transport systems (GalP and MglBAC) is highly dependent on the presence of cAMP-CRP. However, transcription of the gal operon genes, encoding the enzymes involved in galactose utilization, occurs in both the presence and absence of cAMP-CRP when galactose is available (6). The galETKM operon is transcribed from two promoters, P1galE and P2galE. In the absence of cAMP-CRP, P2galE predominantly expresses the promoter-proximal galE gene and very little galK (natural polarity), whereas in the presence of cAMP-CRP, the P1galE promoter expresses all of the structural genes at an equimolar level, whereas P2galE transcription is inhibited (14–16). Because the galE gene product is involved in making substrates for biosynthetic glycosylation reactions, and all of the gal enzymes are needed for catabolism of the sugar d-galactose, it appears that the P2galE transcript serves anabolic requirements, whereas transcription from P1galE provides for catabolism of d-galactose as a carbon source.
Proper timing of gene expression is crucial in the regulation of critical biological phenomena, i.e. cellular adaptation, differentiation, and development (17, 18). Biological systems evolved two fundamentally different mechanisms for the regulation of timing. One possibility is that, after an initial decision, a regulatory network proceeds autonomously, executing a predefined program (18, 19). The other possibility is that the network monitors specific signals in real time and regulates timing accordingly. The goal of this work is to understand the principles governing the timing of gene expression in sugar networks when cells are grown in a batch culture containing a single sugar as a carbon source. We monitored how promoter activities in the galactose utilization network change as the cell population grows and galactose is exhausted from the medium, using specific promoter-uidABC (gus) reporter fusions. Besides responding to system-specific regulators, gene expression is also influenced by global effects, which depend on the physiological state of the cell (20–22). Because cell physiology changed in our experiments, we used a promoter that is not regulated by GalR, GalS, or cAMP-CRP as a control. On the basis of the experimental results, we built a mathematical model to calculate intracellular d-galactose and cAMP-CRP levels.
We found that intracellular reporter protein levels were higher in the stationary phase than in the early exponential phase in all cases. However, the gal regulon promoters differed in the timing of transcription and consequently in the timing of intracellular accumulation of the reporter proteins. Promoter activities were highly dynamic compared with intracellular reporter protein levels, which changed only 2–6-fold depending on the promoter used. We discuss how regulatory structure and signal integration logic affect timing of gene transcription.
EXPERIMENTAL PROCEDURES
Strain Construction
Regulatory regions and promoters of the gal regulon operons were amplified by PCR using E. coli MG1655 chromosomal DNA as a template (GenBankTM accession number NC_000913.2) and inserted between the EcoRI and PstI sites in plasmid pSEM2027. The amplified regulatory regions were the following: spf (4047775–4047954), galR (2974441–2974680), galP (3086041–3086297), the galETKM operon (791519 to 791214), galS (2239921 to 2239711), and the mglBAC operon (2238720 to 2238439). We used a promoter that is not regulated by cAMP-CRP or GalR as a control (GenBankTM accession number GQ872202) (23). pSEM2027 derivatives containing the cloned regulatory regions were digested with BamHI and used as a template for PCR amplification. The amplified region contained the Zeocin cassette, the rrnBT1T2 terminators, the cloned promoter region, and part of the gusA ORF. This region was amplified using the “uidRZdn” (5′-ACCCGGATCCTCAATGCTGCCAGAGAGATTTTTTCAGAAAATGGATTTCACGGAATTCTCAGTCCTGCTCCTCGGCCAC-3′) and “Gusseqdn” (5′-TTCTTGTAACGCGCTTTCCCACCAAC-3′) primers. The ends of the resulting PCR fragment contained sequences of the uid region of the E. coli chromosome (50/128 bp), allowing efficient insertion using recombination-mediated genetic engineering. The PCR product was purified from an agarose gel. Recombination-mediated genetic engineering was performed according to the protocol described by Datsenko and Wanner (24). As a result, the region upstream of the uidA ORF, between chromosomal positions 1694107 and 1694987 (GenBankTM accession number U00096), was replaced with the synthetic construct. Recombinants were selected on LB plates containing 80 μg/ml Zeocin.
