Abstract
Ligation of the lymphotoxin-β receptor (LTβR) by LIGHT (lymphotoxin-related inducible ligand that competes for glycoprotein D binding to herpes virus entry mediator on T cells (TNFSF14)) activates the noncanonical (NC) NF-κB (nuclear factor-κB) pathway and up-regulates CXCL12 gene expression by human umbilical vein endothelial cells (HUVEC). In contrast, TNF only activates classical NF-κB signaling and does not up-regulate CXCL12. To determine whether cross-talk between the classical and NC pathways affects CXCL12 expression, we investigated the effects of TNF on LIGHT signaling in HUVEC. We show here that TNF inhibits both basal and LIGHT-induced CXCL12 expression. Negative regulation by TNF requires the classical NF-κB pathway as inhibition of basal and induced CXCL12 was reversed in HUVEC-expressing dominant negative IκB (inhibitor of NF-κB) kinase (IKK)β (IKKβK44M). TNF did not inhibit the NC NF-κB pathway activation as LIGHT-induced p100 processing to p52 was intact; however, TNF either alone or together with LIGHT up-regulated p100 and RelB expression and induced the nuclear localization of p100-RelB complexes. Enhanced p100 and RelB expression was inhibited by IKKβK44M, which led us to question whether the IκB function of elevated p100 mediates the inhibition of CXCL12 expression by TNF. We retrovirally transduced HUVEC to express p100 at a level similar to that up-regulated by TNF; however, basal and LIGHT-induced CXCL12 expression was normal in the transduced cells. In contrast, ectopic RelB expression recapitulated the effects of TNF on NC signaling and inhibited basal and LIGHT-induced CXCL12 expression by HUVEC. Our findings therefore demonstrate that TNF-induced classical NF-κB signaling up-regulates RelB expression that inhibits both basal and NC NF-κB-dependent CXCL12 expression.
Keywords: Chemokines, Gene Expression, NF-kappa B, Signal Transduction, Tumor Necrosis Factor (TNF), CXCL12, IKK, SDF-1, Cross-talk
Introduction
Activation of vascular endothelial cells (EC)2 by proinflammatory cytokines plays a pivotal role in acute and chronic inflammatory diseases (1). Underlying this function, activated EC express an array of adhesion molecules and chemokines that regulate leukocyte migration into sites of inflammation (1). A major signaling mechanism that regulates proinflammatory gene expression in EC is activation of the NF-κB family of transcription factors (1–3).
The NF-κB family contains five members, named p65 (RelA), RelB, c-Rel, p105/p50 (NF-κB1), and p100/p52 (NF-κB2), and transcriptionally active NF-κB is formed by homo- or heterodimerization of these proteins (2, 3). NF-κB remains inactive in the cytoplasm of resting cells through association with inhibitory IκB proteins. Following cell stimulation, the IκBs are phosphorylated by the IκB kinase (IKK) complex and then ubiquitinated and degraded by the proteasome (2). Free NF-κB translocates to the nucleus to regulate expression of proinflammatory, immune-regulatory, antiapoptotic, and proproliferative genes (2, 3). Notably, genetic deletion of the individual IKK complex subunits revealed two separate NF-κB pathways activated by distinct stimuli that regulate discrete panels of target genes (4–6).
The IKK complex contains two catalytic subunits named IKKα (IKK1) and IKKβ (IKK2) and a noncatalytic regulatory component named NEMO (NF-κB essential modulator) or IKKγ (5–7). Activation of NF-κB by most stimuli, including TNF and ligation of innate immune receptors, absolutely requires NEMO and predominantly IKKβ for phosphorylating IκBα leading to the nuclear translocation of p50·p65 heterodimers (2–6). This NEMO-dependent “classical” NF-κB pathway regulates the expression of many proinflammatory genes in EC including adhesion molecules ICAM-1 (intercellular adhesion molecule 1), VCAM-1, and E-selectin; cytokines IL-1, IL-6, and TNF; and chemokines CCL2, CCL8, CXCL2, and CX3CL1 (1–6).
The second NF-κB pathway is the noncanonical (NC) mechanism that does not require NEMO or IKKβ but depends upon IKKα. NC signaling targets cytoplasmic p100-RelB dimers and analogous to IκB degradation, the inhibitory C terminus of p100 becomes phosphorylated and processed to generate p52 (4, 8, 9). This liberates p52·RelB, which regulates a small panel of genes including the lymphoid and homeostatic chemokines CXCL12, CXCL13, CCL19, and CCL21, and the B cell growth factor BLyS (10). The NC pathway is activated by a subset of stimuli including ligation of the lymphotoxin-β receptor (LTβR) by LTα1β2 and LIGHT and stimulation of BLyS receptor 3 on B cells. In contrast, proinflammatory cytokines such as TNF do not induce IKKα-dependent p100 processing (2, 4). NC NF-κB plays a major role in lymphoid organogenesis and B cell maturation (4, 8, 9), and evidence of a pathophysiological function for this pathway in chronic inflammation is emerging (8, 9).
EC express the LTβR, and we demonstrated recently that LIGHT strongly activates the NC pathway in these cells (11). Furthermore, we showed that CXCL12 is a bona fide NC NF-κB-dependent gene up-regulated in EC by LTβR ligation. LTβR ligation also activates the classical pathway and up-regulates expression of classical NF-κB-dependent genes including CXCL2; however, classical NF-κB activation by LIGHT or LTα1β2 in EC is significantly weaker than that induced by TNF (11). TNF markedly up-regulates p100 levels in EC via classical NF-κB signaling, although it does not activate the NC pathway or induce CXCL12 expression (11). Previous studies demonstrated that TNF enhances expression of the NC gene CXCL13 induced by anti-LTβR in lymph node stromal cells and aortic smooth muscle cells (12–14). Furthermore, antigen receptor ligation on B cells has been shown to increase p100 levels providing a pool of p100 for BLyS-induced NC signaling (15). We therefore questioned whether the classical and NC pathways cross-talk in EC and whether TNF-induced p100 could enhance LIGHT-induced NC signaling and CXCL12 expression in HUVEC.
