Abstract
STAP-2 (signal transducing adaptor protein-2) is a recently identified adaptor protein that contains pleckstrin homology (PH) and Src homology 2-like domains, as well as a STAT3-binding motif in its C-terminal region. STAP-2 is also a substrate of breast tumor kinase (Brk). In breast cancers, Brk expression is deregulated and promotes STAT3-dependent cell proliferation. In the present study, manipulated STAP-2 expression demonstrated essential roles of STAP-2 in Brk-mediated STAT3 activation. STAP-2 interacts with both Brk and STAT3. In addition, small interfering RNA-mediated reduction of endogenous STAP-2 expression strongly decreased Brk-mediated STAT3 activation in T47D breast cancer cells. The PH domain of STAP-2 is involved in multiple steps: the binding between Brk and STAP-2, the activation and tyrosine phosphorylation of STAT3, and the activation of Brk. Notably, a STAP-2 PH-Brk fusion protein exhibited robust kinase activity and increased activation and tyrosine phosphorylation of STAT3. Finally, STAP-2 knockdown in T47D cells induced a significant decrease of proliferation, as strong as that of Brk or STAT3 knockdown. Taken together, our findings are likely to inform the development of a novel therapeutic strategy, as well as the determination of novel prognostic values, in breast carcinomas.
Keywords: Adaptor Proteins, Breast Cancer, Protein Phosphorylation, Signal Transduction, Transcription Factors, Protein-tyrosine Kinase (Tyrosine Kinase), Brk, STAP-2, STAT3
Introduction
The nonreceptor tyrosine kinase breast tumor kinase (Brk)2 was originally isolated from a human breast carcinoma (1). Brk is also known as PTK6, having been identified as a highly expressed protein-tyrosine kinase in human melanocytes (2). In addition, a cDNA for the mouse orthologue, Ptk6 (previously termed Sik), which has 80% amino acid identity to Brk/PTK6, was cloned from mouse intestinal crypt cells (3). Brk contains an Src homology (SH) 3 domain, an SH2 domain, and a tyrosine kinase catalytic domain, but it lacks an N-terminal myristoylation site for membrane targeting (1). Brk is expressed in many malignancies, such as metastatic melanomas and colon and prostate tumors (4–6). Brk expression is also detected in a large proportion of human mammary gland tumors, whereas it is not expressed in the normal mammary gland (1, 7). It is noteworthy that small interfering RNA-mediated down-regulation of Brk expression in breast cancer cells results in their decreased growth capacity (8). Furthermore, it has been demonstrated that overexpression of Brk sensitizes human mammary epithelial cells to EGF and/or heregulin stimuli and increases anchorage-independent growth (9, 10). Down-regulation of Brk can also influence EGF- and heregulin-induced cell proliferation, suggesting a contribution of Brk to signaling induced by members of the EGF receptor family (11). However, the molecular mechanism by which Brk participates in tumorigenesis remains poorly characterized.
STAT3 and STAT5, which play crucial roles in cell proliferation and differentiation, are believed to be activated by Brk (12, 13). Another Brk substrate is STAP-2 (signal transducing adaptor protein-2), whose tyrosine residues are phosphorylated by Brk (14, 15). STAP-2, which we isolated as a c-Fms-interacting protein, contains an N-terminal pleckstrin homology (PH) domain and a region distantly related to the SH2 domain (16). In its C-terminal region, a proline-rich region and a STAT3-binding YXXQ motif are also present (16). Our previous experiments have suggested that STAP-2 interacts with and influences several signaling molecules, including STAT3 and STAT5 (16, 17). The dysregulated activation of STAT3 is a possible mechanism of tumorigenesis in breast cancers; therefore, it would be very informative to analyze the interactions among Brk, STAP-2, and STAT3.
In the present study, we found that STAP-2 acts as an endogenous positive regulator of breast cancer cell growth. Manipulation of STAP-2 expression also indicated that STAP-2 is essential for Brk-mediated STAT3 activation. In addition, we investigate the domains responsible for the effects of STAP-2.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Expression vectors, STAP-2, STAT3, and their mutants were described previously (15–18). Expression vectors for wild-type Brk, a kinase-dead form of Brk (Brk K219M), and STAT3-LUC were provided by Dr. A. J. Harvey (Brunel University, Middlesex, UK) and Dr. T. Hirano (Osaka University, Osaka, Japan), respectively (1, 19). GST-fused Brk mutants were generated by PCR methods and sequenced (primer sequences are available upon request). A STAP-2 PH-Brk fusion construct (PH-Brk), which tagged Brk with STAP-2 PH domain (amino acids 1–147) at the N terminus, was also generated by PCR methods and sequenced (primer sequences are available upon request). Anti-Myc, -Brk, -STAT3, -GST, and -G3PDH antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); anti-FLAG antibody was from Sigma-Aldrich (St. Louis, MO), anti-phosphotyrosine monoclonal antibody (PY20) was from Cosmobio (Tokyo, Japan); anti-phosphoSTAT3 (pSTAT3) Tyr-705 antibody was from Cell Signaling Technology (Beverly, MA); anti-Nucleoporin antibody was from BD Biosciences (Franklin Lakes, NJ); anti-actin antibody was from Chemicon International (Temecula, CA); and anti-STAP-2 antibody was from Everest Biotech (Oxfordshire, UK).
Cell Culture, Transfection, siRNA, and Luciferase Assays
The human embryonic kidney carcinoma cell line 293T was maintained in DMEM containing 10% FCS and transfected by the standard calcium precipitation protocol. Luciferase assay was performed as described (17). The human breast cancer cell lines MCF7 and T47D and the human cervix carcinoma cell line HeLa were maintained in DMEM containing 10% FCS. MCF7/pcDNA3 and MCF7/STAP-2 transfectants were prepared as described previously (20). STAP-2 knockdown T47D cell lines (T47D/shSTAP-2#1 and #2) were established by transfection of with pGPU6/GFP/Neo vector (Shanghai GenePharm, Shanghai, China) bearing shRNA targeting STAP-2 (#1, 5′-CCAGCTGTTGACTATGAGA-3′; #2, 5′-CCAGTCATCCTGAAGCCAA-3′) and then selected with G418 (1 mg/ml; Sigma-Aldrich). Similarly, control shRNA-transfected (nonsilencing, 5′-TTCTCCGAACGTGTCACGT-3′) T47D cell line (T47D/shControl) was also established. siRNAs targeting human STAP-2, STAT3, and Brk used in this study were as follows: STAP-2#1, 5′-GCAGGGUCUCACCAUUUAUTT-3′; STAP-2#2, 5′-GGUGCUAGGCUACGUGGAATT-3′; STAT3, 5′-CCGUCAACAAAUUAAGAAATT-3′; and Brk, 5′-GGGUCCAGGUGGCCAUUAATT-3′. T47D cells were plated on a 24-well plate at 2 × 104 cells/well and then incubated with an siRNA-Lipofectamine 2000 (Invitrogen) mixture at 37 °C for 4 h, followed by the addition of fresh medium containing 10% FCS. MCF-7 and T47D cells were transfected with STAT3-LUC using jetPEI (PolyPlus-transfection, Strasbourg, France) according to the manufacturer's instructions. At 36 h after the cells were transfected, they were harvested and assayed for their luciferase activities, using a dual luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's instructions. Three or more independent experiments were carried out for each assay.