Verifying the DNA Sequence of the gal Regulatory Regions in Strains
Cells were grown overnight, and total genomic DNA was extracted using the Wizard genomic DNA purification kit (Promega). The regulatory region upstream of the uidABC operon was amplified by PCR (Platinum HiFi Supermix, Invitrogen) using the “KpnT1T2” (5′-ATATATGGTACCAAGCTTCTGTTTTGGCGGATGAGA-3′) and Gusseqdn primers. The DNA sequence was determined by BIOMI Ltd. (Gödöllő, Hungary).
Assay of β-Glucuronidase Activity
Cells were grown overnight in LB medium containing 100 μg/ml Zeocin and diluted 1000-fold for further growth in M63 medium containing 10 μg/ml Zeocin and supplemented with 0.4% (w/v) d-galactose, 0.1% (w/v) casamino acids (Sigma A2427), and 0.004% (w/v) vitamin B1. At various times, aliquots of cells were removed, diluted in 1 ml of LB medium to OD600 = 0.2, pelleted, resuspended in M63 medium containing 100 μg/ml chloramphenicol, and stored at −70 °C. To determine the activity of β-glucuronidase in cells, 0.1 mg/ml lysozyme was added to 500 μl of cell suspension. After incubation for 15 min on ice, 250 μl of permeabilization buffer (100 mm Tris (pH 8), 32 mm sodium phosphate, 8 mm dithiothreitol, 8 mm CDTA, and 4% Triton X-100), containing 200 μg/ml polymyxin B (25) was added to the cell suspension. Cells were allowed to permeabilize at room temperature for 15 min before 250-μl aliquots of GUS assay buffer (0.5 mm dithiothreitol, 1 mm EDTA, and 50 mm sodium phosphate (pH 7.0)) containing 1.25 mm α-p-nitrophenyl-β-d-glucuronide were added. The rate of β-glucuronide hydrolysis at 37 °C was determined by measuring absorbance at 405 nm at least five different times.
Proteins
CRP was purified as described by Ryu et al. (26). σ38 was purified using the IMPACT system (New England Biolabs) as described by Shin et al. (27) and was mixed with the RNAP core enzyme (Epicenter) at a 1:1 molar ratio. σ70 RNA polymerase was purchased from U. S. Biochemical Corp.
In Vitro Transcription
Transcription reactions were performed as described previously (5). The reaction mixture (50 μl) contained 20 mm Tris acetate (pH 7.8), 10 mm magnesium acetate, 200 mm potassium glutamate, and 2 nm supercoiled DNA template. CRP was used at 50 nm and cAMP at 100 μm, when present. 20 nm RNA polymerase was added before incubating the reactions at 37 °C for 5 min. Transcription was started by the addition of 1.0 mm ATP, 0.1 mm GTP, 0.1 mm CTP, 0.01 mm UTP, and 5 μCi of [α-32P]UTP (3000 Ci/mmol). Reactions were terminated after 10 min by the addition of an equal volume of transcription loading buffer (0.025% bromphenol blue, 0.025% xylene cyanol, 0.01 m EDTA, and 90% deionized formamide). After heating at 90 °C for 3 min, the samples were loaded onto urea-7% polyacrylamide DNA sequencing gels. RNA bands were quantified using the ImageQuantTM PhosphorImager (Molecular Dynamics).