We show here that TNF does not augment but instead inhibits basal and LIGHT-induced CXCL12 expression. Moreover, inhibition of CXCL12 requires TNF-induced classical NF-κB activation. In addition to increasing the levels of p100, we demonstrate that TNF robustly up-regulates RelB in HUVEC and leads to the formation of a p100·RelB nuclear complex. Moreover, ectopic RelB expression recapitulates the effects of TNF on LIGHT-induced CXCL12 levels. We therefore conclude that classical NF-κB signaling enhances RelB expression that in turn inhibits NC NF-κB-dependent gene expression. Hence, our findings demonstrate that the classical pathway negatively regulates NC NF-κB-dependent gene expression in EC.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human TNF and LIGHT were from R&D Systems (Minneapolis, MN). Rabbit anti-p100/p52 was from Millipore (Billerica, MA), rabbit anti-IκBα and rabbit anti-RelB were from Santa Cruz Biotechnology (Santa Cruz, CA), and rabbit anti-histone-3 was from Cell Signaling Technology (Beverly, MA). Mouse anti-tubulin (clone B-5-1-2) was from Sigma. PCR primers were purchased from Integrated DNA Technologies (Coralville, IA). Collagenase was from Worthington Biochemical (Lakewood, NJ). Real-time reagents Taqman Fast Universal Master Mix, Power SYBR, and Taqman primer-probe sets were purchased from ABI (Foster City, CA).
Cell Culture
HUVEC were isolated from discarded tissue following a protocol approved by the University of Pennsylvania Internal Review Board. Following collagenase digestion (1 mg/ml in PBS) of the canulated umbilical vein, endothelial cells were serially cultured on 1% gelatin-coated (J.T. Baker, Inc., Phillipsburg, NJ) tissue culture plastic (Falcon, Lincoln Park, NJ) in VascuLife® VEGF-Mv medium (Lifeline Cell Technology, Walkersville, MD) containing 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were passaged using trypsin/EDTA (0.05%; Invitrogen), and all experiments were performed using HUVEC at passage 2 or 3.
WT, IKKα−/−, and IKKβ−/− murine embryonic fibroblasts (MEFs) were generously provided by Dr. Inder Verma (The Salk Institute for Biological Studies, La Jolla, CA). MEFs were maintained in DMEM (Invitrogen) supplemented with 10% fetal calf serum, 2 mm l-glutamine, penicillin (50 units/ml), and streptomycin (50 μg/ml).
Retroviral Transduction
Phoenix cells were transiently transfected with LZRS-EGFP, LZRS-IKKβK44M, LZRS-p100, or LZRS-RelB using FuGENE6 (Roche Applied Science) and selected for gene expression 24 h later using puromycin (1 μg/ml). Puromycin-resistant cells were used to derive conditioned medium to provide a retroviral stock for HUVEC transduction. For transduction of primary HUVEC, growth medium was removed, and cells were washed with Hanks' Balanced Salt Solution then incubated for 5–8 h with filtered retroviral conditioned medium containing polybrene (8 μg/ml; Sigma). After incubation, retrovirus was removed and replaced with normal growth medium overnight. The transduction process was repeated a further three to five times with intermittent cell passage as required. Using this protocol, the percentage of HUVEC expressing the transgene is routinely >90%.
Immunoblotting
Each well of a 6- or 12-well plate containing a confluent HUVEC monolayer was washed twice in ice-cold PBS and then lysed by the addition of 50–100 μl of TNT lysis buffer (50 mm Tris·Cl, pH 6.8, 150 mm NaCl, and 1% Triton X-100) containing complete protease inhibitor mixture (Roche Applied Science), NaF (2 mm), and β-glycerophosphate (2 mm). After 20 min on ice, lysates were harvested by scraping. Protein content was determined using Coomassie Plus Reagent (Pierce), and for each sample, an equal amount of protein (10–20 μg) was separated by SDS-PAGE and then transferred electrophoretically to PVDF membrane (Immobilon-P, Millipore) and immunoblotted with the appropriate primary and species-specific HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Westgrove, PA). Detection of the bound antibody was performed using Luminol reagent according to the manufacturer's instructions (Santa Cruz Biotechnology).
Generation of Nuclear Lysates for Immunoblotting
HUVEC monolayers were scraped into PBS at 4 °C and pelleted (425 × g for 10 min). Pellets were resuspended and swollen for 10 min on ice in 100 μl of Buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 2 mm NaF, 2 mm β-glycerophosphate, and complete mini protease inhibitors) plus 0.1% Nonidet P-40 and centrifuged (3800 × g) for 1 min. Supernatants (cytoplasmic fraction) were snap frozen and retained. Pelleted nuclei were washed four times with Buffer A plus 0.1% Nonidet P-40 before being lysed by passing through a 26-gauge 0.5-inch needle in 30 μl of 1% TNT, 1% SDS plus complete mini protease inhibitors. Nuclear lysates were either used immediately or snap frozen and stored at −80 °C. Lysates were immunoblotted as described above.
Immunoprecipitation
For immunoprecipitation, nuclear extracts were prepared using the Nuclear Complex coimmunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. Nuclear samples (25 μg) were precleared for 3–4 h using 25 μl of a 1:1 slurry of protein G-agarose beads (Invitrogen) at 4 °C. After centrifugation at 14,000 rpm for 10 s, the cleared lysates were transferred to another tube and incubated with antibody to RelB (1 μg/sample) or p100/p52 (2 μg/sample) overnight at 4 °C on a rocking platform before the addition of 30 μl of the 1:1 protein G-agarose slurry to each lysate. Incubation was continued at 4 °C for a further 4–6 h. Immune complexes, collected by centrifugation at 16,000 × g for 3 s, were washed in ice-cold PBS and solubilized in 25 μl of sample buffer. The entire sample was subjected to SDS-PAGE and Western blot analysis.
Isolation of mRNA and Quantitative Real-time PCR Analysis
Medium was removed, and RNA was extracted using RNeasy following the manufacturer's instructions (Qiagen, Valencia, CA). Samples were subjected to on column DNase digestion. First strand cDNA was derived from each treatment group using the High Capacity cDNA reverse transcription kit (ABI). For quantitative real-time PCR (qRT-PCR) analysis of p100, IL-6, and β-actin reactions were performed using appropriate primer sets (sequences available upon request) and Power SYBR (ABI). Using an ABI 7500 Real-time PCR system, PCR products were generated in triplicate or quadruplicate and normalized to β-actin levels. Relative quantification (RQ) was derived from the difference in cycle threshold (Ct) between the gene of interest and β-actin using the equation RQ = 2−ΔΔCt and analyzed using SDS software (version 1.3). In each figure, the means of data from at least three separate identical experiments are shown along with the S.E. PCR product specificity was confirmed by performing a dissociation curve at the end of each experiment. For quantitative analysis of CXCL12, CXCL2, and β-actin, TaqMan primer-probes sets were utilized (ABI). PCR product generation and analysis was performed as described above.
