Abstract
Inspection of the complete genome of the yeast Yarrowia lipolytica for the presence of genes encoding homologues of known telomere-binding proteins surprisingly revealed no counterparts of typical yeast Myb domain-containing telomeric factors including Rap1 or Taz1. Instead, we identified a gene, YALIOD10923g, encoding a protein containing two Myb domains, exhibiting a high degree of similarity to the Myb domain of human telomeric proteins TRF1 and TRF2 and homologous to an essential fission yeast protein Mug152 whose expression is elevated during meiosis. The protein, which we named Tay1p (telomere-associated in Yarrowia lipolytica 1), was purified for biochemical studies. Using a model Y. lipolytica telomere, we demonstrate that the protein preferentially binds to Y. lipolytica telomeric tracts. Tay1p binds along the telomeric tract as dimers and larger oligomers, and it is able to remodel the telomeric DNA into both looped structures and synaptic complexes of two model telomere DNAs. The ability of Tay1p to induce dimerization of telomeres in vitro goes in line with its oligomeric nature, where each oligomer can employ several Myb domains to form intermolecular telomere clusters. We also provide experimental evidence that Tay1p may be associated with Y. lipolytica telomeres in vivo. Together with its homologues from Schizosaccharomyces pombe and several basidiomycetous fungi (Sánchez-Alonso, P., and Guzman, P. (2008) Fungal Genet. Biol. 45, S54–S62), Tay1p constitutes a novel family of putative telomeric factors whose analysis may be instrumental in understanding the function and evolution of double-stranded DNA telomeric proteins.
Keywords: DNA-binding Protein, DNA-Protein Interaction, Electron Microscopy (EM), Telomere, Yeast, Schizosaccharomyces pombe, Tay1, Yarrowia lipolytica
Introduction
All linear chromosomes possess two telomeres, specialized nucleoprotein structures that protect the genomic integrity by providing a solution to the “end-replication problem,” prevent inappropriate action of DNA repair machinery, and mask the chromosomal ends against degradation by nucleases (1). Human telomeric chromatin is composed of about 200 different proteins (2) playing various roles at chromosomal termini, including transcription and packaging of TERRA RNA (telomere repeat containing RNA) (3, 4) or in the organization of higher order telomeric chromatin structure (5). Telomere protection is facilitated in large part by proteins that specifically bind to either single-stranded (ss) or double-stranded (ds) regions of telomeric DNA (6). The telomere-binding proteins form a cap that protects chromosome ends against activities of DNA repair machinery (7). Whereas the G-rich 3′ ssDNA telomeric overhang is bound by oligosaccharide binding-fold-containing proteins like Pot1p (8) and Cdc13p (9), the duplex region of the telomere is associated with dsDNA-binding proteins containing a Myb/SANT domain. These include mammalian TRF1 and TRF2 proteins (10), budding yeast proteins Rap1 and Tbf1 (11, 12), and the fission yeast telomeric proteins Taz1 (13) and Tbf1 (14). The telomeric proteins protect telomeres at two levels. First, proteins such as TRF2 and Taz1p mediate formation of a telomeric loop and hide the 3′ telomeric overhang into the ds region of the telomere, thus, preventing inappropriate recognition by the DNA repair machinery (15, 16). Second, the telomere-binding proteins associate with other components and form a special nucleoprotein structure termed shelterin (10). In addition, telomere-binding proteins are important for recruitment of telomerase and other enzymes involved in telomere maintenance at chromosomal ends (7). The central role of telomere-binding proteins in protecting chromosomal ends and regulating DNA transactions, including telomerase-dependent elongation and recombination, underlines the importance of investigating their biochemical properties.
Studies in yeasts, especially in Saccharomyces cerevisiae, Kluyveromyces lactis, Candida albicans, and Schizosaccharomyces pombe, have provided invaluable information about the maintenance and dynamics of nuclear telomeres (17). From these studies it is clear that, whereas some features of telomeres are general and carry over to higher eukaryotes, some telomere-associated components are specific for a given phylogenetic clade. Such findings have provided information about evolutionary trajectories leading to contemporary solutions of telomere-associated problems and substantiated comparative analyses of telomeres in non-conventional yeast species (18, 19).
The species with a potential to open new venues in telomere research is Yarrowia lipolytica. This yeast is an important industrial producer of citric, isocitric, and 2-oxoglutaric acids (20), and it is a well established yeast model for the studies of protein secretion (21), peroxisome biogenesis (22), mitochondrial bioenergetics (23), and cell morphogenesis and differentiation (24, 25). The existence of a sexual cycle as well as a wide variety of genetically well defined strains, molecular tools, and availability of a complete genomic sequence make all peculiarities of Y. lipolytica amenable to detailed analysis.
Y. lipolytica is very distantly related to most yeast model systems, which is exemplified by its position at the basal branches of the phylogenetic tree of hemiascomycetes. In fact, Y. lipolytica shares a number of features with higher eukaryotes, such as dispersion of the rDNA clusters and of the 5 S RNA genes, small nuclear RNA sizes, the protein secretion process, and the signal recognition particle 7 S RNA (26), supporting the idea that it represents an attractive model for studying diverse biological phenomena.
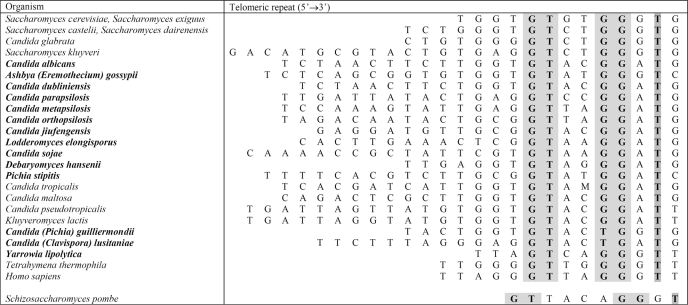
It was recently recognized that Y. lipolytica could also be exploited as an attractive model for telomere biology (19). Its telomeric repeats are represented by an array of 5′-GGGTTAGTCA-3′ sequences,5 whose length (500–1000 bp)6 is slightly larger than in other budding and fission yeasts. As in other yeast models, telomeres in Y. lipolytica seem to be maintained primarily by telomerase, which is backed-up by a potent recombination-dependent mechanism (27). In contrast to S. cerevisiae and S. pombe and similar to Kluyveromyces spp., Candida spp., and humans, the telomeric repeats in Y. lipolytica are highly regular. The repeat can be considered a variant of its human counterpart (5′-TTAGGG-3′), containing a four-base insertion between TTA and GGG. Significantly, the repeat contains a conserved 5′-GTN2–4GG(G/A)T-3′ motif (Table 1), shown to be important for binding of telomere-binding proteins like Rap1p (28–30). This suggests that the telomeres in Y. lipolytica are protected by Myb domain-containing proteins.
TABLE 1.
Telomeric repeats in various organisms exhibit a conserved 5′-GT-N2–4-GG(G/A)T-3′ motif based on Refs. 28–30; newly added organisms and sequences of their telomeric repeats are in bold (see Ref. 18 for more details)
Shaded are the conserved positions. The telomeric repeat of S. pombe is based on Ref. 59; Candida jiufengensis telomere is derived from the template domain of its telomerase RNA (Lukacisin, M., Tomaska, L., and Nosek, J., unpublished results).
When we searched for Y. lipolytica gene-encoding homologues of Rap1p and Taz1p in its genomic sequence, we were surprised to not find any homologues of these major yeast telomere-binding proteins. Therefore, we decided to initiate the studies of Y. lipolytica telomeres in more detail, with the aim of extending our knowledge of the maintenance of its telomeres. In this study we identified an open reading frame encoding an ∼50-kDa protein of a novel family of telomere-binding factors (31) containing two Myb domains exhibiting high similarity to the Myb domain of mammalian TRF1 and TRF2. Our biochemical analyses of the protein, which we named Tay1p (telomere-associated in Yarrowia lipolytica 1), demonstrate that it represents a novel telomeric protein and indicate that investigation of telomeres in Y. lipolytica may be fruitful for understanding the mechanisms implicated in telomere maintenance in general.
EXPERIMENTAL PROCEDURES
Microbial Strains
Y. lipolytica E129 (MAT A, lys11-23, leu2-270, ura3-302, xpr2-322) provided by C. Gaillardin (Institut National de la Recherche Agronomique, Grignon, France) was used as the source of genomic DNA for amplification of the TAY1 coding sequence. Y. lipolytica PO1h (ura3), provided by C. Madzak (Institut National de la Recherche Agronomique, Thiverval-Grignon, France), was used for ChIP experiments. Yeast cultures were grown in YPD medium (1% (w/v) yeast extract (Difco), 2% (w/v) Bacto Peptone (Difco), 2% (w/v) glucose) at 28 °C. Escherichia coli strain BL21-Gold(DE3)pLysS (Stratagene) was used for production of recombinant Tay1–6HN protein.
DNA Manipulations
Recombinant DNA techniques were carried out by standard procedures (32). DNA-modifying enzymes were used according to the instructions provided by the corresponding suppliers. Fungal genomic DNA was isolated as described (20). The oligonucleotides (supplemental Table 1) were synthesized by MWG Operon or Metabion. The plasmid pMH25 carrying ten Y. lipolytica telomeric repeats was constructed by annealing oligonucleotides YlTEL-sense and YlTEL-antisense followed by the digestion of the resulting dsDNA with EcoRI and ligation into the EcoRI-digested and -dephosphorylated plasmid pHIS2 (Clontech). We obtained the desired recombinant plasmid carrying 2 × 5 telomeric repeats separated by an EcoRI site and named it pMH25. Digestion of pMH25 with EcoRI yields a dsDNA fragment containing five telomeric repeats used in initial DNA binding assays.