RNA Isolation, Quantitative Real Time PCR
The cells were harvested, total RNAs were prepared by using Iso-Gen (Nippon Gene, Tokyo, Japan), and quantitative real time PCR analyses of the respective genes as well as the control ACTIN mRNA transcripts were carried out using the assay-on-demand gene-specific fluorescently labeled TaqMan MGB probe in an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) (20).
Immunoprecipitation, Immunoblotting, and in Vitro Phosphorylation
The immunoprecipitation and Western blotting assays were performed as described previously (17). The immunoprecipitates from cell lysates were resolved on SDS-PAGE and transferred to PVDF transfer membrane (PerkinElmer Life Sciences). The filters were then immunoblotted with each antibody. Immunoreactive proteins were visualized using an enhanced chemiluminescence detection system (Millipore, Bedford, MA). In vitro kinase reactions were performed as described (21); briefly, immune complexes of Brk were washed in kinase buffer (10 mm HEPES, pH 7.4, 50 mm NaCl, 0.1 mm sodium orthovanadate, 5 mm MnCl2, 5 mm MgCl2) and mixed with 5 μCi/ml [γ-32P]ATP at 25 °C for 30 min. The products of these reactions were separated by SDS-PAGE. Synthetic STAT3 Y705 peptide of human STAT3, corresponding to residues 695–715, was obtained from Operon Biotechnologies.
Indirect Immunofluorescence Microscopy
To analyze the subcellular localizations of STAP-2 and Brk proteins, Myc-STAP-2 (WT or ΔPH) and FLAG-Brk were transiently transfected into HeLa cells by JetPEI. Immunofluorescence staining procedures were performed as described (20). The primary antibodies used were rabbit anti-FLAG and mouse anti-Myc antibodies. The secondary antibodies used were rhodamine-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG (both from Chemicon). DNA was visualized by staining with DAPI (Wako Chemicals, Osaka, Japan). The staining was visualized by confocal laser scanning microscopy with an LSM510 microscope (Carl Zeiss, Thornwood, NY) equipped with an Apochromat ×63/1.4 oil immersion objective, using excitation wavelengths of 543 nm (rhodamine) and 488 nm (FITC).
Cell Proliferation Assay
The numbers of viable T47D cells after the indicated treatments were measured using a WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) assay (Cell Counting Kit-8; Wako Pure Chemicals) (20). Briefly, 10 μl of WST-8 solution was added to the cells in each well and incubated for 2 h. The absorbances were measured at a test wavelength of 450 nm and a reference wavelength of 650 nm using a microplate reader (Bio-Rad).
RESULTS
STAP-2-enhanced Brk-mediated STAT3 Activation in Breast Cancer Cells
Characterization of Brk showed it to be frequently present in human breast tumors yet absent in normal or fibrocystic mammary tissues (1). In breast tumor cells, Brk promotes growth factor signaling and cell migration (9, 22). STAP-2 is known to be a substrate for Brk, and we previously reported a possible influence of STAP-2 on the function of Brk in 293T and HeLa cells (14, 15). In the present study, we first attempted to clarify roles of STAP-2 on Brk-mediated STAT3 activation in breast cancer cells. We transfected plasmids encoding STAP-2, Brk, and STAT3-LUC into MCF7 breast cancer cells. After 48 h, the cells were harvested, and the STAT3-LUC activities were evaluated. As shown in Fig. 1A, extremely high luciferase activity was observed when both Brk and STAP-2 were expressed simultaneously. However, separate expression of either Brk or STAP-2 failed to induce STAT3-LUC activation. In T47D breast cancer cells, co-expression of Brk and STAP-2 resulted in a significant enhancement of STAT3-LUC activity (Fig. 1B). Interestingly, expression of Brk alone in T47D cells could stimulate STAT3-LUC activation. The different responses to ectopic Brk expression are likely to be dependent on endogenous STAP-2 expression levels. Western blot analysis showed endogenous STAP-2 protein in samples from T47D cells, but not from MCF7 cells (data not shown). To further assess the functional relevance of endogenous STAP-2 to Brk-mediated STAT3 activation in breast cancer cells, we examined the effects of siRNA-mediated reduction of STAP-2. T47D cells were transfected with a specific siRNA for STAP-2 (#1 or #2) or a control siRNA. Total cellular proteins from the transfected cells were subjected to Western blot analysis, which confirmed a reduction of STAP-2 expression. As shown in Fig. 1C, a reduction of STAP-2 expression in T47D cells resulted in decreased Brk-mediated STAT3-LUC activation. Taken together, endogenous STAP-2 is involved in the positive regulation of Brk-mediated STAT3-LUC activation in T47D cells.
FIGURE 1.
STAP-2 regulates Brk-mediated STAT3 activation in breast cancer cells. A, MCF7 cells in a 24-well plate were transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk (100 ng) and expression vector for Myc-tagged STAP-2 WT (30 or 300 ng). At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. **, p < 0.01. An aliquot of each total cell lysate (TCL) was analyzed by immunoblotting (IB) with an anti-FLAG or anti-Myc antibody. B, T47D cells in a 24-well plate were transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk (100 ng) and expression vector for Myc-tagged STAP-2 WT (30 or 300 ng). At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. **, p < 0.01. An aliquot of each TCL was analyzed by immunoblotting with an anti-FLAG or anti-Myc antibody. C, T47D cells in a 24-well plate were transfected with control or STAP-2 (#1 and #2) siRNA, and the cells were then transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk (300 ng) using jetPEI. At 36 h after transfection, the cells were harvested and assayed for luciferase activity using a dual luciferase reporter assay system. The results are indicated as fold induction of luciferase activity from triplicate experiments, and the error bars represent the S.D. **, p < 0.01. An aliquot of each TCL was analyzed by immunoblotting with an anti-Brk, anti-STAP-2, or anti-actin antibody.