RESULTS
TNF Inhibits Basal and LIGHT-induced CXCL12 Expression by HUVEC
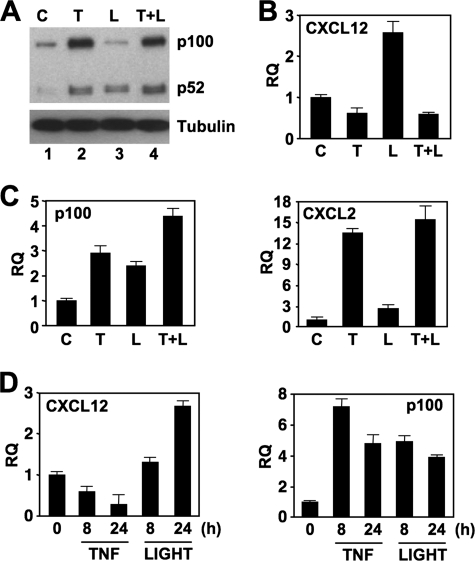
To determine the effects of TNF on NC NF-κB activation and gene expression in EC, we incubated HUVEC with either TNF or LIGHT or a combination of both cytokines. As we reported previously (11), TNF robustly up-regulated p100 levels with an accompanying increase in basally processed p52 (Fig. 1A). Incubation with LIGHT increased the levels of p52 with a concomitant decrease in basal p100 indicating processing of p100 and hence, activation of the NC NF-κB pathway (Fig. 1A) (11). Incubation with both cytokines increased p100 levels and modestly elevated the amount of p52 compared with the effects of either cytokine alone.
FIGURE 1.
TNF reduces basal and LIGHT-induced CXCL12 expression by HUVEC. A, HUVEC were either untreated (C) or incubated with TNF (T; 10 ng/ml), LIGHT (L; 100 ng/ml), or a combination of both cytokines (L+T) for 24 h, and then whole cell lysates were immunoblotted using either anti-p100/p52 (upper panel) or anti-tubulin as a loading control (lower panel). B and C, HUVEC were treated as described for panel A for 24 (B) or 8 (C) h, and then RNA was isolated. Quantitative real-time PCR was performed to determine the expression levels of CXCL12, p100, and CXCL2 as indicated. D, HUVEC were incubated with TNF or LIGHT for the times indicated, and then qRT-PCR was performed to determine the expression levels of CXCL12 or p100 as shown. RQ, relative quantification.
We demonstrated recently that incubation of EC with LIGHT increased CXCL12 expression via the NC NF-κB pathway (11). As LIGHT and TNF increased the amount of p100 and elevated the levels of p52 (Fig. 1A), we hypothesized this combination of cytokines would enhance CXCL12 expression. Surprisingly, however, qRT-PCR analysis revealed that LIGHT-stimulated CXCL12 expression was completely inhibited by TNF (Fig. 1B). In contrast, when we examined the known classical NF-κB gene targets p100 and CXCL2 (Fig. 1C), and others including IL-6 and CX3CL1 (data not shown), the combination of LIGHT and TNF did not inhibit but instead moderately increased expression of some of the genes analyzed.
We consistently observed that HUVEC incubated with TNF alone expressed lower levels of CXCL12 than unstimulated cells (Fig. 1, B and D). These effects of TNF starkly contrast with LIGHT that up-regulates CXCL12 expression over the same time periods (Fig. 1D, left panel). Furthermore, examination of classical NF-κB-dependent genes including p100 (Fig. 1D; right panel), CXCL2, CX3CL1, and IL-6 (data not shown) demonstrated that the negative regulatory effects of TNF on basal expression were limited in our analysis to CXCL12. Hence, these accumulated findings demonstrate that TNF signaling negatively regulates both basal and LIGHT-induced NC NF-κB-dependent CXCL12 expression in HUVEC.
Inhibition of CXCL12 Expression by TNF Requires Classical NF-κB Pathway Activation
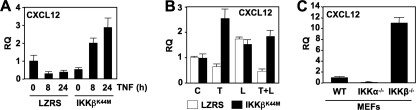
In our earlier study, we demonstrated that stable expression of catalytically inactive IKKα (IKKαSSAA) or IKKβ (IKKβK44M) selectively inhibits the NC and TNF-induced classical NF-κB pathways in EC, respectively (11). Consequently, to determine whether the effects of TNF on basal and LIGHT-induced CXCL12 require classical NF-κB activation, we stably transduced HUVEC with IKKβK44M and examined CXCL12 expression by qRT-PCR. In line with its effects on untransduced EC (Fig. 1), TNF inhibited basal CXCL12 expression in HUVEC stably transduced with the empty LZRS retroviral vector (Fig. 2A). Remarkably, in IKKβK44M-transduced cells, the negative regulation of basal CXCL12 expression by TNF was completely reversed, and CXCL12 levels were increased ∼6-fold over basal after 24 h (Fig. 2A).
FIGURE 2.
Classical NF-κB inhibition reverses the down-regulatory effects of TNF on basal and LIGHT-induced CXCL12. A, LZRS- or LZRS-IKKβK44M-transduced HUVEC were either untreated or incubated with TNF (10 ng/ml) for the times indicated, and qRT-PCR was performed to determine the expression levels of CXCL12. B, LZRS- (open bars) or LZRS-IKKβK44M-transduced (filled bars) HUVEC were either untreated (C) or incubated with TNF (T), LIGHT (L; 100 ng/ml), or a combination of both cytokines (L+T) for 24 h, and then the expression levels of CXCL12 were determined by quantitative real-time PCR. C, CXCL12 expression in WT, IKKα−/−, and IKKβ−/− MEFs was determined by quantitative real-time PCR. RQ, relative quantification.
Consistent with our previous observations (11), LIGHT-induced CXCL12 expression was less robust in retrovirally transduced HUVEC than untransduced cells (Fig. 2B). Nevertheless, incubation with LIGHT up-regulated CXCL12 expression to between 1.5 and 2.5-fold over basal levels, and as expected, this up-regulation was not affected by IKKβK44M (Fig. 2B). However, IKKβK44M completely blocked the negative regulatory effect of TNF on LIGHT-induced CXCL12 expression that was enhanced to the same level as LIGHT stimulation alone (Fig. 2B). Thus, we conclude that activation of the IKKβ-dependent classical NF-κB pathway in HUVEC by TNF negatively regulates basal and LIGHT-induced CXCL12 expression.
To further investigate the role of IKKβ in regulating CXCL12 expression, we examined CXCL12 levels in WT, IKKα-, and IKKβ-deficient MEFs. Unlike HUVEC, CXCL12 expression was not increased by LIGHT in MEFs (data not shown); however, basal CXCL12 levels in IKKα−/− cells were 8-fold less than those in WT MEFs (Fig. 2C). In contrast, CXCL12 levels in cells lacking IKKβ were 10-fold higher than WT. Hence, IKKα is required to maintain normal basal CXCL12 levels, and IKKβ suppresses CXCL12 expression.