Computation of Intracellular d-Galactose and cAMP-CRP Levels
Ref. 28 describes a model that takes a time series of d-Gal and cAMP-CRP levels and provides activities of PgalR, PgalS, PmglB, PgalP, P1galE, and P2galE. These are further incorporated into two differential equations that compute how the levels of GalR and GalS change with time. We extended this model to include similar differential equations for the β-glucuronidase reporters for each promoter: dP/dt = νiAi − γ(t)P, where i = PgalR, PgalE, PgalP, PgalS, PmglB, or Pspf. (As is the case for GalR and GalS, β-glucuronidase is assumed to be stable and diluted only by cell growth.) This model, consisting of these five differential equations in addition to the model of Ref. 28, has the following unknown parameters: the maximal production rates of β-glucuronidase under each promoter (νi), the time series for d-Gal, and the time series for cAMP-CRP. We varied these parameters and observed how well the resulting time series of β-glucuronidase under different promoters fitted the measured β-glucuronidase levels. We found a good fit by minimizing the least-square distance between model and observed β-glucuronidase levels using simulated annealing (29), a general algorithm for solving global optimization problems. The algorithm was implemented using C, and results were visualized in MATLAB.
RESULTS
Changes in the Reporter Protein Content of Cells Grown in a Batch Culture
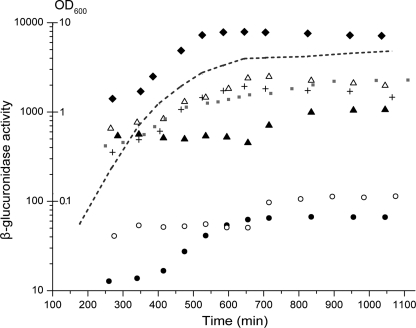
Bacterial growth in batch cultures has six distinct stages: lag phase (growth rate-null), acceleration phase (growth rate increases), exponential phase (growth rate constant), retardation phase (growth rate decreases), stationary phase (growth rate-null), and the phase of decline (“death phase,” growth rate-negative) (30). When diluted in fresh medium, cells first adjust their gene expression pattern to the new conditions. After this initial period, cells start to replicate, resulting in a continuous decrease in the level of available resources in the growth medium. As nutrients become limited, cells undergo an elaborate physiological and morphological differentiation to adapt to starvation. To monitor how the galactose network of E. coli responds when cells meet a finite pool of d-galactose in a limited volume, we created seven promoter-uidABC reporter fusions using the PgalR, PgalE, PgalP, PgalS, PmglB, and Pspf promoter regions and a control promoter that is not regulated by GalR, GalS, or cAMP-CRP (“unregulated” promoter). The fusions were transferred on the chromosome of the reference strain MG1655 separately, resulting in seven strains. Strains were grown in LB medium overnight and then diluted in supplemented M63 minimal medium containing d-galactose as the sole carbon source. The level of the reporter protein (UidA = GusA) was determined at different times by measuring β-glucuronidase activities in 0.5 ml of cells diluted to OD600 = 0.2. The actual level of the reporter protein is affected by production and dilution by cell division. Therefore, for each sample measured, we recorded the optical density of the culture at 600 nm and the time counted from dilution of the culture in the minimal medium (Fig. 1).
FIGURE 1.
β-Glucuronidase content of cells as a function of time. Cells containing promoter-uidABC reporter fusions were grown in M63 minimal medium. At various time points, the optical density of the culture (OD600) was recorded, and the β-glucuronidase enzyme activity was determined. All strains showed similar growth; therefore, only a representative growth curve is shown (dashed line). The β-glucuronidase activity of cells diluted to OD600 = 0.2 was plotted as a function of time for the strains containing the PgalR (●), PgalE (+), PgalP (△), PgalS (○), PmglB (▴),Pspf (♦), and the unregulated (gray squares) promoter fusions. The results shown are the average of three independent measurements. S.D. was <10.6% of the mean value.
All the reporter fusions studied behaved similarly in the sense that the intracellular β-glucuronidase level increased from an initial lower level to a higher level. The change was only 2–6-fold depending on the promoter used. The timing and the speed of the transition between the low and high states varied significantly for the different promoters. In four cases (spf, galE, galP, and galR), the transition occurred in the 400–600-min interval, during the shift from fast growth to very slow growth. However, in the other two cases (galS and mglB), the transition happened later, when cell doubling was almost negligible (650–800 min). In the case of the unregulated promoter, the transition between the low and high states was less sharp.