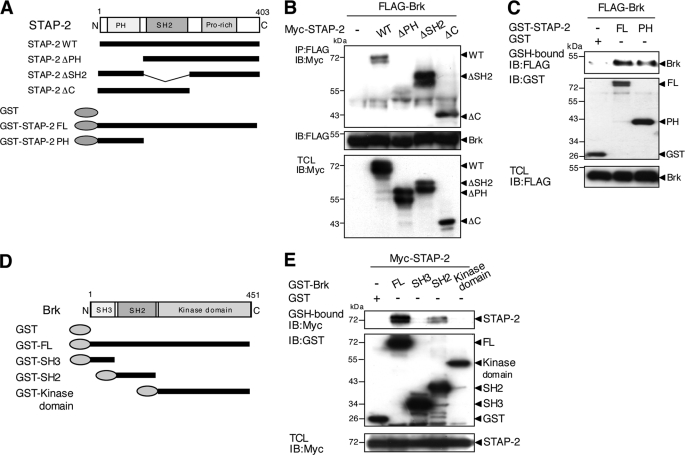
Involvement of the STAP-2 PH Domain in Associations with Brk
To determine the domains of STAP-2 involved in associations with Brk, a series of deletion mutants of Myc-tagged STAP-2 were employed (Fig. 2A). The respective mutants together with FLAG-tagged Brk were transiently expressed in 293T cells. The binding potential of these proteins with FLAG-tagged Brk was examined by immunoprecipitation with an anti-FLAG antibody followed by Western blot analysis with an anti-Myc antibody. As shown in Fig. 2B, a deletion mutant of the PH domain of STAP-2 (STAP-2 ΔPH) failed to interact with Brk, whereas WT and deletion mutants of the SH2-like (STAP-2 ΔSH2) or C-terminal domains (STAP-2 ΔC) of STAP-2 retained binding capacity to Brk. In addition, a fusion protein composed of the PH domain of STAP-2 and GST showed strong association with Brk, similar to that of STAP-2 WT (Fig. 2C). Therefore, the PH domain of STAP-2 is essential and sufficient for binding to Brk. We also determined which domain(s) of Brk were involved in associations with STAP-2 using a series of deletion mutants of Brk fused with GST (GST-FL, GST-SH3, GST-SH2, and GST-kinase domain) (Fig. 2D). Myc-tagged STAP-2 and the respective Brk mutants were transiently expressed in 293T cells, and the binding potential of these proteins to Myc-tagged STAP-2 was examined by pull-down assays with glutathione-Sepharose, followed by Western blot analysis with an anti-Myc antibody. The precipitates of STAP-2 contained both GST-FL and GST-SH2 (Fig. 2E). These results indicate that STAP-2 interacted with the SH2 domain of Brk through its PH domain.
FIGURE 2.
Molecular interactions between STAP-2 and Brk. A, schematic diagrams of the domain structures of the STAP-2 deletion mutant fragments are shown. B, 293T cells (1 × 107) were transfected with FLAG-tagged Brk (10 μg) with or without Myc-tagged STAP-2 deletion mutants (8 μg). At 48 h after transfection, the cells were lysed, immunoprecipitated (IP) with an anti-FLAG antibody, and immunoblotted (IB) with an anti-Myc or anti-FLAG antibody. An aliquot of each TCL was immunoblotted with an anti-Myc antibody. C, 293T cells (1 × 107) were transfected with FLAG-tagged Brk (10 μg) with or without GST, GST-fused STAP-2 WT, or PH (8 μg). At 48 h after transfection, the cells were lysed, pulled down with glutathione-Sepharose, and immunoblotted with an anti-FLAG or anti-GST antibody. An aliquot of each TCL was immunoblotted with an anti-FLAG antibody. D, schematic diagrams of the domain structures of the GST-fused Brk deletion mutant fragments are shown. E, 293T cells (1 × 107) were transfected with Myc-tagged STAP-2 (8 μg) with or without GST-fused Brk deletion mutants (10 μg). At 48 h after transfection, the cells were lysed, pulled down with glutathione-Sepharose, and immunoblotted with an anti-Myc or anti-GST antibody. An aliquot of each TCL was immunoblotted with an anti-Myc antibody.
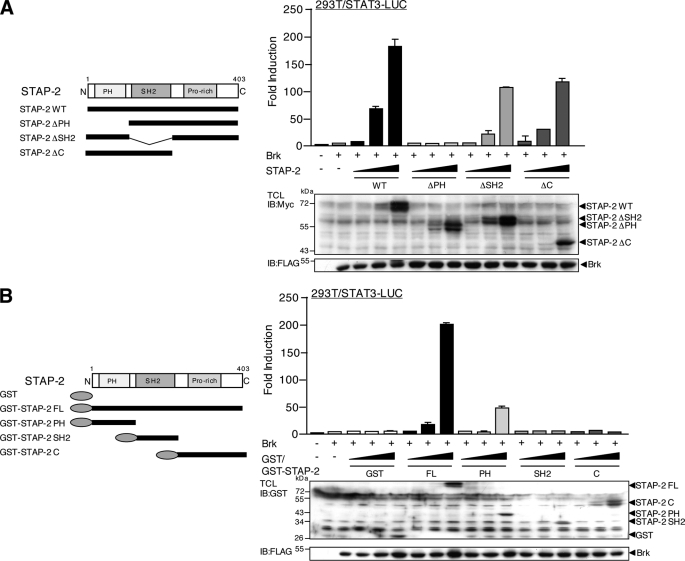
Functional Role of the PH Domain of STAP-2 in Brk-mediated STAT3 Activation
To assess the functional relevance of STAP-2 to Brk-mediated STAT3 activation, we transfected Brk and STAT3-LUC, together with a series of STAP-2 deletion mutants (Fig. 2A), into 293T cells. Brk-mediated STAT3-LUC activity was significantly enhanced by expression of STAP-2 WT, STAP-2 ΔSH2, and STAP-2 ΔC in a dose-dependent manner, although STAP-2 ΔSH2 and STAP-2 ΔC showed slightly lower activation than STAP-2 WT (Fig. 3A). Of importance, STAP-2 ΔPH did not show any enhancing effects on the STAT3-LUC activity mediated by Brk. We also confirmed the above results using a different series of deletion mutants of STAP-2 (Fig. 2A; GST-STAP-2 PH, GST-STAP-2 SH2, and GST-STAP-2 C). Overexpression of STAP-2 PH significantly enhanced Brk-mediated STAT3-LUC activity but failed to completely restore the enhancing effects of STAP-2 WT (Fig. 3B). Both STAP-2 SH2 and STAP-2 C completely lost the enhancing capacity. These results suggest that the PH domain of STAP-2 is essential but not sufficient for the enhancement of Brk-mediated STAT3 activation. In addition, STAP-2 SH2 and STAP-2 C appeared to be involved in Brk-mediated STAT3 activation, although they are not essential. Therefore, full STAT3 activation mediated by Brk is likely to require multidomain interactions with STAP-2 or for STAP-2 to be in its wild-type conformation.
FIGURE 3.