Construction of the Expression Plasmid pTay1–6HN
TAY1, including the flanking sequences for recombinational cloning, was amplified using the primers Tay1-N-S and Tay-N-Anti. PCR was performed using 1 unit of Phusion Hot Start High-Fidelity DNA polymerase (Finnzymes) and contained each dNTP at 200 μm, corresponding primers at 1 μm, and 100 ng of genomic DNA. The initial 5-min denaturation at 95 °C was followed by 34 cycles of 45 s of denaturation at 95 °C, 45 s of annealing at 50 °C, and 3 min of polymerization at 72 °C. Finally, the reaction was incubated for 15 min at 72 °C. The resulting PCR product was gel-purified using the Zymoclean Gel DNA recovery kit (Zymo Research) and cloned into the pEcoli-Nterm vector (Invitrogen) using the In-fusion dry-down reaction according to the supplier's recommendation. The DNA was transformed into the Fusion blue competent cells (Invitrogen) and plated on a solid LB media containing 100 μg/ml ampicillin. Plasmid DNA was isolated using QIAprep Miniprep (Qiagen), and the plasmid DNA exhibiting the predicted restriction map was sequenced using T7 promoter and T7 terminator sequences as primers. The insert, including the flanking regions, perfectly matched the expected sequence, and the plasmid, named pTay1–6HN, was used for production of recombinant Tay1 protein in bacteria.
Purification of Recombinant Tay1 Protein
pTay1–6HN was transformed into BL21-Gold(DE3)pLysS cells, and the transformants were grown on LB plates containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. The cells were then inoculated into 30 ml of 2×YT media (1.6% (w/v) bacto-Tryptone, 1% (w/v) bacto-peptone, 1% (w/v) NaCl (pH 7.1)) containing 2% glucose, 100 μg/ml ampicillin, and 34 μg/ml chloramphenicol and cultivated overnight (15 h) at 37 °C and 225 rpm. The cells were centrifuged for 5 min at 3000 rpm (Sorvall RT 7 Plus) at 25 °C, washed once with 2×YT, inoculated into 1 liter of 2×YT containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol, and cultivated at 37 °C and 275 rpm until the A600 reached a value of 0.7–0.8. The culture was cooled to 28 °C followed by addition of isopropyl β-d-1-thiogalactopyranoside (final concentration 1 mm) and cultivation for additional 3 h at 28 °C. The culture was then centrifuged for 15 min at 5000 rpm at 4 °C (F10–6 × 500y rotor in Sorvall RC 6+), the cells were washed once with 200 ml of ice cold phosphate-buffered saline, and the pellet was frozen at −20 °C.
The pellet was thawed on ice (30–45 min) and resuspended in a final volume of 30 ml of buffer A (20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 1 mm DTT) containing 1× Complete (EDTA-free) protease inhibitors (Roche Applied Science), 10 mm MgCl2, 100 units of DNase I (Promega), and 2 μg of PureLink RNase A (Invitrogen). Lysozyme (United States Biochemicals) was added to a final concentration of 1 mg/ml, and the suspension was incubated for 15 min on ice with occasional shaking. The cells were broken by sonication (5 × 30 s at a setting of 6 (Branson Sonifer 450)). Each cycle of sonication was followed by a 1-min incubation on ice. Triton X-100 was added to a final concentration of 0.1%, and the suspension was sonicated one more time for 30 s and incubated for additional 15 min on ice. The insoluble material was pelleted by 30 min of centrifugation at 12,000 rpm at 4 °C (F21–8 × 50y in Sorvall RC 6+). The supernatant was mixed with a 1-ml bed volume of the TALON Superflow Metal Affinity Resin (Clontech) equilibrated with 3 × 10 volumes of buffer A. The whole suspension was transferred to a 50-ml Falcon tube and incubated for 60–90 min rocking the tube end-over-end at 4 °C. The beads were then washed 3 times with 20 volumes of buffer A containing 0.1% (v/v) Triton X-100 followed by 5 × 10 volumes of the wash buffer (20 mm HEPES-NaOH (pH 7.3), 300 mm NaCl, 10 mm imidazole (pH 7.5)). The beads were then transferred to a chromatographic column, and the bound proteins were eluted with 6 × 1 ml of elution buffer (20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 500 mm imidazole (pH 7.5)). The fractions containing Tay1 protein were pooled and loaded onto a 2-ml HiTrap heparin column (GE Healthcare) connected to an ÄKTA Purifier FPLC system (GE Healthcare). The beads were washed with 10 volumes of the binding buffer (20 mm HEPES-NaOH (pH 7.3), 5% (v/v) glycerol, 1 mm DTT). The bound proteins were eluted with 20 volumes of a linear (0–1.5 m NaCl) gradient, and 0.25-ml fractions were collected. The peak of Tay1p, essentially free of contaminating proteins, was observed between 0.8 and 0.9 m NaCl. The fractions containing purified Tay1p were pooled and dialyzed against 2 liters of 20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 15% (v/v) glycerol, and 1 mm DTT and stored in 100 μl aliquots at −80 °C. The proteins from the peak fractions were subjected to 10% SDS-PAGE (33) and stained with Coomassie Brilliant Blue R-250.
Electrophoretic Mobility Shift Assay (EMSA)
The initial double-stranded YlTEL probe used for EMSA was prepared by digestion of 15 μg of pMH25 plasmid in a final volume of 50 μl using 50 units of EcoRI (New England Biolabs) followed by dephosphorylation using 50 units of calf intestinal phosphatase. The resulting 50-bp fragment was gel-purified using the QIAquick gel extraction kit (Qiagen). The gel-isolated dsDNA YlTEL probe and single-stranded oligonucleotide probes for EMSA were prepared by labeling 200 ng of DNA with 10 units of T4 polynucleotide kinase (New England Biolabs) and 50 μCi of [γ-32P]ATP (final concentration 0.5 μm) for 60 min at 37 °C in a final volume of 20 μl. Reactions were diluted to 50 μl with 0.5 mm EDTA, 10 mm Tris-HCl (pH 8.0), and the labeled oligonucleotide was purified using G-50 Sephadex (Amersham Biosciences). Radiolabeled double-stranded probes were prepared by annealing a 2-fold molar excess of unlabeled complementary oligonucleotide with a corresponding labeled oligonucleotide. Typically, a 100-μl annealing reaction contained 160 ng of labeled and 320 ng of unlabeled oligonucleotide in 20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl. Reactions were boiled for 10 min in a water bath and cooled down until the water reached room temperature.
For the EMSA reactions, the YlTEL probe was diluted 5 times with 1× HN buffer (20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl). The typical DNA binding reaction contained 1 μl of the diluted dsDNA probe, 2–3 μg of recombinant Tay1p, and a DNA competitor as indicated under “Results.” The reactions were carried out in 1× HN buffer at room temperature for 15 min. DNA-protein complexes were resolved by electrophoresis on 5% polyacrylamide gels in 0.5× TBE (45 mm Tris borate, 1 mm EDTA) at 10 V/cm for 90 min. Gels were fixed for 10 min with 10 ml of 10% (v/v) methanol, 10% (v/v) acetic acid, and exposed wet to an x-ray film (Amersham Biosciences) or to Storage Phosphor Screens (Eastman Kodak Co.) for 3–15 h.
Construction of a Y. lipolytica Model Telomere
A plasmid carrying the Y. lipolytica model telomere consisting of 81 5′-GGGTTAGTCA-3′ repeats was constructed as described (34) with the following modifications. First, YlModelTEL_A and YlModelTEL_S2 oligonucleotides were mixed in a 1:1 molar ratio and annealed analogously to the preparation of probes for EMSA. The resulting dsDNA was digested with BamHI and HindIII restriction endonucleases (New England Biolabs) and cloned into the plasmid pUC19 (Stratagene) digested with BamHI and HindIII. To expand the 10-bp-long Y. lipolytica telomeric repeats, the BfuAI and BsmBI restriction sites flanking the telomeric repeats were employed. In addition to the plasmid carrying 81 telomeric repeats (pYLTEL81), the strategy yielded plasmids with shorter telomeric tracts, including pYLTEL41 carrying 41 telomeric repeats, which was used for some experiments.
Ligation of Oligonucleotide Tails to pYLTEL81
The pYTEL81 plasmid (3 μg) was digested with BfuAI and then dephosphorylated using 10 units of calf alkaline intestinal phosphatase. The linearized plasmid was mixed with a 5-fold molar excess of YLTEL3ovhng 40-nt oligo and ligated using 400 units of T4 DNA ligase (New England Biolabs) overnight at 16 °C in 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm dithiothreitol, 1 mm ATP, and 25 μg/ml bovine serum albumin. Ligated products were purified from free oligonucleotides using the QIAquick PCR purification kit (Qiagen).
Gel Filtration of Purified Tay1p
Superdex-200 (GE Healthcare) was packed into a 0.5 × 12-cm column and connected to the BioLogic low pressure chromatography system (Bio-Rad). The column was equilibrated with 5 volumes of buffer B (20 mm Tris-HCl (pH 7.4), 100 mm NaCl, 1 mm EDTA-NaOH) at a flow rate of 0.5 ml/min. The column was then calibrated using (50 μl of 1 mg/ml solution each) blue dextran (void volume), apoferritin (440 kDa), aldolase (147 kDa), and bovine serum albumin (67 kDa). After calibration, 50 μl of 0.2 mg/ml Tay1p purified by TALON Superflow Metal Affinity Resin was loaded on the column, and 0.25-ml fractions were collected starting from the void volume. Fractions adsorbing UV light were analyzed by 10% SDS-PAGE (33).
Electron Microscopy
The typical DNA binding reaction for electron microscopy was performed in 10 μl of 1× HN buffer containing a 5 ng/μl concentration of the substrate DNA and 7–10 ng/μl purified Tay1p. The reactions were carried out at room temperature for 15 min followed by the addition of 10 μl of 1.2% glutaraldehyde and incubation at room temperature for additional 6 min. To remove the unbound proteins and fixative, the samples were diluted to 50 μl in HN buffer and passed over 2-ml columns of 6% agarose beads (ABT inc, Burgos Spain) equilibrated with either TE buffer (10 mm Tris-HCl (pH 7.4), 0.1 mm EDTA-NaOH) for shadowcasting or 10 mm ammonium bicarbonate (pH 7.5) buffer for positive staining. For rotary shadowcasting with tungsten, aliquots of the fractions containing the complexes were mixed with a buffer containing spermidine and adsorbed onto copper grids coated with a thin carbon film glow-charged shortly before sample application. After adsorption of the samples for 2–3 min, the grids were dehydrated through a graded ethanol series and rotary shadowcast with tungsten at 10−7 torr (35). For positive staining, aliquots of the sample in the ammonium bicarbonate buffer were adsorbed to glow-charged thin carbon foils for 3 min followed by staining with 2% unbuffered uranyl acetate for 5 min followed by air drying. Samples were examined in an FEI T12 TEM and a Philips CM12 TEM equipped with Gatan 2kx2k SC200 CCD cameras at 40 kV (shadowcast samples) or at 80 kV (stained samples). Dimensions of particles in the images saved from the CCD cameras were analyzed using Digital Micrograph software (Gatan, Inc.). Adobe Photoshop software was used to arrange images into panels for publication.