TNF Induces Nuclear Localization of p100·RelB Complexes in HUVEC
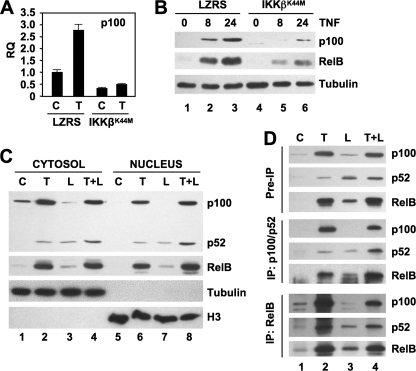
As a crucial substrate for NC NF-κB signaling, p100 is itself a classical NF-κB-dependent gene (16). In agreement with this, we found that p100 levels are enhanced in TNF-stimulated EC (Fig. 1A) (11). To confirm that classical signaling regulates p100 expression in HUVEC, we performed qRT-PCR and immunoblotting in LZRS- and IKKβK44M-transduced cells. As expected, IKKβK44M blocked TNF-induced p100 gene and protein expression (Fig. 3, A and B). Moreover, incubation with TNF up-regulated RelB expression in HUVEC, and this was also inhibited by IKKβK44M (Fig. 3B). These findings therefore indicate that TNF up-regulates expression of both p100 and RelB via the IKKβ-dependent classical NF-κB pathway in HUVEC.
FIGURE 3.
TNF up-regulates p100 and RelB and induces the nuclear localization of p100·RelB complexes. A, LZRS- or LZRS-IKKβK44M-transduced HUVEC were either untreated or incubated with TNF (10 ng/ml) for 8 h, and p100 expression was determined by qRT-PCR. RQ, relative quantification. B, LZRS- or LZRS-IKKβK44M-transduced HUVEC were incubated with TNF for the times shown, and then whole cell lysates were immunoblotted using anti-p100, anti-RelB, or anti-tubulin as indicated. Both p100 and RelB were present in unstimulated cells; however, these are not evident as the blots were underexposed for clearer visualization of the inhibitory effects of IKKβK44M on TNF-stimulated expression. C, HUVEC were either untreated (C) or incubated with TNF (T), LIGHT (L; 100 ng/ml), or a combination of both cytokines (L+T) for 24 h, and cytosolic (lanes 1–4) and nuclear (lanes 5–8) lysates were prepared. The lysates were immunoblotted using either anti-p100/p52 or anti-RelB as indicated. Lysates were also immunoblotted using anti-tubulin or anti-histone H3 (H3) as loading controls and to confirm the integrity of the cytosolic and nuclear lysates, respectively. As in B, RelB was present in control cytosolic extracts, but exposures were adjusted to visualize activation. D, nuclear lysates were prepared from HUVEC treated as described in C. A portion (5%) of each extract was retained as a preimmunoprecipitation sample, and the remainder was divided equally and immunoprecipitated using either anti-p100/p52 or anti-RelB as indicated (left). Preimmunoprecipitation and immunoprecipitation (IP) samples were immunoblotted using the antibodies indicated on the right.
Classical NF-κB-induced p100 augments NC NF-κB signaling and gene expression in B cells (15); however, our data demonstrate that classical signaling inhibits NC NF-κB-dependent gene expression in HUVEC without blocking p100 processing to p52. Recent studies have shown that p100 functions independently of the NC NF-κB pathway as an IκB that binds and retains classical NF-κB complexes in the cytoplasm (17–21). As blocking classical NF-κB activation reverses the negative regulation of CXCL12 expression by TNF (Fig. 2), we questioned whether TNF-up-regulated p100 inhibits the nuclear translocation of p52·RelB NC NF-κB complexes in HUVEC.
Consistent with the effects observed in whole cell lysates (Fig. 1A), TNF increased the levels of p100 and p52 in cytosolic extracts (Fig. 3C). In contrast, cytosolic p100 was decreased by LIGHT, whereas p52 was enhanced indicating processing of p100. A combination of both cytokines increased both p100 and p52 in the cytosol. Furthermore, the level of cytosolic RelB, though expressed basally, was robustly increased by TNF either alone or together with LIGHT, whereas LIGHT alone caused only a modest increase in RelB levels. We detected only low levels of p100, p52, or RelB in the nuclei of unstimulated HUVEC (Fig. 3C); however, as we demonstrated previously (11), incubation with LIGHT induced nuclear accumulation of p52 and RelB but negligible p100, indicating activation of NC NF-κB. In contrast, TNF either alone or together with LIGHT induced nuclear translocation of p100 along with RelB and p52. Notably, the levels of nuclear RelB induced by TNF were significantly higher than those induced by LIGHT. These findings therefore demonstrate that TNF signaling does not block LIGHT-induced nuclear localization of p52 and RelB.
To further investigate the nuclear NF-κB complexes in TNF- and LIGHT-stimulated HUVEC, we performed immunoprecipitations from nuclear extracts using anti-RelB or anti-p100/p52. Both p100 and p52 were coimmunoprecipitated with anti-RelB from cells treated with TNF either alone or together with LIGHT (Fig. 3D, lanes 2 and 4). In contrast, only p52 coprecipitated with RelB from LIGHT-treated cells (Fig. 3D, lane 3). Similarly, p100, p52, and RelB precipitated with anti-p100/p52 from the nuclei of HUVEC treated with TNF alone or together with LIGHT (Fig. 3D, lanes 2 and 4), whereas only p52 and RelB were immunoprecipitated with anti-p100/p52 from LIGHT-stimulated cells (Fig. 3D, lane 3).
In summary, Fig. 3 shows that although TNF strongly up-regulates expression of p100 and RelB, this does not block LIGHT-induced p100 processing to p52 or LIGHT-induced nuclear translocation of p52·RelB NF-κB complexes. However, unlike LIGHT, TNF induces the nuclear accumulation of p100·RelB complexes.
Ectopic p100 Does Not Affect CXCL12 Levels in HUVEC
Ectopic expression of p100 in T cells was demonstrated previously to recapitulate the negative regulation of classical NF-κB signaling that occurs following extended TCR stimulation (20). Although we find that TNF does not block nuclear localization of p52-RelB (Fig. 3), these earlier findings led us to question whether enhanced p100 expression alone was sufficient to mimic the effects of TNF and inhibit LIGHT-induced CXCL12 in HUVEC.
