Abstract
Aminoacyl-tRNA synthetases (ARS) are modular enzymes that aminoacylate transfer RNAs (tRNA) for their use by the ribosome during protein synthesis. ARS are essential and universal components of the genetic code that were almost completely established before the appearance of the last common ancestor of all living species. This long evolutionary history explains the growing number of functions being discovered for ARS, and for ARS homologues, beyond their canonical role in gene translation. Here we present a previously uncharacterized paralogue of seryl-tRNA synthetase named SLIMP (seryl-tRNA synthetase-like insect mitochondrial protein). SLIMP is the result of a duplication of a mitochondrial seryl-tRNA synthetase (SRS) gene that took place in early metazoans and was fixed in Insecta. Here we show that SLIMP is localized in the mitochondria, where it carries out an essential function that is unrelated to the aminoacylation of tRNA. The knockdown of SLIMP by RNA interference (RNAi) causes a decrease in respiration capacity and an increase in mitochondrial mass in the form of aberrant mitochondria.
Keywords: Aminoacyl tRNA Synthetase, Drosophila Metabolism, Mitochondria, Oxidative Stress, Respiration
Introduction
Aminoacyl-tRNA synthetases (ARS)2 are an ancient family of enzymes with an essential role in protein synthesis. ARS catalyze the aminoacylation of transfer RNAs (tRNA) through a two-step catalytic reaction that first activates an amino acid with ATP to form an aminoacyl-adenylate and, second, to transfer the amino acid to the tRNA through the formation of an ester bond with the 3′-terminal ribose of the nucleic acid (1).
Numerous gene duplication events have generated a broad group of proteins generally referred to as ARS-like proteins. The majority of ARS-like proteins are molecules homologous with specific domains of ARS that may or may not carry out functions related to the aminoacylation of tRNA. In addition to ARS-like proteins, ARS domains may, by themselves, carry out additional biological functions after being excised from their original structure. In other cases, proteins that are structurally associated to ARS may also dissociate from these interactions to carry out a number of signaling functions (2).
The evolutionary analysis of ARS-like domains reveals that they have continued to emerge throughout evolution as the consequence of gene duplications, genomic reorganization, or lateral gene transfer events. The capacity of ARS genes to incorporate new functionalities through their structural reorganization is remarkable. This is probably due to the fact that the interaction with tRNA has driven ARS toward highly modular organizations that, secondarily, generate protein domains that are stable by themselves and can evolve to incorporate new cellular functions.
Seryl-tRNA synthetases (SRS) are the enzymes responsible for the serylation of tRNASer. SRS are dimeric enzymes that belong to the subclass IIa of ARS, together with alanyl-, prolyl-, threonyl-, glycyl-, and histidyl-tRNA synthetases. The structure of the SRS monomer comprises an active site domain and an N-terminal domain that generally folds into a long coiled-coil structure (3) that recognizes the long variable arm of tRNASer. The N-terminal coiled-coil domain of SRS is unique among ARS, but similar structures are common in other nucleic acid-binding proteins (4). Whether the N-terminal domain of SRS interacts with other nucleic acids is unknown, but it has been reported that SRS plays a role during the development of zebrafish (5–7). Thus, the coiled-coil domain of SRS may be carrying out functions other than tRNA recognition. The only known exception to this predominant SRS structure is found in methanogenic archaeal SRS, where the coiled-coil domain is replaced by a globular fold and a different type of tRNA recognition mechanism is used (8–10). In metazoans, SRS are among the few enzymes that remain duplicated in the cell; one isoform acts in the cytosol, and the second functions in the mitochondria, where it needs to recognize the highly diverged structures of mitochondrial tRNASer. However, the three-dimensional structure of mitochondrial SRS from Bos taurus reveals a similar structure to cytosolic enzymes, including a coiled-coil motif at its N terminus (11).