Analysis of Tay1p Binding to Telomeres in Vivo
TAY1 fused at the 5′ end with the coding sequence for 3 hemagglutinin (3×HA) peptides (underlined; MYPYDVPDYAGYPYDVPDYAGYPYDVPDYAA) and the 5′-ACCAAA-3′ sequence upstream of the ATG codon (Kozak sequence enabling efficient translation) was prepared as follows. The oligonucleotides 3×HA-sense and 3×HA-antisense were mixed in equimolar amounts, boiled for 10 min, and reassociated by slow cooling. The TAY1 open reading frame was PCR-amplified using the primers Tay1_1H_5′P_S and Tay1_1H_5′P_Anti. The reassociated 3×HA oligonucleotide was ligated with the TAY1 PCR fragment, the product corresponding to 3×HA-TAY1 was gel-purified, and PCR was amplified using the oligonucleotides HA-Amp-S and Tay1_1H_5′P_Anti. TAY1 without the 3×HA coding sequence was PCR-amplified using the oligonucleotides Tay1+ATG+Kozak-S and Tay1_1H_5′P_Anti. The gel-purified PCR fragments (TAY1-3×HA and TAY1) were ligated into the vector pINA1312 (kindly provided by C. Madzak (Institut National de la Recherche Agronomique) and linearized with PmlI, and the resulting plasmid constructs were verified by DNA sequence analysis.
For construction of Y. lipolytica strains expressing Tay1 with or without 3×HA epitope, the plasmids were digested with NotI, and the fragments containing TAY1 and URA3 coding sequences (see Fig. 7A) were transformed into Y. lipolytica strain PO1h (36). The transformants were propagated in SD media (0.67% (w/v) yeast nitrogen base without amino acids and with ammonium sulfate; 2% (w/v) glucose). The expression of 3×HA-Tay1p was verified by immunoblot analysis of the crude protein extracts using an HA probe (F-7; Santa Cruz Biotechnology) and goat-anti-mouse antibody conjugated to alkaline phosphatase (Sigma).
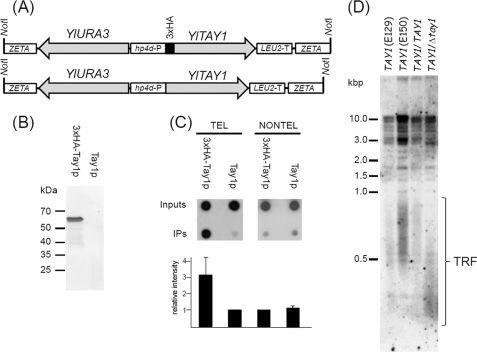
FIGURE 7.
Tay1p binds telomeres of Y. lipolytica in vivo. A, YlTAY1 was cloned with (3×HA-Tay1p) or without (Tay1p) the 3×HA epitope into pINA1312 plasmid vector as described under “Experimental Procedures.” The NotI fragment of the plasmid carrying YlTAY1 and YlURA3 (selection marker) flanked by ZETA sequences facilitating integration into the Y. lipolytica genomic DNA was gel-isolated and transformed into the Y. lipolytica strain PO1h. B, expression of 3×HA-Tay1p was tested by immunoblot analysis of protein extracts prepared from cells transformed with DNA carrying the gene encoding YlTay1p with or without 3×HA epitope. C, ChIP analysis of association of Tay1p with telomeric sequences is shown. Cells expressing 3×HA-Tay1p or Tay1p lacking the epitope were treated with formaldehyde, and 3×HA-Tay1p was immunoprecipitated (IP) with anti-HA antibodies as described under “Experimental Procedures.” The presence of telomeric DNA in immunoprecipitates (IP) was detected by dot-blot analysis using radioactively labeled YlTEL-4x oligonucleotide containing four Y. lipolytica telomeric repeats (TEL) or a mixture of oligonucleotides (YARLI-T1-4x, YARLI-T2-4x, and YARLI-T3-4x) derived from three arrays of non-telomeric tandem repeats (NONTEL). The relative intensity of the spots was determined using ImageJ software (rsb.info.nih.gov). The graph represents results of three independent experiments. D, measurement of TRFs in haploid strains (E129, E150), parental diploid strain (E129xE150; TAY1/TAY1), and heterozygous strain lacking one functional copy of TAY1 gene (TAY1/Δtay1) are shown. In contrast to the lengths of TRFs typical for parental wild-type strains (500–1000 bp), the heterozygote exhibits substantially shorter TRFs (300–500 bp).
The ChIP assay was performed essentially as described (37, 38). The cells were grown to a density of 2–5 × 107 cells/ml and treated for 15 min with 1% formaldehyde followed by quenching with 125 mm glycine. The cells were washed twice with phosphate-buffered saline (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 1.3 mm KCl, 135 mm NaCl (pH 7.2)), transferred to 2-ml tubes, and frozen at −80 °C. Before lysis the cells were converted to protoplasts (39) followed by resuspension in 0.6 ml of lysis buffer (50 mm HEPES-NaOH (pH 7.3), 140 mm NaCl, 1 mm EDTA, 1% (w/v) Triton X-100, 0.1% (w/v) sodium deoxycholate) containing CompleteTM protease inhibitor mixture (Roche Applied Science) and disrupted by glass-bead lysis. The samples were sonicated (Branson Sonifer 450) 4 × 15 s to obtain DNA 500–4000-bp-long fragments. Sonicated lysates were cleared by centrifugation at 7000 × g for 2 min, and input samples (1/20 of the sample) were obtained. The remaining lysate was incubated for 2 h at 4 °C with constant mixing with 40 μl of protein G Dynabeads (Invitrogen) coupled with anti-HA antibodies (Santa Cruz Biotechnology). The beads were washed twice with 0.6 ml of lysis buffer, once with wash buffer (100 mm Tris-HCl (pH 7.6), 250 mm LiCl, 0.5% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 1 mm EDTA-NaOH) containing CompleteTM protease inhibitor mixture and once with 0.6 ml of TE. Cross-linking was reversed by overnight incubation at 65 °C in TE containing 1% sodium dodecyl sulfate. The input and immunoprecipitated samples were treated with proteinase K (0.4 μg/ul) for 2 h at 37 °C and extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and DNA was purified using DNA Clean and Concentrator kit (Zymo Research). The DNA in the samples was detected by dot blot analysis using terminally labeled oligonucleotide YlTEL-4x (for detection of telomeric sequences) or a mixture of YARLI-T1-4x, YARLI-T2-4x, and YARLI-T3-4x derived from three different arrays of non-telomeric tandem repeats from Y. lipolytica genome (supplemental Table 1). The membranes were exposed to an x-ray film (GE Healthcare) or to Storage Phosphor Screens (Kodak).
Preparation of the TAY1/Δtay1 Heterozygous Strain of Y. lipolytica
The TAY1-YlURA3 deletion cassette was prepared by digestion of pTay1–6HN with NheI and AgeI, blunt-ended with Quick blunting kit (New England Biolabs), and ligated with a PvuII fragment of pKSURA plasmid carrying a YlURA3 gene including 5′ and 3′ regulatory sequences. The resulting deletion cassette lacks internally 328 bp of TAY1 ORF and contains YlURA3 flanked by 320- and 630-bp TAY1 sequences, respectively (the orientation of YlURA3 is reversed compared with the orientation of TAY1 ORF). The parental diploid strain was prepared by mating the haploids E129 and E150 on solid YMC medium (0.3% (w/v) Bacto-yeast extract, 0.5% (w/v) Bacto-peptone, 0.3% (w/v) malt extract, 0.05% (w/v) sodium citrate). The diploid strain was transformed with a DNA fragment carrying the TAY1-YlURA3 deletion cassette obtained by PCR amplification using the primers Tay1–1H_5′P_S and Tay1–1H_5′P_Anti. Replacement of the TAY1 gene by the disruption cassette in the transformants was verified by PCR using the Tay1–1H_5′P_S primer and Tay1_3utr primer derived from 3′-UTR of the TAY1 gene. Analysis of the lengths of telomeric restriction fragments was performed as described (27).
RESULTS
In Silico Analysis of the Y. lipolytica Genome Reveals a Novel Putative Telomere Binding Factor Tay1
To identify proteins with potential function(s) at Y. lipolytica telomeres, we first searched for homologues of known telomere-binding proteins, including S. cerevisiae Rap1p and S. pombe Taz1p. As initial attempts did not yield proteins with any significant degree of homology, we used amino acid sequences of Myb domains of various telomeric proteins as queries. This strategy led to the identification of three Myb domain-containing proteins. The first protein we identified corresponded to the ORF YALI0B13992g and was annotated as being weakly similar to the S. cerevisiae Myb-like DNA-binding protein Bas1, which is a transcription factor involved in regulating basal and (induced) expression of genes of the purine and histidine biosynthesis pathways (40–44). An interesting feature of YlBas1p is the presence of three Myb domains instead of two as found in Bas1p of S. cerevisiae. However, experiments described elsewhere (27) did not indicate an involvement of this protein in telomere maintenance. The second protein we identified, encoded by the ORF YALI0B18018g, exhibited homology to S. pombe Tbf1p recently described as an essential telomeric repeat binding factor in fission yeast (14). We will refer to this Y. lipolytica protein as YlTbf1. The third protein we identified was encoded by the ORF YALI0D10923g and exhibited homology with Myb domains from several proteins encoded by Basidiomycetes including Ustilago maydis, Laccaria bicolor, and Coprinopsis cinerea as well as with the Myb domain of human TRF1 and TRF2, respectively. Recently, based on in silico analyses, it was suggested that the U. maydis homologue may represent a TRF1/TRF2 counterpart in this organism (31). We, therefore, decided to characterize this protein using biochemical and electron microscopic analyses, and, based on our results, named it Tay1p. In addition, we found that an ORF in the S. pombe genome (SPAC13G7.10) that had been annotated as transcription factor Mug152 and was up-regulated during meiosis, also exhibits homology to the Y. lipolytica Tay1 protein.