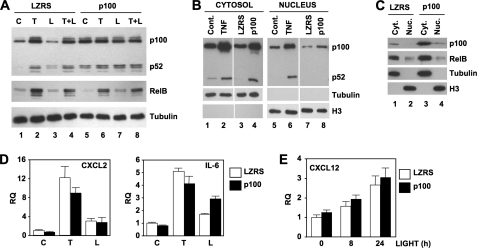
To elevate cellular p100, we retrovirally transduced HUVEC with LZRS-p100 and as shown in Fig. 4A, expression of p100 in transduced cells was similar to that up-regulated by TNF in control EC (compare p100 in lanes 1, 2, and 5). In line with TNF stimulation, exogenous p100 expression increased the basal level of p52; however, unlike the effects of TNF, basal RelB was not up-regulated in LZRS-p100 cells (lane 5). Similarly, RelB was only modestly increased in response to LIGHT in LZRS-p100 transduced HUVEC (Fig. 4A, lane 7). In contrast, TNF alone or together with LIGHT robustly up-regulated RelB expression in LZRS-p100 transduced cells, similar to the effects observed in LZRS vector alone transduced HUVEC.
FIGURE 4.
Elevated p100 levels do not affect CXCL12 expression by HUVEC. A, LZRS- or LZRS-p100-transduced HUVEC were either untreated (C) or incubated with TNF (T; 10 ng/ml), LIGHT (L; 100 ng/ml), or a combination of both cytokines (L+T) for 24 h, and then lysates were immunoblotted using either anti-p100/p52, anti-RelB, or anti-tubulin as a loading control. B, cytosolic (lanes 1–4) and nuclear (lanes 5–8) extracts were made from untransduced HUVEC that were either untreated control (lanes 1 and 5; cont.) or incubated for 24 h with TNF (10 ng/ml) (lanes 2 and 6), and LZRS- (lanes 3 and 7) and LZRS-p100-transduced HUVEC (lanes 4 and 8). Extracts were immunoblotted using anti-p100/p52, anti-tubulin, or anti-histone 3 (H3) as indicated. C, cytosolic (Cyt.) and nuclear (Nuc.) extracts were made from LZRS- and LZRS-p100-transduced HUVEC, and then samples were immunoblotted using either anti-p100/p52, anti-RelB, anti-tubulin, or anti-histone 3 as shown. D, LZRS- (open bar) or LZRS-p100-transduced (filled bar) HUVEC were either untreated (C) or incubated with TNF (T; 10 ng/ml), or LIGHT (L; 100 ng/ml) for 8 h, and then expression of CXCL2 (left) and IL-6 (right) was determined by qRT-PCR. E, LZRS- (open bar) or LZRS-p100-transduced (filled bar) HUVEC were either untreated or incubated with LIGHT (100 ng/ml) for the times indicated, and then CXCL12 expression was determined by qRT-PCR. RQ, relative quantification.
We next compared p100 and RelB levels in cytosolic and nuclear extracts from untreated, TNF-stimulated, LZRS-, and LZRS-p100 transduced HUVEC. As shown in Fig. 4B, cytosolic p100 and p52 levels in LZRS-p100 transduced HUVEC were similar to those in TNF-stimulated control cells (compare lanes 2 and 4). However, although it migrated to the nucleus, the amount of nuclear p100 in LZRS-p100 HUVEC was significantly less than the levels induced by TNF, and we did not detect any nuclear p52 in the LZRS-p100 transduced cells (Fig. 4B, lane 8). Furthermore, the amount of nuclear RelB in p100-transduced HUVEC was no different from nuclear RelB levels in LZRS control cells (Fig. 4C), again indicating that exogenous p100 does not mimic the effects of TNF.
To determine the effects of elevated p100 expression on classical and NC NF-κB-dependent gene expression we performed qRT-PCR analysis. Surprisingly, in light of previous reports that p100 functions as an IκB for classical NF-κB (17–27), exogenous p100 only modestly reduced TNF-induced CXCL2 and IL-6 expression in HUVEC (Fig. 4D). Similar effects were observed with other classical NF-κB-dependent genes including CX3CL1 (data not shown). Furthermore, ectopic p100 did not affect LIGHT-induced CXCL2 and consistently elevated LIGHT-stimulated IL-6 expression (Fig. 4D). Importantly, unlike TNF signaling, exogenously enhanced p100 did not affect basal or LIGHT-induced CXCL12. We therefore conclude that despite being expressed at the same level as TNF-induced p100, ectopic p100 alone does not replicate the negative regulatory effects of TNF on basal and LIGHT-stimulated CXCL12 expression by HUVEC.
Exogenous RelB Recapitulates Effects of TNF on Basal and LIGHT-induced CXCL12
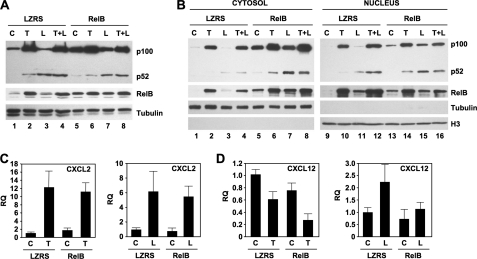
In addition to up-regulating p100, TNF enhanced the levels of RelB in HUVEC via the IKKβ-dependent classical NF-κB pathway (Fig. 3B). Given the lack of effects of ectopic p100 (Fig. 4), we questioned whether up-regulated RelB expression plays a role in TNF-mediated inhibition of basal and LIGHT-induced CXCL12 expression. To investigate this, we retrovirally transduced HUVEC with LZRS-RelB and generated stably transduced cells in which RelB levels were similar to those induced by TNF in control cells (Fig. 5A; compare RelB in lanes 1, 2, and 5). Notably, ectopic RelB robustly increased the amount of p100 and caused a modest rise in p52 levels in resting cells (Fig. 5A; compare p100 in lanes 1 and 5). RelB did not affect p100 processing to p52 in response to LIGHT, although similar to the effects of TNF, the levels of p100 remained elevated compared with LZRS control cells in LIGHT-stimulated LZRS-RelB-transduced cells (Fig. 5A, lane 7). Hence, in this regard, exogenously enhanced RelB expression more closely resembles the effects of TNF than ectopic p100 that did not concomitantly enhance RelB levels (Fig. 4A).
FIGURE 5.
Elevated RelB levels inhibit basal and LIGHT-induced CXCL12 expression by HUVEC. A and B, LZRS- or LZRS-RelB-transduced HUVEC were either untreated (C) or incubated with TNF (T; 10 ng/ml), LIGHT (L; 100 ng/ml), or a combination of both cytokines (L+T) for 24 h, and then either whole cell lysates (A) or cytosolic and nuclear extracts were prepared (B). Samples were immunoblotted using either anti-p100/p52, anti-RelB, anti-tubulin, or anti-histone 3 (H3) as indicated. C and D, LZRS- or LZRS-RelB-transduced HUVEC were either untreated (C) or incubated with TNF (10 ng/ml) or LIGHT (100 ng/ml) for 8 h, and then the expression levels of CXCL2 (C) and CXCL12 (D) were determined by qRT-PCR. RQ, relative quantification.