Functional role of the PH domain of STAP-2 in Brk-mediated STAT3 activation. A, schematic diagrams of the domain structures of the STAP-2 deletion mutant fragments are shown. 293T cells in a 24-well plate were transfected with STAT3-LUC (200 ng) and/or FLAG-tagged Brk (100 ng) and increasing amounts (1.0, 10, and 100 ng) of expression vector for Myc-tagged STAP-2 WT, STAP-2 ΔPH, STAP-2 ΔSH2, or STAP-2 C. At 48 h transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. An aliquot of each TCL was analyzed by immunoblotting (IB) with an anti-Myc or anti-FLAG antibody. B, schematic diagrams of the domain structures of the GST-fused STAP-2 deletion mutant fragments are shown. 293T cells in a 24-well plate were transfected with STAT3-LUC (200 ng) and/or FLAG-tagged Brk (100 ng) and increasing amounts (1, 10, and 100 ng) of expression vector for GST or GST-fused STAP-2 FL, STAP-2 PH, STAP-2 SH2, or STAP-2 C. At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. An aliquot of each TCL was analyzed by immunoblotting with an anti-GST or anti-FLAG antibody.
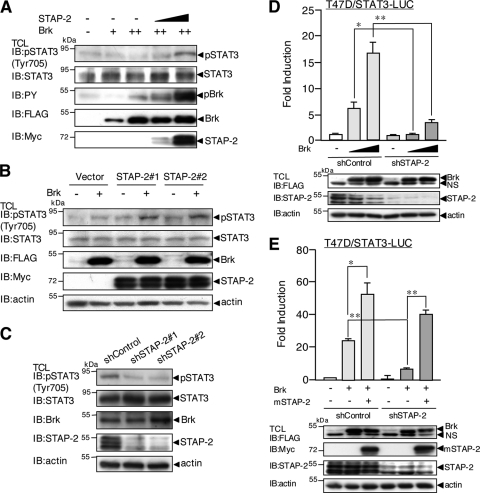
STAP-2 Enhanced Brk-mediated Tyrosine Phosphorylation of STAT3 in Breast Cancer Cells
To further clarify the molecular mechanisms underlying Brk/STAP-2-mediated STAT3 activation in breast cancer cells, we tested the effect of STAP-2 on Brk-mediated tyrosine phosphorylation of STAT3, which is an important step for transcriptional activation. Myc-tagged STAP-2 was transiently expressed with or without FLAG-tagged Brk in MCF7 cells. The cells were lysed, and the lysates were immunoblotted with an anti-pSTAT3 (Tyr-705) antibody. As shown in Fig. 4A, the induction of STAT3 tyrosine phosphorylation by Brk was significantly enhanced in the presence of STAP-2. We then used stable transformants of MCF7 cells expressing STAP-2 (MCF7/STAP-2#1 and #2). As shown in Fig. 4B, tyrosine phosphorylation of STAT3 was markedly enhanced in MCF7 cells overexpressing STAP-2. To examine whether a reduction of endogenous STAP-2 expression affects tyrosine phosphorylation of STAT3 in breast cancer cells, we established STAP-2 knockdown clones (shSTAP-2#1 and #2) in T47D cells. Constitutive tyrosine phosphorylation of STAT3 was observed in control T47D cells. However, tyrosine phosphorylation of STAT3 was markedly reduced in two STAP-2 knockdown cell clones (Fig. 4C). These results indicate that STAP-2 plays important roles in Brk-mediated tyrosine phosphorylation of STAT3 in breast cancer cells.
FIGURE 4.
STAP-2 regulates Brk-induced tyrosine phosphorylation of STAT3 at Tyr-705. A, MCF7 cells in a 12-well plate were transfected with or without Myc-tagged STAP-2 WT (0.3 and 1.0 μg) and/or FLAG-tagged Brk (+, 0.3 μg; ++, 1.0 μg). At 48 h after transfection, the cells were lysed and immunoblotted (IB) with an anti-pSTAT3 (Tyr-705), anti-STAT3, anti-PY, anti-FLAG, or anti-Myc antibody. B, MCF7/pcDNA3, MCF7/STAP-2#1, and MCF7/STAP-2#2 cells in a 12-well plate were transfected with or without FLAG-tagged Brk (1.0 μg). At 48 h after transfection, the cells were lysed and immunoblotted with an anti-pSTAT3 (Tyr-705), anti-STAT3, anti-FLAG, anti-Myc, or anti-actin antibody. C, T47D/shControl, T47D/shSTAP-2#1, and T47D/shSTAP-2#2 cells were lysed and immunoblotted with an anti-pSTAT3 (Tyr-705), anti-STAT3, anti-Brk, anti-STAP-2, or anti-actin antibody. D, T47D/shControl or T47D/shSTAP-2#1 cells in a 24-well plate were transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk (100 or 300 ng). At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. *, p < 0.05; **, p < 0.01. An aliquot of each TCL was analyzed by immunoblotting with an anti-FLAG, anti-STAP-2, or anti-actin antibody. E, T47D/shControl and T47D/shSTAP-2#1 cells in a 24-well plate were transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk (300 ng) and/or expression vector for Myc-tagged mouse STAP-2 WT. At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. *, p < 0.05; **, p < 0.01. An aliquot of each TCL was analyzed by immunoblotting with an anti-FLAG, anti-Myc, anti-STAP-2, or anti-actin antibody.
We also tested the STAT3-LUC activity mediated by Brk in these STAP-2 knockdown T47D cells. As shown in Fig. 4D, Brk-mediated STAT3-LUC activity was markedly reduced in the STAP-2 knockdown cells consistent with the above results. Furthermore, reduced Brk-mediated STAT3-LUC activity in the STAP-2 knockdown cells was restored by overexpression of a mouse STAP-2 cDNA (Fig. 4E). Therefore, Brk-mediated tyrosine phosphorylation of STAT3 increases transcriptional activity of STAT3 in breast cancer cells.