Computation of Growth Rate
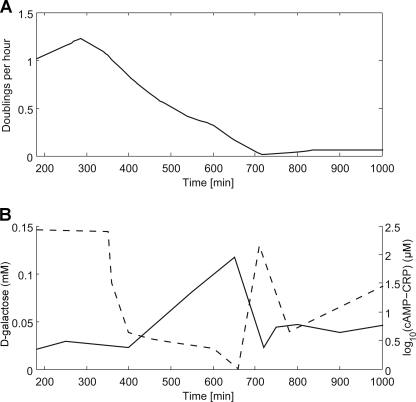
From the measured OD values, we could determine how the growth rate (γ) of bacteria changed with time using the following formula: γ(t) = (d/dt)ln(OD(t)). The data show that the growth rate dropped from a maximum of 1.225 doublings/h at t = 285 min to a minimum of ∼0.011 doublings/h at t = 715 min, after which the growth rate slightly increased again and then remained steady at 0.06 doublings/h until the end of the experiment. Before t = 180 min, we had no OD measurements, but we assume that the growth rate started at zero, remained there for a lag period of ∼100 min, and then rose to its maximum at 285 min (Fig. 2A).
FIGURE 2.
Timing of promoter activities. A, time-dependent growth rate calculated from the experiment shown in Fig. 1. B, intracellular cAMP-CRP and d-galactose time series as found by the simulated annealing procedure (29).
Computation of Promoter Activities
The concentration of the β-glucuronidase protein (P) in the cell is determined by the rate of protein production and by the dilution of the cell contents due to cell division. Assuming that the rate of protein production is proportional to promoter activities (A) and that the optical density of the culture (OD) is proportional to the cell number, we can calculate a time series for the promoter activities using the following equation: A = (Pt+Δt − Pt(ODt)/ODt+Δt)/ΔT.
From the enzyme activity measurements of the unregulated promoter, a promoter activity time series was calculated in the same way as for the other strains. This activity, normalized to 1, was then used to scale all other promoter activities to take growth rate-dependent effects (e.g. gene copy/cell) and the varying resource availability (e.g. abundance of RNA polymerases and ribosomes) into account (Fig. 3). Unlike in previous reports using balanced growth conditions (20–22), in our experiments, the growth rate and the computed activity of the unregulated promoter did not show a direct correlation. One possible explanation for this observation is that besides production of ATP, the galactose metabolism pathway also affects other cellular processes. For example, availability of the intermediate metabolite UDP-galactose affects peptidoglycan synthesis and transcription (31).
FIGURE 3.
Calculated activities of PgalR (●), PgalE (+), PgalP (△), PgalS (○), PmglB (▴), and Pspf (♦) (solid black lines). The activity of the unregulated promoter, which was used to scale all other promoter activities to take the varying resource availability into account, is shown by the dotted line in the upper left panel. The scaled activities are shown in gray for all promoters. Promoter activities were individually normalized to the maximal activities observed (= 1).
The results show that, in the galactose system, promoter activities have different timing patterns, and the scaled activities follow two different trajectories (Fig. 3, gray lines). The activities of PgalR, PgalE, and Pspf can be characterized by a fast initial increase, followed by a sharp drop, resulting in a single peak. In the case of PgalR and PgalE, the peak is broader for the non-scaled activities. The scaled PgalP, PgalS, and PmglB start expression patterns are similar, having two separate activity peaks, the first at ∼250 min and the second at ∼600 min for PgalP and 700 min for PgalS and PmglB. The scaled and non-scaled timing patterns (Fig. 3, gray versus black lines) show only moderate differences, suggesting that the timing of gene expression is mostly determined by the system-specific regulators (GalR, GalS, and cAMP-CRP).