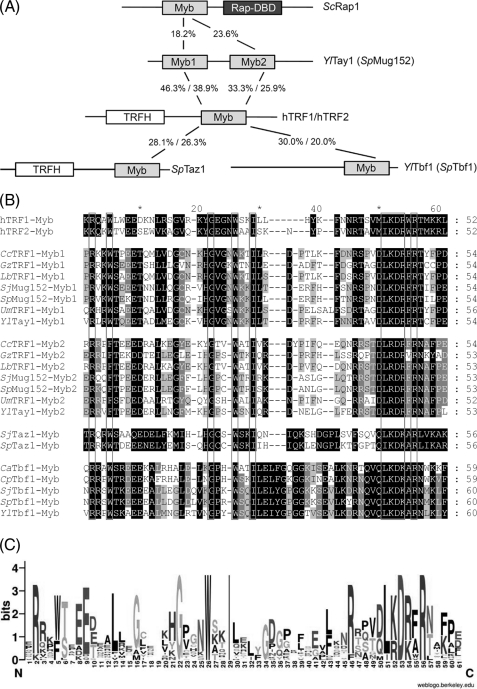
Tay1p contains two centrally located Myb/SANT domains exhibiting a high degree (about 40% identity and 65% similarity) of homology to each other (Fig. 1A). Importantly, the Myb domains in both proteins are highly similar to Myb domains of human TRF1 and TRF2, respectively. The Tay1-Myb1 domain exhibits 51% identity to the Myb domain of TRF1 and 43% to the Myb domain of TRF2 (Fig. 1A). The Tay1-Myb1 domain, thus, exhibits higher homology with the Myb domain of mammalian TRF1 and TRF2 proteins than with any other yeast telomere-binding protein, including Taz1 and Tbf1 (both about 30% identity, Fig. 1, A and B). The degree of identity between both Tay1p Myb domains and the Myb domains of ScRap1, SpTaz1, and YlTbf1 is in all cases less than 30%. In contrast, the Myb domains of Y. lipolytica Tay1p and S. pombe Mug152p are much more identical to each other (67% identity between Myb1 domains and 69% identity between Myb2 domains).
FIGURE 1.
Tay1p is a member of a novel family of telomere-binding proteins. A, shown is a schematic representation of the known telomere-binding proteins, Tay1p, and other known dsDNA telomere-binding proteins (the lengths of the proteins are not in scale). The numbers indicate the proportion of identical amino acids. X%/X%, comparison of the corresponding Myb domain with the TRF1 (first number) and TRF2 (second number) Myb domain. B, amino acid sequence alignment of the Myb/SANT domains extracted from fungal homologs of Tay1, Taz1, Tbf1, and TRF1 and human TRF1,2 proteins. The Myb/SANT domains were predicted using the SMART utility (61). The sequence alignment was calculated using the MUSCLE utility of the Geneious Pro 4.8.3 package (62) and manually adjusted. The shading was performed using the GeneDoc program (63). The following proteins were included in the alignment: C. albicans Tbf1 (CaTbf1; orf19.801), Candida parapsilosis Tbf1 (CpTbf1; CPAG00784), C. cinerea TRF1 (CcTRF1; CC1G_02158), Gibberella zeae TRF1 (GzTRF1; FG08713.1), human TRF1 (hTRF1; U40705.1), human TRF2 (hTRF2; U95970.1), L. bicolor TRF1 (LbTRF1; LACBIDRAFT_307744), Schizosaccharomyces japonicus Taz1 (SjTaz1; SJAG_01777), S. japonicus Mug152 (SjMug152; SJAG_01916), S. japonicus Tbf1 (SjTbf1; SJAG_02212), S. pombe Taz1 (SpTaz1; SPAC16A10.07c), S. pombe Mug152 (SpMug152; SPAC13G7.10), S. pombe Tbf1 (SpTbf1; SPBC19G7.13), U. maydis TRF1 (UmTRF1; UM02326.1), Y. lipolytica Tay1 (YlTay1; YALI0D10923g), Y. lipolytica Tbf1 (YlTbf1; YALI0B18018g). Note that Myb1 and Myb2 represent two different Myb domains identified within the corresponding protein. C, the sequence logo generated from the alignments shown in Fig. 1B using the WebLogo tool (60) displays highly conserved amino acid residues in the Myb/SANT domains of distant telomeric DNA-binding proteins.
We compared the Myb domains of known telomeric proteins and found that the Tay1p Myb1 domain clusters with the Myb1 domain of its homologues from Basidiomycetes and S. pombe as well as with the Myb domain of both mammalian TRF1 and TRF2 (Fig. 1, A–C, supplemental Fig. 1). However, the Myb2 domain and the Myb domains of Taz1p and Tbf1p seem to form separate clusters (supplemental Fig. 1). The fact that the Myb1 domain of Tay1p is closely related to mammalian TRF1 and TRF2 strongly argues that Tay1p is a telomeric dsDNA-binding protein in Y. lipolytica.
Purified Tay1p Binds Telomeric DNA in Vitro
To analyze the DNA binding properties of Tay1p in vitro, we constructed an expression vector (pTay1–6HN) for production of the protein in E. coli and purification using affinity chromatography, taking advantage of the 6×HN tag located at the N terminus of the recombinant protein. The plasmid construct, verified by both restriction enzyme analysis and DNA sequencing, was transformed into the BL21-Gold (DE3)-pLysS bacterial host, and the expressed protein was purified to near homogeneity in a two-step purification employing TALON affinity chromatography and subsequent heparin affinity chromatography (Fig. 2). Although the predicted molecular mass of Tay1–6HN is ∼47 kDa, purified Tay1p migrated as a band corresponding to ∼65 kDa on 10% SDS-PAGE gels. The reason for slower migration is unknown (see “Discussion”) but is analogous to a similar situation with recombinant Taz1p (16).
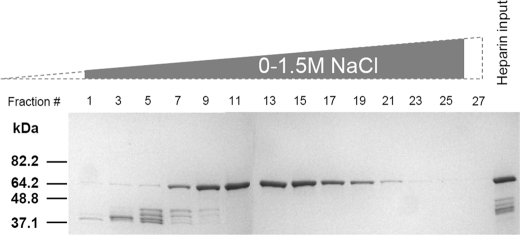
FIGURE 2.
Purification of the recombinant Tay1 protein. Tay1p-6HN was purified by TALON Superflow metal affinity chromatography (heparin input), loaded onto a heparin column, and eluted by a linear 0–1.5 m NaCl gradient, and the proteins were analyzed by 10% SDS-PAGE. Shown are peak fractions eluted from the heparin column. Fractions 10–16 were pooled, dialyzed, and used for subsequent experiments.
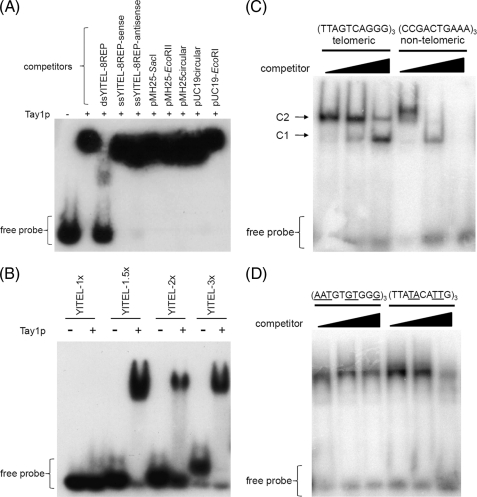
EMSA were used to examine the binding of Tay1p to duplex DNAs. As an initial probe we used a cloned double-stranded EcoRI fragment (YlTEL) containing five Y. lipolytica telomeric repeats (5′-GGGTTAGTCA-3′) in the plasmid pMH25 (see Fig. 4A for the restriction map of pMH25). When incubated with Tay1p, the labeled YlTEL probe was retarded in its migration in the polyacrylamide gel, indicating that Tay1p and YlTEL formed a stable complex (Fig. 3A). We then tested the ability of various DNAs to compete with the YlTEL probe for Tay1p binding and found that whereas the dsDNA containing eight telomeric repeats was an effective competitor (Fig. 3A, third lane), dsDNA containing three telomeric repeats and single-stranded oligonucleotides carrying the sequence of G-rich and C-rich strand as well as various non-telomeric dsDNAs were ineffective (Fig. 3A, fourth through tenth lanes). To probe the minimum binding site of Tay1p, we performed EMSA using dsDNA probes carrying 1, 1.5, 2, and 3 telomeric repeats. We found that the ability of Tay1p to bind DNA is limited to the probes containing >1 telomeric repeat (Fig. 3B).
FIGURE 4.
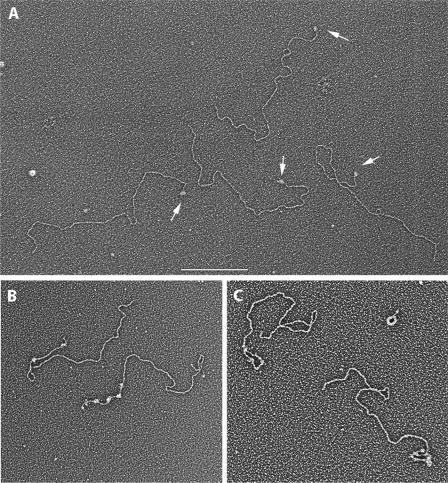
Visualization of Tay1p bound to a Y. lipolytica model telomeric DNA. Model telomere DNA templates were generated that contain 100 bp (A) or 810 bp (B and C) of the 10-bp Y. lipolytica telomeric repeat. The DNAs in B and C also contain a 40-nt-long single strand 3′ overhang on the G-rich strand. The DNAs were complexed with Tay1p at a ratio of 1.7:1 (A) or 2.3:1 (B and C) μg of protein/μg of DNA for 20 min at 21 °C, then fixed and prepared for visualization by EM (“Experimental Procedures”) including adsorption to thin carbon foils, dehydration, and rotary shadowcasting with tungsten. The field in A shows four model DNAs each with a single Tay1p complex at one end (arrows). The examples in B and C show model telomeres bound by Tay1p in an open (B) or looped configuration (C). Results are shown in reverse contrast. The bar in A is equivalent to 500 nm (A) and 200 nm (B and C).