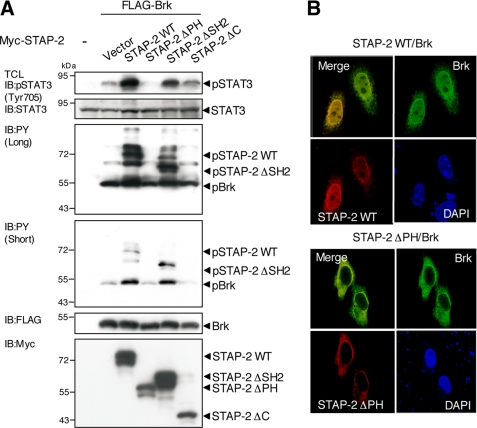
The PH Domain of STAP-2 Is Required for Brk Activation
To further assess the functional relationship among Brk, STAP-2, and STAT3, we examined Brk-induced tyrosine phosphorylation of STAP-2 and STAT3. Following transfection of 293T cells with expression vectors for FLAG-tagged Brk and a series of Myc-tagged STAP-2 deletion mutants, the cells were lysed, and the lysates were immunoblotted with anti-pSTAT3 (Tyr-705), anti-STAT3, anti-phosphotyrosine (PY), anti-FLAG, and anti-Myc antibodies. As shown in Fig. 5A, the Brk-enhanced tyrosine phosphorylation of STAT3 disappeared in STAP-2 ΔPH cells and decreased in STAP-2 ΔSH2 and STAP-2 ΔC cells compared with that in STAP-2 WT cells. This pattern correlates well with the results of the STAT3-LUC experiments (Fig. 3A). STAP-2 WT and STAP-2 ΔSH2 were strongly phosphorylated when Brk was co-expressed. However, STAP-2 ΔPH failed to be tyrosine-phosphorylated by Brk. This might be because STAP-2 ΔPH could not bind to Brk, as shown in Fig. 2B. STAP-2 ΔC also failed to be tyrosine-phosphorylated by Brk. This may be explained by our previous report showing that Tyr-250 in the C-terminal region is a major Brk-induced phosphorylation site on STAP-2 (15). On the other hand, phosphorylation of Brk was enhanced when STAP-2 WT or STAP-2 ΔSH2 was co-transfected. STAP-2 ΔPH and STAP-2 ΔC did not have such enhancing effects. These results are likely to indicate that both the PH domain and the C-terminal region of STAP-2 play a role in the activation of Brk.
FIGURE 5.
STAP-2 PH domain regulates Brk-induced tyrosine phosphorylation of STAT3 at Tyr-705 and nuclear co-localization with Brk. A, 293T cells in a 12-well plate were transfected with FLAG-tagged Brk (1.0 μg) with or without Myc-tagged STAP-2 deletion mutants (1.0 μg). At 48 h after transfection, the cells were lysed and immunoblotted (IB) with an anti-pSTAT3 (Tyr-705), anti-STAT3, anti-PY (short exposure and long exposure for ECL detection), anti-FLAG, or anti-Myc antibody. B, HeLa cells in 12-well plates were transfected with FLAG-tagged Brk (1.0 μg), together with Myc-tagged STAP-2, or Myc-tagged STAP-2 ΔPH (1.0 μg) using jetPEI. At 48 h after transfection, the cells were fixed, incubated with anti-FLAG or anti-Myc antibodies, and visualized with FITC- and rhodamine-conjugated secondary antibodies. The same slides were also stained with DAPI to detect the nuclei.
The difference between STAP-2 WT and STAP-2 ΔPH was also confirmed with confocal microscopy experiments. As shown in Fig. 5B, STAP-2 WT was distributed throughout the cytoplasm and nucleus as previously described (14), but STAP-2 ΔPH was mainly located in the cytoplasm. Of importance, nuclear localization of Brk was observed in STAP-2 WT transfectants but not in STAP-2 ΔPH transfectants. Therefore, STAP-2 affects Brk distribution, probably depending on the activation state of Brk.
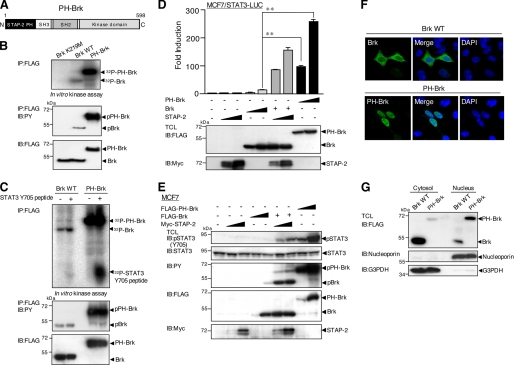
To clarify the effect of the STAP-2 PH domain on Brk activation, we used a STAP-2 PH-Brk fusion protein (PH-Brk), in which Brk was fused to the N terminus of STAP-2 PH (Fig. 6A). As shown in Fig. 6 (B and C), PH-Brk protein was strongly expressed and showed a robust kinase activity compared with Brk WT. Furthermore, PH-Brk alone induced marked activation of STAT3-LUC activity and tyrosine phosphorylation of STAT3 in breast cancer cells (Fig. 6, D and E). PH-Brk was mainly localized in the nucleus, although Brk was localized throughout the cytoplasm and nucleus (Fig. 6F). Nuclear fractionation also showed nuclear accumulation of PH-Brk proteins in the nucleus (Fig. 6G). Therefore, the STAP-2 PH domain controls the kinase activity of Brk and Brk-mediated STAT3 activation by influencing the localization of Brk.
FIGURE 6.
A STAP2 PH-Brk fusion protein exhibited robust kinase activity and increased activation and tyrosine phosphorylation of STAT3. A, schematic diagrams of the domain structures of the STAP2 PH-Brk fusion protein PH-Brk are shown. B, kinase activity of PH-Brk. FLAG-tagged Brk K219M (a kinase-dead form of Brk), WT, or PH-Brk (10 μg) was transiently transfected into 293T cells. The cell were lysed and immunoprecipitated (IP) with anti-FLAG antibody and assayed for in vitro kinase activity (top panel). The amounts of Brk proteins were shown to be same by Western blotting with anti-FLAG antibody (bottom panel). Tyrosine phosphorylation of each Brk protein was also monitored by Western blotting with anti-PY (middle panel). C, FLAG-tagged Brk WT or PH-Brk (10 μg) were transiently transfected into 293T cells. The cell were lysed and immunoprecipitated with anti-FLAG antibody and assayed for in vitro kinase reactions using synthetic STAT3 Y705 peptide (10 μg) (top panel). The amounts of Brk proteins were shown to be same by Western blotting with anti-FLAG antibody (bottom panel). Tyrosine phosphorylation of each Brk protein was also monitored by Western blotting with anti-PY (middle panel). D, MCF7 cells in a 24-well plate were transfected with STAT3-LUC (100 ng) and/or FLAG-tagged Brk WT or PH-Brk (100 and 300 ng) and/or expression vector for Myc-tagged STAP-2 WT (30 and 300 ng). At 48 h after transfection, the cells were harvested, and the luciferase activities were measured. At least three independent experiments were carried out for each assay. The error bars represent the S.D. **, p < 0.01. An aliquot of each TCL was analyzed by immunoblotting (IB) with an anti-FLAG or anti-Myc antibody. E, MCF7 cells in a 12-well plate were transfected with or without Myc-tagged STAP-2 (0.2 and 1.0 μg) and/or FLAG-tagged Brk WT or PH-Brk (0.2 and 1.0 μg). At 48 h after transfection, the cells were lysed and immunoblotted with an anti-pSTAT3 (Tyr-705), anti-STAT3, anti-PY, anti-FLAG, or anti-Myc antibody. F, HeLa cells in 12-well plates were transfected with FLAG-tagged Brk or PH-Brk (1.0 μg) using jetPEI. At 48 h after transfection, the cells were fixed, incubated with anti-FLAG antibody, and visualized with FITC-conjugated secondary antibodies. The same slides were also stained with DAPI to detect the nuclei. G, HeLa cells in a 12-well plate were transfected with FLAG-tagged Brk or PH-Brk (1.0 μg) using jetPEI. At 48 h after transfection, the cells were lysed and fractionated into cytosol and nuclear fractions. An aliquot of both fractions were immunoblotted with anti-FLAG, anti-G3PDH, or anti-nucleoporin antibodies.