Computation of Intracellular cAMP-CRP and d-Galactose Levels
The measured promoter activities are functions of the intracellular cAMP-CRP and/or d-galactose levels and the gene expression capacity of cells. Conversely, given time series of the promoter activities, it is, in principle, possible to deduce the time development of cAMP-CRP and d-galactose. Ref. 28 describes a model that, given the intracellular cAMP-CRP and d-galactose levels, computes the activities of all gal regulon promoters. We used this model to find the cAMP-CRP and d-galactose time series that best fit the reporter protein activities we measured (normalized to the unregulated control to eliminate the effects of cell physiology). We found that the d-Gal level started rising significantly from t ∼ 400 min onward, until it peaked at t ∼ 650 min (Fig. 2B). After that, the level quickly decreased until t = 720 (the time at which the growth rate reached its minimum) before moderately increasing again. This increase is consistent with the production of the Mgl transport system at 700 min. As expected from previous observations (13), the cAMP-CRP level is inversely related to the d-Gal level; however, the biochemical background of this observation is not yet understood. The cAMP-CRP level started off high, decreasing to a low level at t = 400 min before rising again at t ∼ 670 min and then dropping again when d-Gal rose at t ∼ 720 min (Fig. 2B).
Changes in P1galE and P2galE Transcription
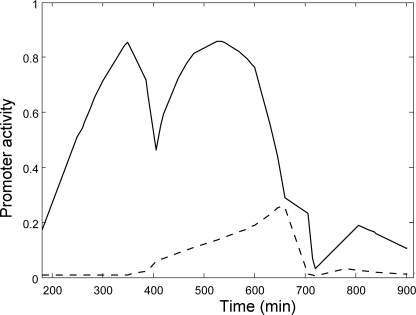
The model of Ref. 28 also enabled us to compute the relative contributions of the P1galE and P2galE promoters to galETKM transcription. We found that the main contribution to galETKM transcription throughout the growth curve came from P1galE. P2galE rose for only a relatively short duration around the time when the cAMP-CRP level reached its minimum at t = 670 min (Fig. 4).
FIGURE 4.
Predicted activities of the P1galE (solid line) and P2galE (dashed line) promoters, normalized such that the maximum of the total activity is 1.
Transcription of the gal Regulon Promoters by Stationary Phase (σ38) RNA Polymerase
The transition from exponential to stationary phase is accompanied by replacement of the RNA polymerase-associated σ70 subunit with σ38, the process of which is facilitated by sequestration of σ70 by the Rsd protein (32). It was previously known that both the vegetative phase (σ70) and stationary phase (σ38) RNA polymerases can transcribe the galETKM operon, although they show different preferences for the P1galE and P2galE promoters (33). Our results show that the PmglB promoter is active at the later stages of growth, showing ∼45% of its maximal expression at 800 min. To test whether σ38 RNA polymerase can support transcription of PmglB at a rate comparable to σ70 RNA polymerase, we analyzed transcription of the gal regulon promoters by σ38 and σ70 RNA polymerases in vitro in the presence and absence of cAMP-CRP (Fig. 5). Both the σ70 RNA polymerase (RNAP) and σ38 RNAP transcribed only three of the gal regulon promoters, P1galE, P2galE, and PgalR, in the absence of cAMP-CRP. Our results confirmed the previous observation that, as opposed to σ70 RNAP, σ38 RNAP shows strong preference for P1galE and transcribes P2galE weakly (33). Like P2galE, PgalR showed lower activity when transcribed by σ38 RNAP compared with σ70 RNAP. Similar to σ70 RNAP transcription, cAMP-CRP activated σ38 RNAP transcription of P1galE, PmglB, PgalP, and PgalS. However, significant differences were obtained in the cAMP-CRP-activated levels of promoter activities. Although the σ38 RNAP-transcribed PmglB activity reached ∼70% of the transcription level obtained with σ70 RNAP, PgalS reached only ∼20% (Fig. 5, lanes 2 and 4). Both the strong autoregulation of GalS (8) and the weak transcription of PgalS by σ38 RNAP could explain the fast drop in PgalS activity (compared with PmglB) between 700 and 750 min (Fig. 3). Although P1galE was more efficiently transcribed by σ38 RNAP than by σ70 RNAP in the absence of cAMP-CRP, σ38 RNAP transcription was only slightly increased compared with the activation of σ70 RNAP transcription in the presence of cAMP-CRP. Because the galR and galS genes are poorly transcribed by σ38 RNAP, we suggest that repression of the gal regulon promoters (primarily P1galE and PmglB) in the stationary phase is maintained by the repressor proteins accumulated earlier.