FIGURE 3.
DNA binding properties of Tay1 analyzed by EMSA. A, shown is EMSA analysis using radiolabeled YlTEL (50-bp EcoRI fragment of pMH25) as a probe. The DNA competitors indicated above the lanes were used at a 1000-fold excess over the YlTEL probe. B, Tay1p is able to bind dsDNA probes carrying ≥1.5 telomeric repeats. C, EMSA analysis using radiolabeled dsDNA oligonucleotides containing three telomeric repeats (telomeric' YlTEL_EMSA-1) and three non-telomeric repeats (nontelomeric YlTEL_EMSA-C), respectively. 100, 300, and 500 ng of poly(dI-dC) DNA were used as competitor DNA. The two bands (C1 and C2) most likely represent complexes containing one and two Tay1 protein molecules, respectively. D, shown is an EMSA analysis of Tay1p binding to radiolabeled mutated telomeric probes containing either conserved GT and GG dinucleotides with other residues of the telomeric repeat mutated ((AATGTGTGGG)3; YlTEL_EMSA-M5) or vice versa ((TTATACATTG)3; YlTEL_EMSA-M6). Mutated residues are underlined. 300, 500, and 1000 ng of linearized pUC19 were used as a competitor. In all EMSA experiments the dsDNA probes were prepared by annealing ss oligonucleotides as described under “Experimental Procedures.”
Next, we tested the specificity of Tay1p binding to telomeric versus non-telomeric sequences. Whereas the binding of Tay1p to the dsDNA probe containing three telomeric repeats (YlTEL_EMSA-1) was not substantially affected by an excess of competitor DNA, the control dsDNA probe containing three non-telomeric repeats (YlTEL_EMSA-C) was out-competed by relatively low concentrations of either pUC19 plasmid DNA or poly(dI-dC) (Fig. 3C). The observation of two bands (Fig. 3C; C1 and C2) indicates that the probe is bound by an increasing number of Tay1p molecules.
Tay1p Recognizes a Conserved Motif in Telomeric Repeat
Comparison of the telomeric repeats from a wide variety of organisms including yeast, protozoa, and mammals indicates that there is a conserved motif important for binding of ss- and dsDNA telomeric proteins such as Rap1p and Cdc13 (28–30). To this list we have now added the sequence of Y. lipolytica as well as the sequences identified in our comparative analysis of telomerase RNAs (18) (Table 1). Indeed, all telomeric repeats exhibit a conserved motif 5′-GTNNGG(G/A)T-3′. The only exceptions are Candida (Pichia) guilliermondii and Candida (Clavispora) lusitaniae (5′-GTNNTG-3′) and S. pombe (5′-GTTACAGG-3′), where the spacer between GT and GG is four instead of two nucleotides. In any case, the sequence of Y. lipolytica telomeric repeat contains the 5′-GTNNGGNT-3′ motif, suggesting that the conserved nucleotides are important for the binding of Tay1p.
To test this possibility we prepared a dsDNA probe (YlTEL_EMSA-M5) carrying three repeats that contain a non-telomeric sequence, except for GT and GG dinucleotides in the conserved positions. The YlTEL_EMSA-M5 probe was retarded to a similar extent as the wild-type (YlTEL_EMSA-1) telomeric probe (Fig. 3D). Conversely, we tested a probe (YlTEL_EMSA-M6), where the conserved GT and GG dinucleotides were mutated, whereas the rest of the repeat remained unchanged. In this case, the YlTEL_EMSA-M6 exhibited stronger binding than the control non-telomeric probe (YlTEL_EMSA-C) and only slightly weaker binding compared with YlTEL_EMSA-M5 (Fig. 3D). Therefore, it seems that not only the conserved GTNNGGNT motif, but also the rest of the telomeric repeat sequence, plays an important role in Tay1p binding. Overall, the EMSA experiments indicated that Tay1p exhibits a preference for dsDNA containing telomeric repeats of Y. lipolytica with a minimum binding site ≥1.5 telomeric repeats (see “Discussion”).
Preparation of a Model Telomere
To visualize Tay1p binding to DNA by EM, we employed two DNA substrates. The first was based on pMH25 (supplemental Fig. 2A) linearized by SacI and PstI, and the 3.7-kb fragment (p3.7kb+TEL) carrying the telomere at one end was gel-purified and used for EM analysis. The substrate used for most of the EM experiments was created by expansive cloning (see “Experimental Procedures”) and produced a plasmid construct (pYLTEL81) carrying 81 repeats (810 bp) (supplemental Fig. 2B). For the DNA binding experiments, this plasmid was linearized with BfuAI, leaving the entire telomeric block at one end of the DNA. In addition, digestion by BfuAI results in a 5′ recessed end enabling the addition of a 3′ overhang to the telomeric end by ligation of an oligonucleotide carrying 4 telomeric repeats (supplemental Fig. 2B). The length of the telomeric block in pYLTEL81 (810 bp) is within the range of native Y. lipolytica telomeres (500–1000 bp, (27)), and thus, it represents an ideal model telomere for in vitro studies. Using E. coli SSB protein as a marker for the presence of the ligated overhang, EM analysis revealed that >90% of the model telomere molecules contained ssDNA overhangs.
EM Analysis of Tay1p DNA Complexes
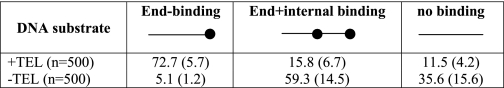
To directly visualize Tay1p interactions with telomeric DNA in vitro, we generated three different model telomere DNA templates carrying 10 or 81 telomeric repeats and the latter with a 40-nt 3′ overhang on the G-rich strand (“Experimental Procedures”). To begin the studies, Tay1p was incubated with the p3.7kb+TEL DNA containing 10 repeats at one end at a ratio of 2 Tay1p monomers per 1.5 repeat for 15 min at room temperature then fixed and prepared for EM including dehydration and rotary shadowcasting with tungsten (“Experimental Procedures”). In fields of Tay1p-bound DNAs, when 500 molecules were scored, 70% contained 1–2 protein particles located at one end of the DNA (arrows, Fig. 4A, Table 2). Molecules with protein particles bound at both ends were never observed, and infrequently at this protein:DNA ratio, protein was bound internally along the DNA. Furthermore, parallel incubation of similar-sized linear DNAs lacking the telomeric tract exhibited a much lower (∼5%) frequency of end-bound molecules. To examine the binding of Tay1p to DNA with a telomeric tract close to the size present in vivo, Tay1p was incubated with the linear pYLTEL81 DNA template containing 810 bp of telomeric DNA at one end. The concentration of Tay1p was adjusted to the number of telomeric repeats. As our previous EMSA analyses indicated that the minimal binding site for Tay1p is represented by 1.5 repeats (Fig. 3B), we used protein:DNA ratios ranging from 0.3 to 3 Tay1 monomers per 1.5 repeat. Optimal binding was observed at 2 molecules of Tay1p per 1.5 telomeric repeat (data not shown), a ratio that was then used for most of the subsequent experiments. Examination of Tay1p bound to pYLTEL81 DNA revealed arrays of particles along a 810-bp region at 1 end of the model DNA (Fig. 4B, supplemental Fig. 3). Tay1p appeared to bind along the telomeric DNA forming a linear structure that, at saturating Tay1p concentrations, became more and more filled in with Tay1p particles (Fig. 4C, supplemental Fig. 3) reminiscent of protein arrays formed by hTRF1 on human telomeric repeats (45). Inspection showed no evidence of the DNA being bent as it passed through the protein particle. With enough protein added, the telomeric tracts were fully covered, and the length of the covered segments was similar to the length of the bare telomeric tract. From this we can deduce that the protein binds non-cooperatively and does not appreciably shorten or bend the DNA upon binding, even at saturating concentrations. Importantly, based on the comparison of contour lengths of the entire plasmid versus its portion bound by Tay1p, all the protein particles were bound within the 810-bp long telomeric tract. Of more than 300 molecules scored, 79% were bound by Tay1p within the telomeric tract, less than 1% showed Tay1p bound internally, and about 20% DNA templates were protein-free. This corresponds to a very high binding specificity for the telomeric repeats. However, at elevated concentrations of Tay1p, protein binding could be driven onto the non-telomeric regions. Similar results were obtained when the pYLTEL41 plasmid was used as a substrate (data not shown). In some of the model telomere preparations, not all the DNAs had been linearized, and furthermore, some of the plasmids were present as dimers. Inspection of Tay1p binding to these DNAs showed single tracts of Tay1particles on the relaxed or supertwisted monomer circles or two polar tracts on the relaxed or supertwisted plasmid dimers (supplemental Fig. 4), providing further confirmation of the observations with the linear model DNAs.
TABLE 2.
Summary of the binding of Tay1p to linear DNA with (+TEL) or without (−TEL) an array of telomeric repeats located at the ends
The percentages of the corresponding species of molecules are expressed as a fraction of the total molecules. The numbers in parentheses represent the S.D. from one experiment to another (n = no. of inspected molecules per corresponding DNA substrate).