Involvement of STAP-2 in the Formation of a Complex between Brk and STAT3
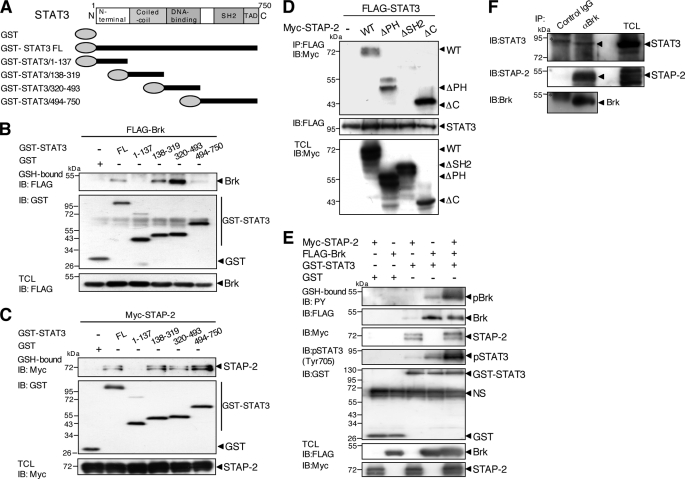
To further understand the detailed molecular interactions among Brk, STAP-2, and STAT3, we tried to determine the domains of STAP-2 involved in its associations with STAT3. The respective mutants together with FLAG-tagged STAT3 (Fig. 7A) were transiently expressed in 293T cells. As shown in Fig. 7B, precipitates for STAT3/138–319, corresponding to the coiled-coil domain, as well as STAT3/320–493, corresponding to the DNA-binding domain, contained Brk proteins. As for the binding regions between STAP-2 and STAT3, STAT3/138–319 (the coiled-coil domain), STAT3/320–493 (the DNA-binding domain), and STAT3/494–750 (the C-terminal region) individually interacted with STAP-2 (Fig. 7C). Therefore, STAT3 has multiple regions for interaction with Brk and/or STAP-2.
FIGURE 7.
Molecular interactions among STAP-2, STAT3, and Brk. A, schematic diagrams of the domain structures of STAT3 and a deletion mutant are shown. B, 293T cells (1 × 107) were transfected with FLAG-tagged Brk (10 μg) with or without GST-fused STAT3 FL or a deletion mutant (10 μg). At 48 h after transfection, the cells were lysed, pulled down with glutathione-Sepharose, and immunoblotted (IB) with an anti-FLAG or anti-GST antibody. An aliquot of each TCL was immunoblotted with an anti-FLAG antibody. C, 293T cells (1 × 107) were transfected with Myc-tagged STAP-2 (10 μg) with or without GST-fused STAT3 FL or a deletion mutant (10 μg). At 48 h after transfection, the cells were lysed, pulled down with glutathione-Sepharose, and immunoblotted with an anti-Myc or anti-GST antibody. An aliquot of each TCL was immunoblotted with an anti-Myc antibody. D, 293T cells (1 × 107) were transfected with FLAG-tagged STAT3 (10 μg) with or without Myc-tagged STAP-2 deletion mutants (8 μg). At 48 h after transfection, the cells were lysed, immunoprecipitated (IP) with an anti-FLAG antibody, and immunoblotted with an anti-Myc or anti-FLAG antibody. An aliquot of each TCL was immunoblotted with an anti-Myc antibody. E, 293T cells (1 × 107) were transfected with or without FLAG-tagged Brk (10 μg) with or without GST-STAT3 FL (10 μg) and/or Myc-tagged STAP-2 (8 μg). At 48 h after transfection, the cells were lysed, pulled down with glutathione-Sepharose, and immunoblotted with an anti-PY, anti-Myc, anti-pSTAT3 (Tyr-705), anti-FLAG, or anti-GST antibody. An aliquot of each TCL was immunoblotted with an anti-FLAG or anti-Myc antibody. F, human breast cancer T47D cells (3 × 107) were lysed, immunoprecipitated with control IgG or anti-Brk antibody, and immunoblotted with anti-STAT3, anti-STAP-2, or anti-Brk antibody. The asterisk shows a nonspecific band.
We also determined which domain(s) of STAP-2 were involved in associations with STAT3. 293T cells were transfected with FLAG-tagged STAT3 and/or a series of Myc-tagged STAP-2 deletion mutants. The transfectants were lysed, immunoprecipitated with an anti-FLAG antibody, and then immunoblotted with an anti-Myc antibody. As shown in Fig. 7D, STAP-2 ΔSH2 failed to interact with STAT3, whereas both STAP-2 ΔPH and STAP-2 ΔC did bind to STAT3. These results indicate that STAP-2 may interact with STAT3 through its SH2 domain.
We further examined whether STAP-2 forms an efficient complex with Brk and STAT3 to activate Brk and STAT3. Expression vectors for FLAG-tagged Brk and GST-fused STAT3 together with or without Myc-tagged STAP-2 were transfected into 293T cells. The cells were lysed and subjected to pull-down assays with glutathione-Sepharose, followed by Western blot analysis with anti-PY, anti-FLAG, anti-Myc, anti-pSTAT3 (Tyr-705), or anti-GST antibodies. The precipitates for STAT3 contained Brk and STAP-2 (Fig. 7E). Of importance, tyrosine phosphorylation of STAT3 and Brk was greatly enhanced by co-expression of STAP-2. These results might indicate that Brk, STAT3, and STAP-2 form a complex in vivo. Finally, with regard to endogenous interactions among these three molecules, the immunoprecipitates for endogenous Brk contained significant levels of endogenous STAT3 and STAP-2 in T47D cells (Fig. 7F). Therefore, Brk complex formation in the presence of STAP-2 and STAT3 is efficient.