The mitochondrial tRNA serylation system is relevant in biomedical terms because mutations in human tRNASer have been described to cause several mitochondrial diseases. Mitochondrial diseases, such as mitochondrial myopathies, DEAF (maternally inherited deafness), or SNHL (sensorineural hearing loss), constitute a loose family of illnesses often caused by mutations in mitochondrial tRNAs (12). Similarly, some mutations in nuclear-encoded mitochondrial ARS have been related to mitochondrial diseases (13–15).
During the process of constructing a model for human disorders caused by mitochondrial tRNA aminoacylation deficiencies in Drosophila melanogaster, we realized that the genome of this species contains three genes coding for SRS homologous sequences. All three proteins encoded by these genes contain the canonical class II ARS motifs and share a significant level of sequence identity among each other. Here we show that one of these three genes codes for an SRS-like protein termed SLIMP (seryl-tRNA synthetase-like insect mitochondrial protein).
The gene coding for SLIMP is present in several invertebrates and, more specifically, in all available insect genomes. We determine that SLIMP is not a functional SRS, although it retains a significant affinity for tRNAs. We show that SLIMP is expressed in species of Diptera and Coleoptera and that, in flies, its expression is developmentally regulated and kept silent in early embryo stages.
SLIMP localizes to the mitochondria through a signal peptide that is processed upon translocation. The function of the protein is essential to D. melanogaster, and its knockdown by RNAi causes severe morphological defects in mitochondria. However, lethality can be strongly suppressed by supplementing the diet of the flies with known anti-oxidant molecules, suggesting that depletion of the protein causes oxidative stress in the affected animals.
EXPERIMENTAL PROCEDURES
Bioinformatics Analyses
The sequences used for the phylogenies were retrieved from UniProtKB (16), GenBankTM (17), of RefSeq (18) or found by BLAST searches (19) and were aligned using CLUSTAL_X (20). The N-terminal coiled-coil extensions were excluded from the analysis to focus on the putative catalytic domain of SLIMP. The phylogenetic analysis was performed by parsimony, distance, and maximum likelihood methods (using PHYLIP 3.63 (21) and PhyML (22)). The programs SEQBOOT and CONSENSE (21) were used to calculate the confidence limits of each cluster from 100 bootstrap replicates. tRNA sequences were retrieved from the Transfer RNA database (tRNAdb) (23). Subcellular localization and mitochondrial signal peptides were predicted using MitoProt (24) and iPSORT (25) software programs. Homology-based three-dimensional models were constructed using the Phyre server (26) and analyzed with PyMOL (27). The identification of SLIMP residues potentially involved in the interaction with seryl-adenylate was based on the analysis of multiple sequence alignments of SLIMP with canonical mitochondrial SRS sequences and by visual analysis of the three-dimensional SLIMP model overlapped with the B. taurus SRS2 structure (11).
Protein Cloning and Purification
SLIMP and DmSRS1 cDNAs (FlyBase ID CG31133 and CG17259, respectively) were obtained from the Drosophila Genomics Resource Center (DGRC). The cDNAs were cloned into pQE-70 vector (Qiagen) for E. coli expression and purification. The N-terminal putative mitochondrial signal peptide of SLIMP was deleted from the construction by PCR site-directed mutagenesis (Stratagene) (ΔNt SLIMP). Purification of ΔNt SLIMP was performed by nickel affinity standard chromatography. Protein concentration was determined by the Bradford protein assay (Bio-Rad).
Insect Stocks
UAS and GAL4 lines used in this study were: w; UAS-dicer-2, yw; actin5C-GAL4/TM6B, w; patched-GAL4, and the yw; nubbin-GAL4; UAS-dicer-2/CyO-TM6B that ensures co-segregation of nubbin-GAL4 with UAS-dcr2 (kindly provided by Dr. Andreu Casali, Institut de Biologia Molecular de Barcelona (IBMB)-CSIC/IRB). Tribolium castaneum and Blattella germanica were kindly provided by Dr. Xavier Bellés (CSIC-Universitat Pompeu Fabra (UPF)), Bombyx mori larvae were a kind gift from Gloria Sanahuja, and Lepisma sp. individuals were collected locally.