To determine the effects of exogenous RelB on nuclear localization of NF-κB proteins, we immunoblotted cytoplasmic and nuclear extracts from untreated HUVEC and cells stimulated with TNF, LIGHT, or a combination of both cytokines. Agreeing with the observations in whole cell extracts (Fig. 5A), ectopic RelB enhanced the basal level of cytoplasmic p100 (Fig. 5B, lane 5). Furthermore, the amount of cytoplasmic p100 in LIGHT-stimulated cells remained elevated in LZRS-RelB transduced HUVEC compared with control cells, and this was accompanied by an increase in cytoplasmic p52 (Fig. 5B, lane 7). Similar to the effects of TNF in control cells, RelB and p100 were both present in the nucleus of unstimulated LZRS-RelB transduced HUVEC (Fig. 5B, lane 13). Moreover, unlike control transduced cells, nuclear p100 levels remained high in LIGHT-stimulated HUVEC-expressing ectopic RelB, in which p100 processing to p52 was intact (Fig. 5B, lane 15). These findings therefore indicate that the effects of exogenously elevating RelB levels alone resemble those of TNF on both RelB and p100 nuclear translocation in the absence and presence of LIGHT stimulation.
Analysis of classical NF-κB-dependent CXCL2 expression in control and LZRS-RelB-transduced HUVEC revealed no effects of ectopic RelB on basal, TNF-, or LIGHT-induced CXCL2 (Fig. 5C). A similar lack of effect of RelB was observed when we examined basal or cytokine-induced expression of IL-6 (data not shown). In contrast, basal CXCL12 expression was reduced in LZRS-RelB-transduced HUVEC similar to the decreased basal expression in TNF-treated control cells (Fig. 5D, left panel). Moreover, treatment of LZRS-RelB-transduced HUVEC with TNF further reduced basal CXCL12 expression in these cells. Importantly, and consistent with the effects of TNF, LIGHT-induced CXCL12 expression was inhibited in LZRS-RelB cells (Fig. 5D, right panel). These findings therefore establish that ectopic expression of RelB alone recapitulates the negative regulatory effects of TNF signaling on basal and NC NF-κB-dependent CXCL12 expression by HUVEC.
DISCUSSION
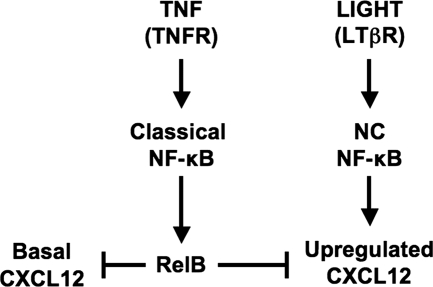
We showed recently that CXCL12 is up-regulated in EC by LIGHT or LTα1β2 via activation of the NC NF-κB pathway (11). In contrast, TNF only activates the classical NF-κB pathway and does not enhance CXCL12 expression in HUVEC (11). Several recent studies demonstrated that p100 and RelB function as regulators of classical pathway activation (16–18, 21, 25, 27, 28) suggesting interplay between the NC and classical pathways. Furthermore, studies in B cells established that classical NF-κB-dependent up-regulation of p100 provides the substrate pool required for NC signaling (15). We therefore initiated this study to determine whether classical NF-κB activation functionally cross-talks with NC signaling in EC and augments NC NF-κB activation and CXCL12 expression in HUVEC. Surprisingly, although TNF robustly up-regulated p100 and RelB expression, it completely blocked LIGHT-induced CXCL12 expression. Furthermore, TNF significantly reduced basal CXCL12, and these inhibitory effects were mimicked by ectopic expression of RelB. Our findings therefore reveal a model for classical and NC NF-κB signaling cross-talk in EC, in which classical NF-κB activation negatively regulates both basal and NC-NF-κB-dependent CXCL12 expression via up-regulation of RelB (Fig. 6).
FIGURE 6.
Classical NF-κB signaling inhibits LIGHT-induced CXCL12 expression via up-regulation of RelB. LIGHT stimulation up-regulates CXCL12 expression in HUVEC, whereas both basal and LIGHT-induced CXCL12 levels are inhibited by TNF. Our findings demonstrate that classical NF-κB-dependent up-regulation of RelB negatively regulates basal and NC NF-κB-dependent CXCL12 expression HUVEC.
CXCL12 is constitutively expressed by vascular, stromal, and hematopoetic cells and is a key regulator of hematopoesis and myelopoesis (29, 30). In addition, CXCL12 regulates the recruitment and migration of hematopoetic progenitors, monocytes, and lymphocytes and plays important roles in the development of chronic inflammation, tumorigenesis, and metastasis of distinct solid tumors (29–31). Consequently, understanding the mechanisms that regulate CXCL12 expression is crucial for potentially targeting its pathophysiological function. Several extracellular stimuli including sheer stress, hypoxia, and various growth factors and cytokines up-regulate CXCL12 expression (29–31), and we and others (10, 11) established that CXCL12 is enhanced in response to LTβR ligation. Signaling pathways that play a role in up-regulating CXCL12 include JNK, hypona inducible factor, and NC NF-κB activation (10, 11, 32, 33); however, the role of classical NF-κB activation in regulating CXCL12 expression has not been directly addressed. Intriguingly, we found that the down-regulatory effects of TNF on CXCL12 expression by HUVEC were reversed by inhibition of the classical NF-κB pathway, demonstrating a novel role for classical NF-κB signaling as a negative regulator of CXCL12 expression. Moreover, the enhanced level of CXCL12 expression we observed in IKKβ-deficient MEFs strongly suggests that basal classical NF-κB signaling suppresses constitutive CXCL12 expression.
Consistent with the inhibitory effects of TNF on basal CXCL12 expression by HUVEC, previous studies demonstrated reduction of constitutive CXCL12 in dermal fibroblasts (34), bone marrow stromal cells (35), and brain microvascular EC (36) following incubation with proinflammatory cytokines. Our findings further establish that TNF inhibits up-regulated CXCL12 expression, and we show that the inhibitory effect on basal and induced CXCL12 requires classical NF-κB pathway activation. Notably, although inhibition of classical signaling rescued the ability of LIGHT to up-regulate CXCL12 in the presence of TNF (Fig. 2B), this also robustly increased CXCL12 levels in cells incubated with TNF alone (Fig. 2A). As TNF does not activate the NC NF-κB pathway (2, 4), it is possible that a separate TNF-induced signal such as JNK or hypona inducible factor activation up-regulates CXCL12 in the absence of classical NF-κB signaling (32, 33). Further work is required to determine the mechanism that regulates CXCL12 expression in the absence of classical NF-κB activation; however, our data indicate that selective therapeutic targeting of classical NF-κB may enhance the expression of genes such as CXCL12 that are not normally up-regulated by proinflammatory cytokines.