FIGURE 5.
Transcription of the gal regulon promoters by stationary phase (σ38) RNA polymerase. In vitro transcription assays were performed on a supercoiled pRPGSM DNA (6) template in the presence of the heat unstable nucleoid protein HU (80 nM) and cAMP. The presence or absence of CRP is indicated at the bottom of each lane. Experiments in lanes 1 and 2 were performed with σ70 RNAP holoenzyme, and those in lanes 3 and 4 with reconstituted σ38 RNAP holoenzyme.
DISCUSSION
Enteric bacteria are evolved to deal with varying nutritional conditions in nature. Nutrients are often limited in the environment (famine), but there are nutrient-rich periods as well (feast). The metabolic and morphological changes accompanying the transitions in the feast-famine cycles have been studied in detail (34). In this work, we analyzed how the galactose utilization system of E. coli reacts to the availability of a large but limited galactose pool. We found that galactose metabolism has a global effect on gene expression (e.g. through influencing nucleotide triphosphate levels) and also a specific effect on the transcription of genes belonging to the galactose regulon (through the intracellular d-galactose and cAMP levels, influencing the activity of GalR, GalS, and CRP). We computed changes in the promoter activities of the galactose regulon genes and in the intracellular d-galactose and cAMP-CRP levels. The galactose system monitors the intracellular d-galactose and cAMP-CRP levels in real time and computes the promoter activities accordingly. Because the proteins of the galactose system are assumed to be stable, similar to the reporter protein used, decisions of the system are limited to protein production. The actual intracellular levels of proteins depend on the history of protein production (promoter activity) and dilution (cell division). This is why promoter activities are more dynamic than protein levels. We found that timing of promoter expression depends on the current intracellular signal levels (d-Gal and cAMP-CRP), which are determined by the current extracellular galactose level and the levels of the proteins (and small RNA), affecting galactose transport and utilization (which depend on the history of the cell). Accordingly, decisions in the system are based on the current state of the cell and not on monitoring the level of extracellular galactose in real time.
Coordination of Galactose Transport and Utilization
Enzymes encoded by the galETKM operon are responsible for the amphibolic utilization of galactose. All of the Gal enzymes are required for galactose metabolism; however, only GalE is needed for making substrates for biosynthetic glycosylation reactions in the absence of external galactose. Transcription from the P1galE promoter results in equimolar expression of the Gal enzymes, serving the catabolic requirements, whereas transcription from the P2galE promoter results in discoordinated expression (more production of GalE and GalT than the promoter-distal GalK and GalM), which suits the biosynthetic requirements (15, 16, 35, 36). Our result that P2galE promoter activity is negligible when cells are grown in a galactose batch culture is consistent with this model. However, production of the proteins encoded by the gal operon can also be discoordinated by the Spot42 small RNA, which specifically inhibits GalK production in the absence of cAMP-CRP (37). In our experiments, transcription of the Spot42 promoter (Pspf) coincided with the increase in the intracellular d-Gal level and with the decrease in the cAMP level. We suggest that inhibition of GalK production by the Spot42 small RNA is involved in balancing galactose transport and metabolism and that this regulation suits cellular needs in an environment where the extracellular galactose concentration is decreasing. Limitation in galactose transport due to the decreasing extracellular galactose concentration can be compensated by increasing the concentration of transporters. To obtain higher expression of transporters, cells need to build up a higher level of intracellular galactose, which in turn also allows increased production of the enzymes involved in galactose utilization. If the system did not reduce the rate of galactose utilization when the galactose influx decreases, the cell would quickly use up the intracellular galactose pool, leading to switching off of promoters by reactivated GalR and GalS and therefore reduced galactose transport and utilization. This effect can be overcome by changing the relative strengths of the transport and metabolism feedback loops (1). Limiting GalK production is an efficient way to block the metabolic feedback loop without compromising transport, allowing accumulation of intracellular galactose. However, Spot42 is produced at a high rate at low cAMP, regardless of whether the extracellular galactose concentration is decreasing or not. Increasing the transport capacity is likely unnecessary in a high-galactose environment; therefore, we suggest that uncoupling of the transport and metabolism loops by post-transcriptional regulation is a result of adaptation to a fluctuating galactose environment. Spot42 production allows more efficient utilization of galactose at lower extracellular concentrations, and because of the abundance of the Spot42 small RNA, this effect can last for a few cell generations even after its synthesis is stopped (38).