Remodeling of Y. lipolytica Telomeric DNA by Tay1p
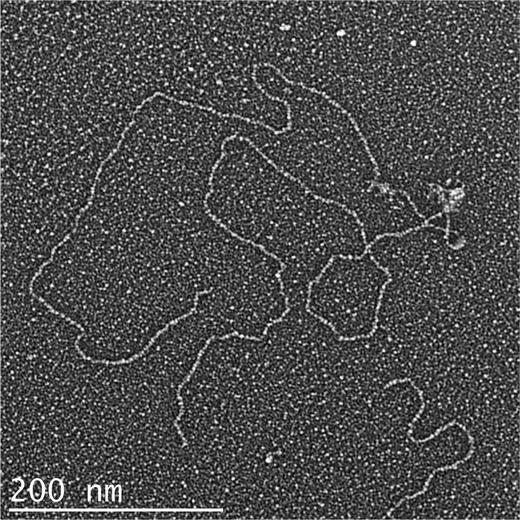
When Tay1p binds to the linear pYLTEL81 template, a frequent observation is that two and sometimes three or more DNAs bound together by Tay1p particles in a head-to-head configuration (Fig. 5). This was observed whether or not the model DNAs contained a 40-nt 3′ ssDNA overhang. These structures seem analogous to the head-to-head dimers formed by hTRF1 with templates carrying human telomeric repeats (45). When the pYLTEL81 DNA contained the 40-nt 3′ overhang, Tay1p exhibited increased binding at the terminal part of the model telomere, corresponding to the ss/dsDNA junction (data not shown). Furthermore, with the overhang present, the DNA was frequently remodeled into a lasso or telomeric loop-like structure (Fig. 4C, supplemental Fig. 5). In one experiment, scoring 303 DNA templates containing the overhang, 79.9% showed Tay1p bound, and 20.1% were protein-free. Of the protein-bound molecules, 76% had one or more Tay1p particles at one end, 23% of the DNAs were arranged into loops by Tay1p, and 1% showed Tay1p particles internally along the plasmid sequences. In contrast, without the overhang, the fraction of looped DNA molecules was negligible (1–3%). The loops were always ∼800 bp or smaller, and in some cases the telomeric tract was fully complexed with Tay1p, whereas in others the telomeric tract was only sparsely bound by Tay1p. In all cases, however, a Tay1p complex could be observed at the base of the loop (supplemental Fig. 5). These observations parallel studies of human TRF2 and fission yeast Taz1p, both of which promote formation of telomeric loops in vitro (15, 16) and exhibit a preference for ss/dsDNA telomeric junctions (16, 34).
FIGURE 5.
Visualization of Tay1p induced synapsis of two model Y. lipolytica telomere DNAs. The model telomere DNA described in Fig. 4 containing 810 bp of Y. lipolytica telomeric repeats and a 40-nt single strand overhang in the G-rich strand was complexed with Tay1p and prepared for EM as described in Fig. 5. A frequent occurrence observed is the synapsis of two-model telomere DNAs by Tay1p at the telomeric tracts in a head-to-head fashion.
Size and Nature of the Tay1p Oligomers Bound to DNA
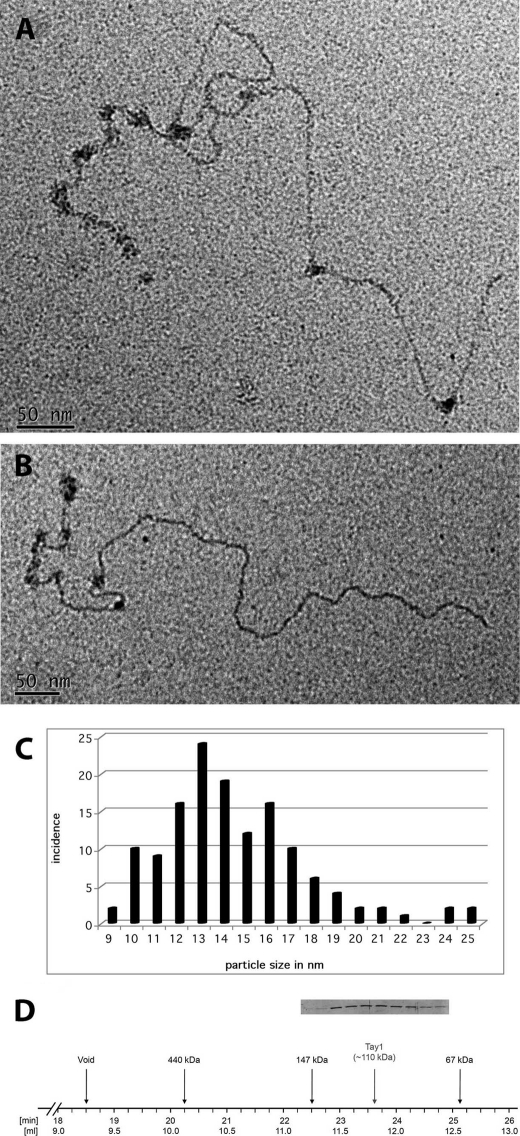
To estimate the size of Tay1 particles, we examined the tungsten shadowcast Tay1p complexes. The observed Tay1p particles were of varied sizes, ranging from some that were smaller than E. coli SSB tetramers (68 kDa), added during mounting as a simple size marker (images not shown), to many particles that were somewhat larger than SSB and others that were clearly larger by significant amounts. To obtain a more quantitative estimate of the size of the bound particles, linear pYLTEL81 DNA was complexed with Tay1p under the conditions described above including fixation and gel chromatography to remove unbound protein and then directly stained with uranyl acetate for EM (Fig. 6, A and B). This avoids any increase in size due to metal shadowcasting. Measurement of the diameter of 127 particles bound along the telomeric tract showed a range of sizes centering about 10, 13, and 16 nm (Fig. 6C). These sizes are most consistent with dimers and higher oligomers of Tay1p bound to DNA (see “Discussion”). These observations were in accord with our analysis of purified Tay1p subjected to gel-filtration chromatography on Superdex-200. The major peak of Tay1p was observed around 110 kDa corresponding to the dimeric form of the protein (Fig. 6D). However, Tay1p was relatively spread around the peak, indicating the presence of monomers and oligomers larger than dimers.
FIGURE 6.
Estimation of the size of the Tay1p particles bound to DNA by positive staining. Complexes of Tay1p on the model Y. lipolytica telomere DNA were formed as described in Fig. 4 (see also “Experimental Procedures”) and prepared for EM by adsorption to thin carbon foils and staining with 2% uranyl acetate. A and B, Tay1p binds preferentially along the 810-bp telomeric tract but occasionally elsewhere on the DNA. Measurement of the diameter of 127 particles from examples such those shown in A and B generated a size distribution shown in C. D, when analyzed by gel filtration on Superdex-200, the peak of Tay1p is eluted in the fractions corresponding to dimeric forms of the protein. The lanes in the inset correspond to the fractions indicated on the horizontal line.
Association of Tay1p with Y. lipolytica Telomeres in Vivo
To investigate association of Tay1p with telomeres of Y. lipolytica, we performed two types of experiments. First, we prepared a tagged version of Tay1p by placing a sequence encoding three human influenza hemagglutinin (3×HA) epitope tags at the 5′ end of the TAY1 ORF (Fig. 7A). 3×HA-Tay1p and Tay1p without the tag were produced in Y. lipolytica PO1h strain, and the expression of epitope-tagged Tay1p was verified by immunoblot analysis using anti-HA antibodies (Fig. 7B). The clones expressing 3×HA-Tay1p were used for ChIP experiments (the strain expressing Tay1p without the epitope was used as a control). We reproducibly observed a severalfold enrichment of telomeric DNA (in contrast to non-telomeric repetitive DNA) in the samples obtained by anti-HA immunoprecipitation of 3×HA-Tay1p producing strains (Fig. 7C), indicating that at least part of Tay1p binds to telomeres in vegetatively growing cells.
Second, in addition to ChIP experiments, we tried to delete the TAY1 gene from the Y. lipolytica genome. Our attempts to knock-out TAY1 from haploid strains were not successful, indicating that similarly to Mug152p in S. pombe, Tay1p is an essential protein. We, therefore, deleted one copy of TAY1 from a diploid strain of Y. lipolytica and compared the lengths of telomeric restriction fragments (TRF) of the TAY1/Δtay1 heterozygote with those of parental strains. Our results indicate that the absence of one TAY1 copy results in shorter telomeres in the diploid strain (300–500 bp in the mutant versus 500–1000 bp in the wild type; Fig. 7D) suggesting a role for Tay1p in maintenance of Y. lipolytica telomeres.
DISCUSSION
Yeast duplex telomeric DNA is bound by distinct types of proteins. In budding yeast, Rap1p is the major component of the telomeric chromatin (other than the histones) and binds the DNA via two Myb/SANT domains (46). In addition, Tbf1p, containing a single Myb domain, binds 5′-TTAGGG-3′ repeats located within the subtelomeric regions (47). Both Rap1p and Tbf1p are essential proteins involved in regulation of transcription at several loci (47). Tbf1p was also identified in S. pombe, although its precise function at telomeres is not known (14) and is probably identical to the protein Teb1 shown to bind vertebrate-like telomeric repeats in vitro (48). An additional dsDNA telomere-binding protein is Taz1p, whose presence seems to be restricted to S. pombe and closely related species. Based on its biochemical properties and phenotypic analyses of null mutants, Taz1p is considered to be a functional orthologue of mammalian TRF1 and TRF2 (13, 16).
While searching for homologues of yeast telomeric proteins in Y. lipolytica, we identified only a putative Tbf1p encoded by YALI0B18018g. Surprisingly, the genome of Y. lipolytica does not contain any open reading frame encoding putative homologues of either Rap1p or Taz1p. Instead, we found an open reading frame encoding a putative protein (Tay1p) carrying two Myb domains. In this respect, Tay1p is similar to the budding yeast Rap1 protein; however, the similarity between the Tay1p and Rap1p Myb domains is very low. Furthermore, the Myb domains of Tay1p share only a weak homology with the Myb domains of Tbf1p or Taz1p. Instead, they are very similar to the Myb domains of putative telomere-binding proteins identified in basidiomycetous fungi. Another interesting homologue of Tay1p is an essential protein, Mug152, encoded by fission yeast open reading frame SPAC13G7.10, which was shown to be up-regulated in meiosis (49–51). Interestingly, crude protein lysates of Δtaz1 S. pombe cells were shown to contain a protein able to bind the fission yeast dsDNA telomeric sequence, suggesting that in addition to Taz1p, fission yeast possess yet another uncharacterized telomere-binding protein (48). It seems that this activity cannot be attributed to Tbf1p, as it exhibits specificity for vertebrate-like 5′-TTAGGG-3′ repeats instead of the fission yeast telomeric repeats (14, 48). The possible connection of Mug152p to meiosis is an interesting possibility, as telomere clustering at meiotic prophase seems to be a prerequisite for successful recombination and/or spindle pole body formation (52–54). The ability of Tay1p to promote formation of head-to-head dimers of DNA molecules through the telomeric tracts (Fig. 5) supports the hypothesis that Tay1p (and possibly Mug152p) employ their two Myb domains for mediating telomere clustering in vivo.