Generation of Transgenic UAS-RNAi Lines
A 566-bp fragment from SLIMP cDNA was subcloned into the pWIZ vector in an inverted repeat manner (28). Transgenic fly lines were obtained by microinjection of the construction into w1118 embryos using standard procedures (29). One homozygous line was obtained carrying the UAS-RNAi transgene (RNAiSLIMP strain 8). The same line containing a UAS-dicer2 transgene (RNAiSLIMP strain 8-dcr2) was also used. An extra RNAiSLIMP line was purchased from the Vienna Drosophila RNAi Center (ID 33774) (30). Transcription of RNAi fragments was based on the UAS-GAL4 system (31). Crosses with actin5C-GAL4 driver were maintained at 25 or 29 °C, and progeny was counted up to n > 220. For the anti-oxidant experiments, a mixture of anti-oxidant molecules in the form of the micronutrient supplement K-PAX (K-PAX Inc.) was added at a final concentration of 6.9 mg/ml, and progeny was counted up to n > 150. Crosses with the heterozygous actin5C-GAL4 strain are expected to produce progeny with an active RNAi transgene in 50% of the cases. The remaining progeny will not produce dsRNA and is used as an internal negative control. The proportion of SLIMP-silenced adults was calculated relative to the total progeny. Results were represented by setting the theoretical maximum viability of RNAi active flies at 100%. Statistical significance was determined by χ-square test.
Mitochondrial Separation
Mitochondrial and cytosolic fractions of Drosophila Schneider 2 cells (S2) were separated as described previously (32).
S2 Stable Transfection
Sequences coding for SLIMP or ΔNt SLIMP were subcloned into the pRmHa-3 vector for expression in S2 cells (33). S2 cells were maintained at 25 °C on Schneider's Drosophila medium (Lonza) supplemented with 10% FBS and l-Gln. 2–4 × 106 cells/ml were co-transfected with 30 μg of each pRmHa-3 construction and 500 ng of pHSP70PL vector containing a puromycin resistance cassette, by standard calcium phosphate method (34). Puromycin was used as selection agent.
S2 Induction and Immunostaining
SLIMP expression was induced during 72 h by the addition of 625 μm CuSO4 into log phase transfected S2 cells. Cells were inoculated on a plate with concanavalin-treated coverslips, incubated with 200 nm MitoTracker Red CMXRos (Invitrogen) for 30 min, fixed with 4% paraformaldehyde, incubated with the anti-His(C-term) antibody (Invitrogen) (1:200), Alexa Fluor® 488 anti-mouse secondary antibody (Invitrogen) (1:400), and 0.04 ng/μl DAPI. They were mounted on Mowiol and visualized in a Leica SPE confocal microscope.
Immunoblotting
Insect total protein extracts were obtained by pestle homogenization in 5× protein loading buffer (35). Samples were fractionated by 10% SDS-PAGE and transferred to PVDF membranes. Membranes were incubated with two polyclonal antibodies (Innovagen AB): the anti-Nt SLIMP against the putative mitochondrial peptide and the anti-SLIMP against an internal peptide. A polyclonal serum against Drosophila β-ATPase (36) (gift from Dr. Rafael Garesse, Instituto de Investigaciones Biomédicas-Universidad Autónoma de Madrid (IIB-UAM)) was used as mitochondrial marker and loading control.
RNA Extraction and Semiquantitative RT-PCR
w1118 larvae, pupae, adults, and embryos were subjected to total RNA extraction with TRIzol (Invitrogen). Samples were digested with DNase I and cleaned with the RNeasy MinElute cleanup kit (Qiagen). 1 μg of total RNA was retrotranscribed, and semiquantitative PCRs were performed using adjusted dilutions of the cDNAs and TaqDNA polymerase (Invitrogen). Oligodeoxynucleotide pairs were designed to amplify 200–300-bp cDNA regions for actin5C, which was used as a loading control, and for SLIMP. Reactions with non-retrotranscribed RNA samples were performed in parallel to discard genomic DNA contamination.