To elucidate the mechanism of TNF-induced down-regulation of CXCL12 expression, we first questioned the role of elevated p100. Although p100 is crucial for NC NF-κB activation, a growing body of evidence has identified p100 as an inhibitor of classical NF-κB activity (16–23, 25–27). In resting cells, p100 functions as an IκB that binds to classical NF-κB heterodimers (e.g. p50-p65) blocking their nuclear translocation (16, 17, 19, 22, 23, 25, 26). Furthermore, up-regulated p100 provides feedback inhibition of classical NF-κB signaling in T cells, fibroblasts, and osteoclasts (18, 20, 21, 25, 27). However, when we overexpressed p100 in HUVEC to the same levels as that induced by TNF, it did not block nuclear translocation of NC NF-κB or expression of CXCL12. Notably, ectopic p100 did not inhibit TNF-induced classical NF-κB-dependent gene expression in HUVEC, suggesting that the inhibitory function of p100 in this regard is cell type-specific. Finally, exogenously enhanced p100 did not stabilize the levels of RelB as described previously in fibroblasts (24), again suggesting that the effects of elevated p100 are cell type-specific. Hence, our findings demonstrate that elevated p100 expression alone does not recapitulate the effects of TNF on basal or NC-NF-κB-dependent CXCL12 expression by HUVEC.
In contrast to the effects of p100, ectopically expressed RelB reduced both basal and LIGHT-induced CXCL12 expression in HUVEC. Moreover, exogenous RelB expression was accompanied by an increase in p100 levels, and qRT-PCR analysis demonstrated that this requires, at least in part, transcriptional up-regulation of p100 (data not shown). Similar to TNF stimulation, ectopic RelB promoted the nuclear localization of p100·RelB complexes, whereas this was not observed with expression of p100. These findings therefore establish that expression of RelB alone fully recapitulates the effects of TNF on CXCL12 expression, endogenous p100 levels, and the nuclear localization of NF-κB complexes. Hence, we conclude that classical NF-κB-dependent up-regulation of RelB negatively regulates both basal and NC-NF-κB dependent CXCL12 expression in HUVEC (Fig. 6).
RelB has been shown to function as both an activator and repressor of classical NF-κB-dependent gene transcription (18, 37–42), and the expression levels of classical NF-κB-regulated genes are markedly enhanced in RelB-deficient cells (41, 42). To date, however, the only role described for RelB in NC NF-κB signaling is functioning as the transcriptionally active partner of p52 (2, 4). Our findings now demonstrate that elevation of RelB levels by either activation of the classical NF-κB pathway or exogenous RelB expression, transcriptionally represses basal and NC-NF-κB-dependent CXCL12 expression. Consequently, our data establish RelB as a mechanistic link underlying negative regulatory cross-talk between the classical and NC NF-κB pathways (Fig. 6).
The mechanism for the repressive function for RelB is not clear; however, it has been shown to form inhibitory nuclear NF-κB complexes with p65, p50, and p100 (18, 38, 39, 43). We found that both TNF up-regulated and ectopically expressed RelB associates with p100 in the nucleus, and it is possible that these complexes block the ability of active p52-RelB to transcriptional regulate CXCL12. Intriguingly, recent studies have also shown that RelB can associate directly or indirectly with DNA and histone methyltransferases, which block transcription at target genes (37, 40, 44). Due to the current lack of any known NC or classical NF-κB consensus binding sites in the CXCL12 promoter, it is not yet possible to determine how RelB functions at the level of DNA-binding, co-factor recruitment, and transcriptional activation of CXCL12. Nevertheless, our findings clearly expand the negative regulatory function for RelB to include effects on basal and NC NF-κB-dependent expression of CXCL12.
Contrasting with the negative regulation of CXCL12 expression in HUVEC, studies in lymph node stromal cells (12, 14) and smooth muscle cells (13) showed that TNF stimulation enhanced LTβR ligation-induced expression of CXCL13. We did not detect CXCL13 in HUVEC or dermal EC (11), suggesting that the expression profiles of NC NF-κB-dependent genes are cell type-specific. Moreover, as TNF blocks LTβR-induced CXCL12 but augments CXCL13, the negative regulation of NC NF-κB-dependent gene expression by classical NF-κB-induced RelB may only affect a subset of NC genes. It will therefore be important to identify the panels of genes that are either augmented or inhibited by combined classical and NC NF-κB activation and to determine the mechanisms underlying the cell type and gene specificity of these distinct outcomes.
Combining LTβR ligation with TNF stimulation did not affect or in some cases enhanced classical NF-κB-dependent gene expression in HUVEC. Similar up-regulation of classical NF-κB regulated genes by concomitant TNF stimulation and LTβR ligation was reported previously in fibroblasts (45) and smooth muscle cells (13). The mechanism of this up-regulation is not yet known; however, it is possible that additive activation of classical NF-κB by both stimuli may enhance the overall transcription of target genes. Alternatively, stabilization of NF-κB inducible kinase by LTβR signaling, which can, in turn, augment classical NF-κB activation, may contribute to this enhanced expression (45). Further investigation of the combined effects of LIGHT and TNF on classical gene expression in HUVEC is necessary; however, our results demonstrate that activation of the NC pathway does not inhibit classical NF-κB signaling and gene expression. Hence, the negative regulatory cross-talk between the classical and NC NF-κB pathways that we have identified is selective and one-directional (Fig. 6).
Acknowledgment
We thank the staff of the Labor and Delivery Department at the Hospital of the University of Pennsylvania for collecting umbilical cords.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1HL080612 and RO1HL080612-04S1. This work was also supported by W. W. Smith Charitable Trust Grant H0703.