In conclusion, we propose that the galactose system of E. coli was adapted to function in an environment where galactose levels vary in time. However, the mechanisms of decision suit environments where future galactose levels are unpredictable rather than environments with regular feast and famine cycles. If the galactose system were adapted for the latter kind of environment, with regular fluctuations, we would expect the system to give higher preference to the expression of the MglBAC transport system when the intracellular d-Gal level starts to decrease, thus avoiding the dip in the growth rate at 700 min (Fig. 2A).
Determinants of Transcription Patterns of Promoters in the Galactose System
Transcription of the gal regulon promoters (at any given time point in the experiment, shown in Fig. 1) depends on (i) the availability of resources for transcription, (ii) combinations of the intracellular cAMP and d-Gal levels, (iii) the concentration of regulators sensing these signals, and (iv) the structure and function of the regulatory region and the promoter. Previously, we described how transcription of the gal regulon promoters depends on cAMP and d-galactose concentrations and found that integration of the two signals at the promoters resembles Boolean logic functions (6, 28). Here, we found a correlation between the shapes of the scaled promoter activity curves (Fig. 3, gray lines) and the logic of signal integration at the promoters. We observed one activity peak in the case of the “d-Gal” and the “NOT cAMP” logic and two activity peaks in the case of AND-like gates (Fig. 3). However, the positions of the peaks in the latter case depend on the strength of GalR/GalS- and cAMP-CRP-binding sites in the regulatory regions. For example, for PgalP, which has the strongest GalR-binding sites and the highest sensitivity to cAMP-CRP activation, the second peak appears earlier compared with PgalS, which needs much higher cAMP-CRP levels to become activated and less d-Gal to lift GalR/GalS-mediated repression.
Acknowledgments
We thank our colleagues in the laboratory for various inputs, in particular, Thomas Soares and Mofang Liu for purification of CRP; Takácsné Botond Judit for excellent technical assistance; and Kim Sneppen, Sankar Adhya, László Orosz, Mogens H. Jensen, and András Holczinger for useful discussions and support. We also thank Hyon E. Choy for the plasmid and instructions for σ38 purification.
This work was supported by the Hungarian Scientific Research Fund (OTKA) Grant PD75496 (to S. S.) and by the Danish National Research Foundation.
- CRP
- cAMP receptor protein
- RNAP
- RNA polymerase.