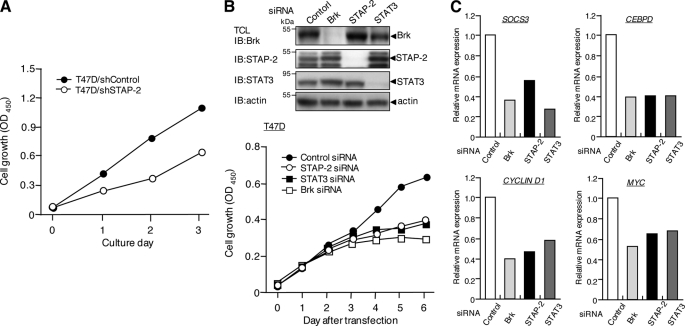
STAP-2 Acts as an Endogenous Positive Regulator of Breast Cancer Cell Growth
Because STAP-2 positively regulated Brk-mediated STAT3 activation in breast cancer cells, we tested whether STAP-2 expression affected cell growth. As shown in Fig. 8A, STAP-2 knockdown T47D clones grew more slowly than control T47D cells. We could not detect any difference in cell viability between the two cells types (data not shown). We then knocked down Brk, STAT3, or STAP-2 individually in T47D cells. Western blot analysis confirmed a great reduction of each protein on day 3 (Fig. 8B, upper panel), and the decrease of protein levels continued for at least 6 days (data not shown). As shown in Fig. 7B, the siRNA-mediated reduction of Brk, STAT3, or STAP-2 expression in T47D cells caused a significant decrease in cell growth, indicating that they play important roles in T47D cell growth. It is noteworthy that growth was decreased to similar extents in these three T47D cell types. This might suggest that Brk-induced cell growth in T47D cells is largely dependent on both STAT3 and STAP-2. Taken together, Brk/STAT3-mediated proliferation is a major mechanism for breast cancer cell growth, and STAP-2 plays essential roles in this process. Notably, the expression of several genes in the STAT3-mediated signaling, including SOCS3, C/EBPδ, cyclin D1, and c-Myc, was also reduced by STAP-2 knockdown in T47D cells (Fig. 8C).
FIGURE 8.
STAP-2 regulates breast cancer cell growth. A, T47D/shControl and T47D/shSTAP-2 (1 × 104 cells/well) cells were cultured in 96-well plates for the indicated periods. The cell numbers were measured using a Cell Counting Kit-8. The data are the means of triplicate experiments, which generally varied by <10%. Similar results were obtained in three independent experiments. B, T47D cells were transfected with a control siRNA, STAP2 siRNA (#1), STAT3 siRNA, or Brk siRNA. The cells were then cultured in 96-well plates for the indicated periods. The cell numbers were measured using a Cell Counting Kit-8. The data are the means of triplicate experiments, which generally varied by <10%. Similar results were obtained in three independent experiments. An aliquot of each TCL after siRNA transfection (day 3) was analyzed by immunoblotting (IB) with an anti-Brk, anti-STAP-2, anti-STAT3, or anti-actin antibody. Knockdown of each protein was confirmed until 6 days after siRNA transfection. C, T47D cells were transfected with a control siRNA, STAP2 siRNA (#1), STAT3 siRNA, or Brk siRNA. Total RNA samples isolated from these cells were also subjected to quantitative real time PCR analysis using SOOC3, CEBPD, CYCLIN D1, MYC, or ACTIN primers. The data represent the levels of mRNA normalized to that of an ACTIN internal control and are expressed relative to the value of control siRNA-treated samples. Shown is a representative experiment that was repeated at least three times with similar results.
DISCUSSION
The activation of STAT3 by Brk is a critical event during the process of Brk-induced proliferation in breast cancer cells. Our manipulation of STAP-2 expression demonstrated essential roles of STAP-2 in this process through complex interactions among Brk, STAP-2, and STAT3. In particular, experiments using STAP-2 deletion mutants indicated that the PH domain of STAP-2 is involved in multiple steps; the binding between Brk and STAP-2, the activation and tyrosine phosphorylation of STAT3, and the activation of Brk. These results suggest that STAP-2 cooperates with Brk to enhance breast cancer growth.
Brk is an intracellular tyrosine kinase that is distantly related to Src family tyrosine kinases (1, 23). Like Src family kinases, Brk is negatively regulated by phosphorylation of a C-terminal tyrosine residue (24). However, unlike Src family kinases, Brk is not myristoylated or specifically targeted to the membrane (1). Therefore, Brk may be localized to different cellular compartments, including the nucleus, where it might have a distinct set of substrates and interacting proteins. The finding that the PH domain of STAP-2 interacts with and activates Brk is very attractive, because the PH domain is required for STAP-2 translocation to the membrane after EGF stimulation (16). The PH domain of STAP-2 may function to achieve proper activation, as myristoylation does for Src. Indeed, a STAP-2 PH and Brk fusion protein on its own exhibited a robust kinase activity and tyrosine phosphorylation and activation of STAT3. Furthermore, a STAP-2 PH and Brk fusion protein was mainly localized in the nucleus, indicating that STAP-2 PH controls not only kinase activity of Brk but also localization of Brk.
Several Brk-interacting proteins or Brk substrates, such as the RNA-binding protein, Sam68, and the polypyrimidine tract-binding (PTB) protein-associated splicing factor (PSF), have been reported (24, 25). However, the molecular mechanism by which Brk cooperates with these proteins and participates in tumorigenesis remains poorly characterized. Therefore, the Brk/STAP-2 interactions described here may be helpful for breast cancer treatment, because the oncogenic potential of activated STAT3 in breast cancer is well known (26, 27).
With regard to STAT5, another target of Brk, STAP-2 also augmented Brk-mediated STAT5 activation in breast cancer cells.3 However, we previously reported that STAP-2 negatively regulated STAT5 activation in erythropoietin-, IL-2-, and IL-3-induced signaling (17). At the present time, we cannot explain the different effects of STAP-2 on STAT5 activation, although it is possible that unknown signal-specific or cell type-specific factors may influence the effects of STAP-2.
Brk promotes cell migration and tumor invasion by phosphorylating the focal adhesion protein paxillin, followed by activation of the small GTPase, Rac1, via CrkII (22). It has also been demonstrated that Brk regulates the small GTPases RhoA and Ras by phosphorylating p190RhoGAP-A, thereby promoting breast malignancy (28). These observations are interesting, because STAP-2 associates with Vav1, the guanine-nucleotide exchanging factor for Rac1, and activates Rac1 signaling during the stromal cell derived factor (SDF)-1α-induced chemotaxis of T cells (29).
Brk and STAP-2 are highly expressed in breast cancer cells, suggesting that this linkage plays an important role in the dysregulated activation of STAT3. Our work presented here has clarified the molecular mechanisms underlying Brk/STAP-2-mediated modification of STAT3 and will provide insights toward the development of a novel therapeutic strategy for breast cancers. Brk expression in breast cancer cells has been reported to indicate poor prognosis (30); however, synergistic effects of Brk and STAP-2 on STAT3 activation suggest that knowledge of Brk expression together with that of STAP-2 might give more valuable prognostic scores for the outcome of breast carcinomas than knowledge of Brk expression alone.