The binding properties of Tay1p seem to be reminiscent of budding yeast Rap1 protein binding to its cognate sequences. The Rap1 minimal binding site is known to be ∼12 bp, where each of the two Myb domains independently contacts 5–6 bp (55). The minimal binding site for Tay1p was 15 bp (1.5 repeat), which roughly corresponds to the 12 bp observed for Rap1p (Fig. 3B). It is likely that each of the two Myb domains in Tay1p also contacts 5–6 bp, and the remaining sequence might represent a spacer between the “half-sites.” Alternatively, the observation that Tay1p bound more strongly to 1.5, as contrasted to 1.0 repeats of 5′-TTAGTCAGGG-3′, may reflect the fact that the minimal binding site for Tay1p is represented by a different permutation of the 5′-TTAGTCAGGG-3′ sequence. Detailed analysis of this using the approach taken in the studies of Bianchi et al. (56, 57) would determine whether this is the case. In some experiments where we used probes containing three telomeric repeats, we observed two bands (C1 and C2; Fig. 3C) of the retarded probe, presumably corresponding to two different Tay1p-DNA species.
The more detailed mapping of residues involved in Tay1p binding to the DNA substrate was based on the studies aimed at characterization of binding sites of budding yeasts ss- and dsDNA telomere-binding proteins (28–30, 58). When we included the sequences of recently identified telomeric repeats from several yeast species (18), we confirmed that they contain a conserved 5′-GTNNGGNT-3′ motif (Table 1). Importantly, the Y. lipolytica telomeric repeat, whose sequence was obtained from native chromosomal ends,6 also contained the conserved motif (this is in contrast to the telomeric repeat sequence previously reported in the literature (19, 59)). Our EMSA experiments led to ambiguous results. We observed that mutation of the conserved GT and GG dinucleotides decreases but does not abolish the binding of Tay1p to DNA (Fig. 3D). It is likely that in addition to the conserved residues, the remaining bases, most likely a stretch of 4–5 base pairs constituting a core recognition sequence, are also involved in binding, and their presence can compensate for the loss of the conserved nucleotides, similar to the case of TRF1, TRF2, or Rap1p (55, 57).
Inspection of the images of Tay1p particles bound along the DNA revealed many examples in which the DNA took a path along one side of the particle and other examples where the DNA in projection appeared to pass either through, over, or under the protein particle. This appearance is consistent with Tay1p complexes binding along one face of the DNA as contrasted to forming a sliding clamp or donut with the DNA going through the middle of the particle. Furthermore, the DNA did not appear appreciably bent by Tay1p binding. Given that the telomeric repeat is 10 bp, any bending, even modest, would be in phase with the next and, thus, amplified in an array of bound particles, and this was not observed.
EM analysis revealed that Tay1p binding to telomeric sequences resembles the mode of binding of hTRF1 to human telomeric repeats. Apart from forming protein particles consisting of dimers and tetramers, hTRF1 forms higher order intra- or intermolecular structures (45). Binding of hTRF1 is not restricted to adjacent telomeric repeats, and the monomeric subunits of hTRF1 dimers or tetramers can bind to repeats separated by a stretch of non-telomeric DNA or to telomeric repeats on two distinct DNA molecules. This results in forming either intramolecular loops (56) or head-to-head DNA dimers (45). Similar to hTRF1, when bound to the model telomere containing 81 telomeric repeats, Tay1p formed protein particles of various sizes localized within telomeric tracts. In addition, we observed head-to-head dimeric structures, i.e. two DNA molecules paired by multiple Tay1p particles. There are two possible ways of Tay1p-mediated DNA dimer formation. First, one monomer of Tay1p may employ its two Myb domains for binding to two distinct DNA molecules. Second, Tay1p may form protein dimers, where each Tay1p monomer would bind to one DNA molecule. The first possibility would require the presence of a highly flexible hinge region between the two Myb domains. However, in silico structure analysis did not predict a flexible hinge structure in this particular region of the protein. It, therefore, seems more likely that Tay1p employs its di- or multimeric structure for mediating the formation of DNA dimers. This possibility is supported by the observation of Tay1p particles larger than the size of a monomer by both EM and gel filtration (Fig. 6). It will be interesting to investigate the possibility that Tay1p promotes telomere clustering in vivo.
The remodeling of the model telomeres containing a 40-nt 3′ single-stranded telomeric overhang into looped forms by Tay1p (Fig. 4, supplemental Fig. 5) was striking and was absent in the absence of the overhang. This indicates that Tay1p, similar to fission yeast Taz1p and mammalian TRF2, promotes the formation of telomeric loops (16, 34). It is possible that Tay1p promotes strand invasion of the 3′ overhang back into the repeating duplex segment to stabilize the loop as TRF2 is believed to do. However, it is also possible that the preferential binding of Tay1p to the ss/ds junction at the end of the DNA optimally positions the protein to then fold the telomere into a loop by binding back along the duplex segment of the telomere without formation of a strand invasion D-loop. Further physical and EM experiments will be required to distinguish these two possibilities. Even if Tay1p were to not initiate strand invasion itself, the generation of such looped structures could place the 3′ end in a position optimal for the subsequent action of other factors such as the Rad51 proteins that would catalyze strand invasion.
As recently discussed by Lue (19), the mechanisms of telomere protection in Y. lipolytica have undergone dramatic changes. Its genome not only lacks discernable Rap1p or Taz1p homologues, but it also does not contain proteins homologous to known factors involved in stabilization of the 3′ single-stranded telomeric overhang, including Pot1p and members of the CST complex (Cdc13p, Stn1p, Ten1p). Tay1p and its homologues in S. pombe (Mug152p) and basidiomycetous fungi may represent a novel class of dsDNA telomere-binding proteins. Our experiments presented in this paper (Fig. 7) indicate that at least a fraction of Tay1p is associated with telomeres in vivo and participates in their maintenance. Our recent results demonstrate that Y. lipolytica is able to overcome loss of telomeric sequences caused by the absence of the catalytic subunit of telomerase through circularization of all six chromosomes (27). However, Tay1p (similarly to Mug152p in S. pombe) is an essential protein. It is, therefore, very likely that Tay1p is not dedicated solely to telomeres but binds to additional loci within the Y. lipolytica genome. Tay1p may, thus, represent a multifunctional protein analogous to the major telomere-binding protein Rap1 in S. cerevisiae, whose essential nature is not related to its telomere-associated functions (46).
Future studies will be aimed at detailed analysis of biochemical properties of Tay1p. For example, it will be interesting to investigate the DNA binding characteristics of its two Myb domains and analysis of their contribution to specific binding to telomeric sequences. In addition, the exact role(s) of Tay1p at telomeres (in both vegetative and meiotic cells) as well as other parts of the Y. lipolytica genome will be examined in more detail. The replacement of the native YlTAY1 promoter with an inducible promoter would be instrumental in circumventing the lethality of the null mutant and studying the effect of Tay1p on telomeres in vivo. This together with the analysis of a Tay1p homologue in S. pombe with well characterized telomeres and powerful molecular-genetic tools will be a major focus of future studies on physiological roles of Tay1p-like proteins in fungi. The fact that Tay1p exhibits similar biochemical properties with both TRF1 and TRF2 indicates that such studies on Tay1p may prove highly fruitful in extending our knowledge in general about the origin and evolution of telomeres.
Supplementary Material
Acknowledgments
We thank Ladislav Kovac (Comenius University, Bratislava, Slovak Republic) for inspiration and continuous support, members of our laboratories for discussions, Claude Gaillardin (Institut National de la Recherche Agronomique, Thiverval-Grignon, France) for providing sequences of Y. lipolytica chromosomal ends, Catherine Madzak (Institut National de la Recherche Agronomique, Thiverval-Grignon, France) for the plasmid pINA1312 and the strain PO1h, Richard Rachubinski (University of Alberta, Edmonton, Canada) for providing pTC3 plasmid used in preliminary experiments, Janka Kordulakova (Comenius University) for expert advice, and Janka Tomaskova (Institute of Virology, Slovak Academy of Sciences) and Darinka Luknarova (Comenius University) for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31819, ES013773, and TW005654 (to J. D. G.). This work was also supported by Howard Hughes Medical Institute Grant 55005622 (to J. N.) and Slovak grant agencies Agentura na podporu vyskumu a vyvoja (20-001604 (to L. T.) and 0024-07 (J. N.)) and Vedecta grantova agentura 1/0132/09 (to L. T.) and 1/0219/08 (to J. N.). J. K. was supported by a travel fellowship from SAIA (Slovak Academy Information Agency). This contribution/publication is the result of the project implementation: “Centre of excellence for exploitation of informational biomacromolecules in the disease prevention and improvement of quality of life” supported by the Research and Development Operational Programme funded by the Europsky fond regionalneho rozvoja.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–5.
This paper is dedicated to the 70th anniversary of the Faculty of Natural Sciences, Comenius University in Bratislava.
C. Gaillardin, personal communication.
- bp
- base pair.