REFERENCES
- 1.Krishna S., Semsey S., Sneppen K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20815–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sneppen K., Krishna S., Semsey S. (2010) Annu. Rev. Biophys. 39, 43–59 [DOI] [PubMed] [Google Scholar]
- 3.Monod J., Changeux J. P., Jacob F. (1963) J. Mol. Biol. 6, 306–329 [DOI] [PubMed] [Google Scholar]
- 4.Weickert M. J., Adhya S. (1993) Mol. Microbiol. 10, 245–251 [DOI] [PubMed] [Google Scholar]
- 5.Geanacopoulos M., Adhya S. (1997) J. Bacteriol. 179, 228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semsey S., Krishna S., Sneppen K., Adhya S. (2007) Mol. Microbiol. 65, 465–476 [DOI] [PubMed] [Google Scholar]
- 7.Weickert M. J., Adhya S. (1993) J. Bacteriol. 175, 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semsey S., Krishna S., Erdossy J., Horváth P., Orosz L., Sneppen K., Adhya S. (2009) J. Bacteriol. 191, 4487–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park Y. H., Lee B. R., Seok Y. J., Peterkofsky A. (2006) J. Biol. Chem. 281, 6448–6454 [DOI] [PubMed] [Google Scholar]
- 10.Reddy P., Kamireddi M. (1998) J. Bacteriol. 180, 732–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein W., Rothman-Denes L. B., Hesse J. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 2300–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogema B. M., Arents J. C., Inada T., Aiba H., van Dam K., Postma P. W. (1997) Mol. Microbiol. 24, 857–867 [DOI] [PubMed] [Google Scholar]
- 13.Death A., Ferenci T. (1994) J. Bacteriol. 176, 5101–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidi-Rontani C., Danchin A., Ullmann A. (1984) Mol. Gen. Genet. 195, 96–100 [DOI] [PubMed] [Google Scholar]
- 15.Lee H. J., Jeon H. J., Ji S. C., Yun S. H., Lim H. M. (2008) J. Mol. Biol. 378, 318–327 [DOI] [PubMed] [Google Scholar]
- 16.Ullmann A., Joseph E., Danchin A. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 3194–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudet J., Muttumu S., Horner M., Mango S. E. (2004) PLoS Biol. 2, e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amir A., Kobiler O., Rokney A., Oppenheim A. B., Stavans J. (2007) Mol. Syst. Biol. 3, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson C. H. (2004) Curr. Issues Mol. Biol. 6, 103–110 [PubMed] [Google Scholar]
- 20.Klumpp S., Hwa T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20245–20250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klumpp S., Zhang Z., Hwa T. (2009) Cell 139, 1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang S., Bipatnath M., Xu Y., Chen S., Dennis P., Ehrenberg M., Bremer H. (1999) J. Mol. Biol. 292, 19–37 [DOI] [PubMed] [Google Scholar]
- 23.Hunziker A., Tuboly C., Horváth P., Krishna S., Semsey S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schupp J. M., Travis S. E., Price L. B., Shand R. F., Keim P. (1995) BioTechniques 19, 18–20 [PubMed] [Google Scholar]
- 26.Ryu S., Kim J., Adhya S., Garges S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin M., Song M., Rhee J. H., Hong Y., Kim Y. J., Seok Y. J., Ha K. S., Jung S. H., Choy H. E. (2005) Genes Dev. 19, 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishna S., Orosz L., Sneppen K., Adhya S., Semsey S. (2009) J. Mol. Biol. 391, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otten R. H. J. M., van Ginneken L. P. P. P. (1989) The Annealing Algorithm, Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 30.Monod J. (1949) Annu. Rev. Microbiol. 3, 371–394 [Google Scholar]
- 31.Lee S. J., Trostel A., Le P., Harinarayanan R., Fitzgerald P. C., Adhya S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19515–19520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jishage M., Ishihama A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4953–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolb A., Kotlarz D., Kusano S., Ishihama A. (1995) Nucleic Acids Res. 23, 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolter R., Siegele D. A., Tormo A. (1993) Annu. Rev. Microbiol. 47, 855–874 [DOI] [PubMed] [Google Scholar]
- 35.Semsey S., Virnik K., Adhya S. (2006) J. Mol. Biol. 358, 355–363 [DOI] [PubMed] [Google Scholar]
- 36.Adhya S. (2003) Sci. STKE. 2003, pe22. [DOI] [PubMed] [Google Scholar]
- 37.Møller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P. (2002) Genes Dev. 16, 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitarai N., Benjamin J. A., Krishna S., Semsey S., Csiszovszki Z., Massé E., Sneppen K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10655–10659 [DOI] [PMC free article] [PubMed] [Google Scholar]