This work was supported in part by the Suhara Memorial Foundation and Grant-in-Aid for scientific research from Ministry of Education, Culture, Sports, Science and Technology of Japan.
O. Ikeda, A. Mizushima, Y. Sekine, C. Yamamoto, R. Muromoto, A. Nanbo, K. Oritani, A. Yoshimura, and T. Matsuda, unpublished observation.
- Brk
- breast tumor kinase
- SH
- Src homology
- PH
- pleckstrin homology
- PY
- phosphotyrosine
- TCL
- total cell lysate
- STAP-2
- signal transducing adaptor protein-2.
REFERENCES
- 1.Mitchell P. J., Barker K. T., Martindale J. E., Kamalati T., Lowe P. N., Page M. J., Gusterson B. A., Crompton M. R. (1994) Oncogene 9, 2383–2390 [PubMed] [Google Scholar]
- 2.Lee S. T., Strunk K. M., Spritz R. A. (1993) Oncogene 8, 3403–3410 [PubMed] [Google Scholar]
- 3.Siyanova E. Y., Serfas M. S., Mazo I. A., Tyner A. L. (1994) Oncogene 9, 2053–2057 [PubMed] [Google Scholar]
- 4.Easty D. J., Mitchell P. J., Patel K., Flørenes V. A., Spritz R. A., Bennett D. C. (1997) Int. J. Cancer 71, 1061–1065 [DOI] [PubMed] [Google Scholar]
- 5.Llor X., Serfas M. S., Bie W., Vasioukhin V., Polonskaia M., Derry J., Abbott C. M., Tyner A. L. (1999) Clin. Cancer Res. 5, 1767–1777 [PubMed] [Google Scholar]
- 6.Derry J. J., Prins G. S., Ray V., Tyner A. L. (2003) Oncogene 22, 4212–4220 [DOI] [PubMed] [Google Scholar]
- 7.Barker K. T., Jackson L. E., Crompton M. R. (1997) Oncogene 15, 799–805 [DOI] [PubMed] [Google Scholar]
- 8.Harvey A. J., Crompton M. R. (2003) Oncogene 22, 5006–5010 [DOI] [PubMed] [Google Scholar]
- 9.Kamalati T., Jolin H. E., Mitchell P. J., Barker K. T., Jackson L. E., Dean C. J., Page M. J., Gusterson B. A., Crompton M. R. (1996) J. Biol. Chem. 271, 30956–30963 [DOI] [PubMed] [Google Scholar]
- 10.Kamalati T., Jolin H. E., Fry M. J., Crompton M. R. (2000) Oncogene 19, 5471–5476 [DOI] [PubMed] [Google Scholar]
- 11.Ostrander J. H., Daniel A. R., Lofgren K., Kleer C. G., Lange C. A. (2007) Cancer Res. 67, 4199–4209 [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Gao Y., Qiu H., Miller W. T., Poli V., Reich N. C. (2006) Oncogene 25, 4904–4912 [DOI] [PubMed] [Google Scholar]
- 13.Weaver A. M., Silva C. M. (2007) Breast Cancer Res. 9, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell P. J., Sara E. A., Crompton M. R. (2000) Oncogene 19, 4273–4282 [DOI] [PubMed] [Google Scholar]
- 15.Ikeda O., Miyasaka Y., Sekine Y., Mizushima A., Muromoto R., Nanbo A., Yoshimura A., Matsuda T. (2009) Biochem. Biophys. Res. Commun. 384, 71–75 [DOI] [PubMed] [Google Scholar]
- 16.Minoguchi M., Minoguchi S., Aki D., Joo A., Yamamoto T., Yumioka T., Matsuda T., Yoshimura A. (2003) J. Biol. Chem. 278, 11182–11189 [DOI] [PubMed] [Google Scholar]
- 17.Sekine Y., Yamamoto T., Yumioka T., Sugiyama K., Tsuji S., Oritani K., Shimoda K., Minoguchi M., Yoshimura A., Matsuda T. (2005) J. Biol. Chem. 280, 8188–8196 [DOI] [PubMed] [Google Scholar]
- 18.Sekine Y., Ikeda O., Hayakawa Y., Tsuji S., Imoto S., Aoki N., Sugiyama K., Matsuda T. (2007) Oncogene 26, 6038–6049 [DOI] [PubMed] [Google Scholar]
- 19.Nakajima K., Yamanaka Y., Nakae K., Kojima H., Ichiba M., Kiuchi N., Kitaoka T., Fukada T., Hibi M., Hirano T. (1996) EMBO J. 15, 3651–3658 [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda O., Sekine Y., Yasui T., Oritani K., Sugiyma K., Muromoto R., Ohbayashi N., Yoshimura A., Matsuda T. (2008) Mol. Cell. Biol. 28, 5027–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda T., Feng J., Witthuhn B. A., Sekine Y., Ihle J. N. (2004) Biochem. Biophys. Res. Commun. 325, 586–594 [DOI] [PubMed] [Google Scholar]
- 22.Chen H. Y., Shen C. H., Tsai Y. T., Lin F. C., Huang Y. P., Chen R. H. (2004) Mol. Cell. Biol. 24, 10558–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell P. J., Barker K. T., Shipley J., Crompton M. R. (1997) Oncogene 15, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 24.Derry J. J., Richard S., Valderrama Carvajal H., Ye X., Vasioukhin V., Cochrane A. W., Chen T., Tyner A. L. (2000) Mol. Cell. Biol. 20, 6114–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukong K. E., Huot M. E., Richard S. (2009) Cell Signal. 21, 1415–1422 [DOI] [PubMed] [Google Scholar]
- 26.Bowman T., Garcia R., Turkson J., Jove R. (2000) Oncogene 19, 2474–2488 [DOI] [PubMed] [Google Scholar]
- 27.Bromberg J. (2000) Breast Cancer Res. 2, 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen C. H., Chen H. Y., Lin M. S., Li F. Y., Chang C. C., Kuo M. L., Settleman J., Chen R. H. (2008) Cancer Res. 68, 7779–7787 [DOI] [PubMed] [Google Scholar]
- 29.Sekine Y., Ikeda O., Tsuji S., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A., Matsuda T. (2009) J. Immunol. 183, 7966–7974 [DOI] [PubMed] [Google Scholar]
- 30.Aubele M., Walch A. K., Ludyga N., Braselmann H., Atkinson M. J., Luber B., Auer G., Tapio S., Cooke T., Bartlett J. M. (2008) Br. J. Cancer 99, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]