- EC
- endothelial cells
- HUVEC
- human umbilical vein endothelial cells
- IKK
- IκB kinase
- LTβR
- lymphotoxin-β receptor
- NC
- noncanonical
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1.Pober J. S., Sessa W. C. (2007) Nat. Rev. Immunol. 7, 803–815 [DOI] [PubMed] [Google Scholar]
- 2.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 3.Hayden M. S., West A. P., Ghosh S. (2006) Oncogene 25, 6758–6780 [DOI] [PubMed] [Google Scholar]
- 4.Bonizzi G., Karin M. (2004) Trends Immunol. 25, 280–288 [DOI] [PubMed] [Google Scholar]
- 5.Häcker H., Karin M. (2006) Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 6.Scheidereit C. (2006) Oncogene 25, 6685–6705 [DOI] [PubMed] [Google Scholar]
- 7.Solt L. A., May M. J. (2008) Immunol. Res. 42, 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejardin E. (2006) Biochem. Pharmacol. 72, 1161–1179 [DOI] [PubMed] [Google Scholar]
- 9.Xiao G., Rabson A. B., Young W., Qing G., Qu Z. (2006) Cytokine Growth Factor Rev. 17, 281–293 [DOI] [PubMed] [Google Scholar]
- 10.Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. (2002) Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 11.Madge L. A., Kluger M. S., Orange J. S., May M. J. (2008) J. Immunol. 180, 3467–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katakai T., Suto H., Sugai M., Gonda H., Togawa A., Suematsu S., Ebisuno Y., Katagiri K., Kinashi T., Shimizu A. (2008) J. Immunol. 181, 6189–6200 [DOI] [PubMed] [Google Scholar]
- 13.Lötzer K., Döpping S., Connert S., Gräbner R., Spanbroek R., Lemser B., Beer M., Hildner M., Hehlgans T., van der Wall M., Mebius R. E., Lovas A., Randolph G. J., Weih F., Habenicht A. J. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suto H., Katakai T., Sugai M., Kinashi T., Shimizu A. (2009) Int. Immunol. 21, 467–476 [DOI] [PubMed] [Google Scholar]
- 15.Stadanlick J. E., Kaileh M., Karnell F. G., Scholz J. L., Miller J. P., Quinn W. J., 3rd, Brezski R. J., Treml L. S., Jordan K. A., Monroe J. G., Sen R., Cancro M. P. (2008) Nat. Immunol. 9, 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basak S., Shih V. F., Hoffmann A. (2008) Mol. Cell. Biol. 28, 3139–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basak S., Kim H., Kearns J. D., Tergaonkar V., O'Dea E., Werner S. L., Benedict C. A., Ware C. F., Ghosh G., Verma I. M., Hoffmann A. (2007) Cell 128, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derudder E., Dejardin E., Pritchard L. L., Green D. R., Korner M., Baud V. (2003) J. Biol. Chem. 278, 23278–23284 [DOI] [PubMed] [Google Scholar]
- 19.Ishimaru N., Kishimoto H., Hayashi Y., Sprent J. (2006) Nat. Immunol. 7, 763–772 [DOI] [PubMed] [Google Scholar]
- 20.Legarda-Addison D., Ting A. T. (2007) J. Immunol. 178, 7767–7778 [DOI] [PubMed] [Google Scholar]
- 21.Novack D. V., Yin L., Hagen-Stapleton A., Schreiber R. D., Goeddel D. V., Ross F. P., Teitelbaum S. L. (2003) J. Exp. Med. 198, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dejardin E., Bonizzi G., Bellahcène A., Castronovo V., Merville M. P., Bours V. (1995) Oncogene 11, 1835–1841 [PubMed] [Google Scholar]
- 23.Dejardin E., Deregowski V., Chapelier M., Jacobs N., Gielen J., Merville M. P., Bours V. (1999) Oncogene 18, 2567–2577 [DOI] [PubMed] [Google Scholar]
- 24.Fusco A. J., Savinova O. V., Talwar R., Kearns J. D., Hoffmann A., Ghosh G. (2008) J. Biol. Chem. 283, 12324–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih V. F., Kearns J. D., Basak S., Savinova O. V., Ghosh G., Hoffmann A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speirs K., Lieberman L., Caamano J., Hunter C. A., Scott P. (2004) J. Immunol. 172, 752–756 [DOI] [PubMed] [Google Scholar]
- 27.Yao Z., Xing L., Boyce B. F. (2009) J. Clin. Invest. 119, 3024–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basak S., Hoffmann A. (2008) Cytokine Growth Factor Rev. 19, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rot A., von Andrian U. H. (2004) Annu. Rev. Immunol. 22, 891–928 [DOI] [PubMed] [Google Scholar]
- 30.Sallusto F., Baggiolini M. (2008) Nat. Immunol. 9, 949–952 [DOI] [PubMed] [Google Scholar]
- 31.Vandercappellen J., Van Damme J., Struyf S. (2008) Cancer Lett. 267, 226–244 [DOI] [PubMed] [Google Scholar]
- 32.Loh S. A., Chang E. I., Galvez M. G., Thangarajah H., El-ftesi S., Vial I. N., Lin D. A., Gurtner G. C. (2009) Plast. Reconstr. Surg. 123, 65S-75S [DOI] [PubMed] [Google Scholar]
- 33.Sung M. L., Wu C. C., Chang H. I., Yen C. K., Chen H. J., Cheng J. C., Chien S., Chen C. N. (2009) Circ. Res. 105, 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fedyk E. R., Jones D., Critchley H. O., Phipps R. P., Blieden T. M., Springer T. A. (2001) J. Immunol. 166, 5749–5754 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q., Guo R., Schwarz E. M., Boyce B. F., Xing L. (2008) Arthritis Res. Ther. 10, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K. K., Dorovini-Zis K. (2009) J. Neuroimmunol. 215, 49–64 [DOI] [PubMed] [Google Scholar]
- 37.Croxton R., Puto L. A., de Belle I., Thomas M., Torii S., Hanaii F., Cuddy M., Reed J. C. (2006) Cancer Res. 66, 9026–9035 [DOI] [PubMed] [Google Scholar]
- 38.Jacque E., Tchenio T., Piton G., Romeo P. H., Baud V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14635–14640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marienfeld R., May M. J., Berberich I., Serfling E., Ghosh S., Neumann M. (2003) J. Biol. Chem. 278, 19852–19860 [DOI] [PubMed] [Google Scholar]
- 40.Puto L. A., Reed J. C. (2008) Genes Dev. 22, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y., Chen S., Wang Y., Mackman N., Ku G., Lo D., Feng L. (1999) Mol. Cell. Biol. 19, 7688–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y., Pauza M. E., Feng L., Lo D. (1997) Am. J. Pathol. 151, 375–387 [PMC free article] [PubMed] [Google Scholar]
- 43.Yoza B. K., Hu J. Y., Cousart S. L., Forrest L. M., McCall C. E. (2006) J. Immunol. 177, 4080–4085 [DOI] [PubMed] [Google Scholar]
- 44.Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarnegar B., Yamazaki S., He J. Q., Cheng G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]