REFERENCES
- 1.McEachern M. J., Krauskopf A., Blackburn E. H. (2000) Annu. Rev. Genet. 34, 331–358 [DOI] [PubMed] [Google Scholar]
- 2.Déjardin J., Kingston R. E. (2009) Cell 136, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. (2007) Science 318, 798–801 [DOI] [PubMed] [Google Scholar]
- 4.Schoeftner S., Blasco M. A. (2008) Nat. Cell Biol. 10, 228–236 [DOI] [PubMed] [Google Scholar]
- 5.Blasco M. A. (2007) Nat. Chem. Biol. 3, 640–649 [DOI] [PubMed] [Google Scholar]
- 6.Linger B. R., Price C. M. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palm W., de Lange T. (2008) Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 8.Baumann P., Cech T. R. (2001) Science 292, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 9.Pennock E., Buckley K., Lundblad V. (2001) Cell 104, 387–396 [DOI] [PubMed] [Google Scholar]
- 10.de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 11.Kanoh J., Ishikawa F. (2003) Cell. Mol. Life Sci. 60, 2295–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cockell M. M., Lo Presti L., Cerutti L., Cano Del Rosario E., Hauser P. M., Simanis V. (2009) Eukaryot. Cell 8, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper J. P., Nimmo E. R., Allshire R. C., Cech T. R. (1997) Nature 385, 744–747 [DOI] [PubMed] [Google Scholar]
- 14.Pitt C. W., Valente L. P., Rhodes D., Simonsson T. (2008) J. Biol. Chem. 283, 2693–2701 [DOI] [PubMed] [Google Scholar]
- 15.Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 16.Tomaska L., Willcox S., Slezakova J., Nosek J., Griffith J. D. (2004) J. Biol. Chem. 279, 50764–50772 [DOI] [PubMed] [Google Scholar]
- 17.Cohn M., Liti G., Barton D. B. H. (2006) in Topics Curr. Genet. (Sunnerhagen P., Piskur J. eds) pp. 101–130, Springer, Berlin [Google Scholar]
- 18.Gunisova S., Elboher E., Nosek J., Gorkovoy V., Brown Y., Lucier J. F., Laterreur N., Wellinger R. J., Tzfati Y., Tomaska L. (2009) RNA 15, 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lue N. F. (2010) Trends Biochem. Sci. 35, 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth G., Gaillardin C. (1997) FEMS Microbiol. Rev. 19, 219–237 [DOI] [PubMed] [Google Scholar]
- 21.Pertuiset B., Beckerich J. M., Gaillardin C. (1995) Curr. Genet. 27, 123–130 [DOI] [PubMed] [Google Scholar]
- 22.Nuttley W. M., Brade A. M., Eitzen G. A., Veenhuis M., Aitchison J. D., Szilard R. K., Glover J. R., Rachubinski R. A. (1994) J. Biol. Chem. 269, 556–566 [PubMed] [Google Scholar]
- 23.Brandt U., Abdrakhmanova A., Zickermann V., Galkin A., Dröse S., Zwicker K., Kerscher S. (2005) Biochem. Soc. Trans. 33, 840–844 [DOI] [PubMed] [Google Scholar]
- 24.Szabo R. (1999) Folia Microbiol. 44, 19–24 [DOI] [PubMed] [Google Scholar]
- 25.Szabo R. (2001) Study of Cell Morphogenesis and Differentiation on Yeast Yarrowia Lipolytica. Ph.D. thesis, Department of Biochemistry, Faculty of Natural Sciences, Comenius University, Bratislava [Google Scholar]
- 26.Casaregola S., Neuvéglise C., Lépingle A., Bon E., Feynerol C., Artiguenave F., Wincker P., Gaillardin C. (2000) FEBS Lett. 487, 95–100 [DOI] [PubMed] [Google Scholar]
- 27.Kinsky S., Mihalikova A., Kramara J., Nosek J., Tomaska L. (2010) Curr. Genet. 56, 413–425 [DOI] [PubMed] [Google Scholar]
- 28.Cohn M., McEachern M. J., Blackburn E. H. (1998) Curr. Genet. 33, 83–91 [DOI] [PubMed] [Google Scholar]
- 29.Rhodin-Edsö J. (2009) Functional Studies of Telomere-binding Proteins in Saccharomyces castelli. Ph.D. thesis, Department of Cell and Organism Biology, Lund University [Google Scholar]
- 30.Rhodin J., Astromskas E., Cohn M. (2006) J. Mol. Biol. 355, 335–346 [DOI] [PubMed] [Google Scholar]
- 31.Sánchez-Alonso P., Guzman P. (2008) Fungal Genet. Biol. 45, S54–S62 [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34.Stansel R. M., de Lange T., Griffith J. D. (2001) EMBO J. 20, 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffith J. D., Christiansen G. (1978) Annu. Rev. Biophys. Bioeng. 7, 19–35 [DOI] [PubMed] [Google Scholar]
- 36.Chen D. C., Beckerich J. M., Gaillardin C. (1997) Appl. Microbiol. Biotechnol. 48, 232–235 [DOI] [PubMed] [Google Scholar]
- 37.Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. (1997) Genes Dev. 11, 83–93 [DOI] [PubMed] [Google Scholar]
- 38.van Attikum H., Cobb J. (2010) Epigenomics www.epigenome-noe.net/WWW/researchtools/protocol.php?protid=27
- 39.Kovac L., Bednarova H., Greksak M. (1968) Biochim. Biophys. Acta 153, 32–42 [Google Scholar]
- 40.Arndt K. T., Styles C., Fink G. R. (1987) Science 237, 874–880 [DOI] [PubMed] [Google Scholar]
- 41.Daignan-Fornier B., Fink G. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6746–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denis V., Boucherie H., Monribot C., Daignan-Fornier B. (1998) Mol. Microbiol. 30, 557–566 [DOI] [PubMed] [Google Scholar]
- 43.Denis V., Daignan-Fornier B. (1998) Mol. Gen. Genet. 259, 246–255 [DOI] [PubMed] [Google Scholar]
- 44.Springer C., Künzler M., Balmelli T., Braus G. H. (1996) J. Biol. Chem. 271, 29637–29643 [DOI] [PubMed] [Google Scholar]
- 45.Griffith J., Bianchi A., de Lange T. (1998) J. Mol. Biol. 278, 79–88 [DOI] [PubMed] [Google Scholar]
- 46.Shore D. (1994) Trends Genet. 10, 408–412 [DOI] [PubMed] [Google Scholar]
- 47.Hogues H., Lavoie H., Sellam A., Mangos M., Roemer T., Purisima E., Nantel A., Whiteway M. (2008) Mol. Cell 29, 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassetzky N. S., Gaden F., Brun C., Gasser S. M., Gilson E. (1999) Nucleic Acids Res. 27, 4687–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mata J., Lyne R., Burns G., Bähler J. (2002) Nat. Genet. 32, 143–147 [DOI] [PubMed] [Google Scholar]
- 50.Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., Horinouchi S., Yoshida M. (2006) Nat. Biotechnol. 24, 841–847 [DOI] [PubMed] [Google Scholar]
- 51.Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., Basham D., Bowman S., Brooks K., Brown D., Brown S., Chillingworth T., Churcher C., Collins M., Connor R., Cronin A., Davis P., Feltwell T., Fraser A., Gentles S., Goble A., Hamlin N., Harris D., Hidalgo J., Hodgson G., Holroyd S., Hornsby T., Howarth S., Huckle E. J., Hunt S., Jagels K., James K., Jones L., Jones M., Leather S., McDonald S., McLean J., Mooney P., Moule S., Mungall K., Murphy L., Niblett D., Odell C., Oliver K., O'Neil S., Pearson D., Quail M. A., Rabbinowitsch E., Rutherford K., Rutter S., Saunders D., Seeger K., Sharp S., Skelton J., Simmonds M., Squares R., Squares S., Stevens K., Taylor K., Taylor R. G., Tivey A., Walsh S., Warren T., Whitehead S., Woodward J., Volckaert G., Aert R., Robben J., Grymonprez B., Weltjens I., Vanstreels E., Rieger M., Schäfer M., Müller-Auer S., Gabel C., Fuchs M., Düsterhöft A., Fritzc C., Holzer E., Moestl D., Hilbert H., Borzym K., Langer I., Beck A., Lehrach H., Reinhardt R., Pohl T. M., Eger P., Zimmermann W., Wedler H., Wambutt R., Purnelle B., Goffeau A., Cadieu E., Dréano S., Gloux S., Lelaure V., Mottier S., Galibert F., Aves S. J., Xiang Z., Hunt C., Moore K., Hurst S. M., Lucas M., Rochet M., Gaillardin C., Tallada V. A., Garzon A., Thode G., Daga R. R., Cruzado L., Jimenez J., Sánchez M., del Rey F., Benito J., Domínguez A., Revuelta J. L., Moreno S., Armstrong J., Forsburg S. L., Cerutti L., Lowe T., McCombie W. R., Paulsen I., Potashkin J., Shpakovski G. V., Ussery D., Barrell B. G., Nurse P., Cerrutti L. (2002) Nature 415, 871–880 [DOI] [PubMed] [Google Scholar]
- 52.Cooper J. P., Watanabe Y., Nurse P. (1998) Nature 392, 828–831 [DOI] [PubMed] [Google Scholar]
- 53.Nimmo E. R., Pidoux A. L., Perry P. E., Allshire R. C. (1998) Nature 392, 825–828 [DOI] [PubMed] [Google Scholar]
- 54.Tomita K., Cooper J. P. (2007) Cell 130, 113–126 [DOI] [PubMed] [Google Scholar]
- 55.Konig P., Giraldo R., Chapman L., Rhodes D. (1996) Cell 85, 125–136 [DOI] [PubMed] [Google Scholar]
- 56.Bianchi A., Smith S., Chong L., Elias P., de Lange T. (1997) EMBO J. 16, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bianchi A., Stansel R. M., Fairall L., Griffith J. D., Rhodes D., de Lange T. (1999) EMBO J. 18, 5735–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhodin, Edsö J., Tati R., Cohn M. (2008) FEMS Yeast Res. 8, 1289–1302 [DOI] [PubMed] [Google Scholar]
- 59.Teixeira M. T., Gilson E. (2005) Chromosome Res. 13, 535–548 [DOI] [PubMed] [Google Scholar]
- 60.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I., Doerks T., Bork P. (2009) Nucleic Acids Res. 37, D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drummond A. J., Ashton B., Cheung M., Heled J., Kearse M., Moir R., Stones-Havas S., Thierer T., Wilson A. (2009) Geneious Version 4.8, Biomatters, Ltd., Auckland, New Zealand [Google Scholar]
- 63.Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., 2nd (1997) EMBNEW. News 4, 14 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.