Wing Preparation and Microscopy Image Analyses
Flies were kept in 75% ethanol, 25% glycerol for >24 h. 12–20 wings were excised in cold PBS and mounted in Fauré's medium. Images were taken in a Nikon E600 microscope with an Olympus DP72 camera. L3-L4 areas were measured from males and females separately with the ImageJ software (37), and statistics were performed by a two-way analysis of variance test. Images from flies were taken at 30× with an MZ 16F Leica stereomicroscope equipped with a DFC 300FX camera.
ATP Photocross-linking
Reactions were performed following published protocols (38) with 2.4 μm pure DmSRS1, ΔNt SLIMP, BSA, or E. coli protein extracts. Radioactive signal was detected using a PhosphorImagerTM with Typhoon scanner control software.
Electrophoretic Gel Mobility Shift Assay (EMSA)
tRNAs were in vitro transcribed by standard methods (39), labeled with γ-32P using T4 polynucleotide kinase, and incubated for 20 min at 4 °C with 15 mm MgCl2, 15 mm KCl, 0.5 mm DTT, 10% (w/v) glycerol, 75 mm Tris buffer pH 7.0, 20 ng/μl oligo(dT)25, and 5 μm ΔNt SLIMP. Competition assays were performed, adding to the reaction 10× molar concentration of each non-radiolabeled D. melanogaster in vitro transcribed tRNASer, tRNAArg, or a heterologous tRNA (E. coli tRNALys (Sigma)). Reactions were separated by electrophoresis onto a 6% (w/v) polyacrylamide gel in 0.5× Tris-borate-EDTA. Signal was digitalized using a PhosphorImagerTM from a gel-exposed storage phosphor screen and was quantified using the ImageJ software (37).
Relative mtDNA Copy Number Quantification
mtDNA was quantified by real-time quantitative PCR using Power SYBR Green and a StepOnePlus real-time PCR system (Applied Biosystems). Genomic DNA templates from third instar larvae were amplified with a pair of primers to detect the mitochondrial gene ATPase6 (5′-CCCGCTATTCTTATACCTTTTATAGT-3′ and 5′-TGTCCAGCAATTATATTAGCAGTTA-3′) and the gene mRp110 used as nuclear gene standard (5′-TCGAACAGGCGGTGAAGAA-3′ and 5′-TGCAATGATTGGAGTGGAACA-3′). Primer sequences were kindly provided by Dr. Aurelio Teleman (German Cancer Research Center (DKFZ)). Standard curves were calculated for both primer pairs to ensure a high efficiency level. 20-μl reactions were prepared following the manufacturer's instructions, using carboxy-X-rhodamine (ROX) as reference dye and the following conditions: 50 °C for 2 min; 95 °C for 10 min; 40 cycles (95 °C for 15 s; 60 °C for 1 min). Results were statistically analyzed by Student's t test.
Electron Microscopy (EM)
Fat bodies were fixed in 2% glutaraldehyde in 0.1 m cacodylate buffer pH 7.2. Postfixation was performed with 2% OsO4 and 1.6% K3Fe(CN)6 in cacodylate buffer. Sections were contrasted with uranyl acetate to be later visualized in a Jeol JEM 1010 electron microscope. Mitochondrial density and areas were measured with the ImageJ software (37). Student's t test was calculated to determine significant differences.
Oxygen Consumption Measurements
4–8 larvae were dissected and permeabilized with 25 μm digitonin, and oxygen consumption was measured using an Oxygraph-2k (Oroboros) after serial addition of 10 mm glutamate and 2 mm malate, 2.5 mm ADP-Mg2+, 10 μm cytochrome c, 10 mm succinate, 0.5 μm steps of carbonyl cyanide p-trifluoromethoxyphenylhydrazone, 0.5 μm rotenone, and 2.5 μm antimycin A. Oxygen consumption was normalized, dividing the obtained values by the mitochondrial rate (from the relative mtDNA copy number (40) and the mitochondrial density determined by EM), considering the rate for control larvae as 1. Results were subjected to Student's t test.
RESULTS
SLIMP Is a Fast Evolving SRS Paralogue Universally Distributed in Insecta
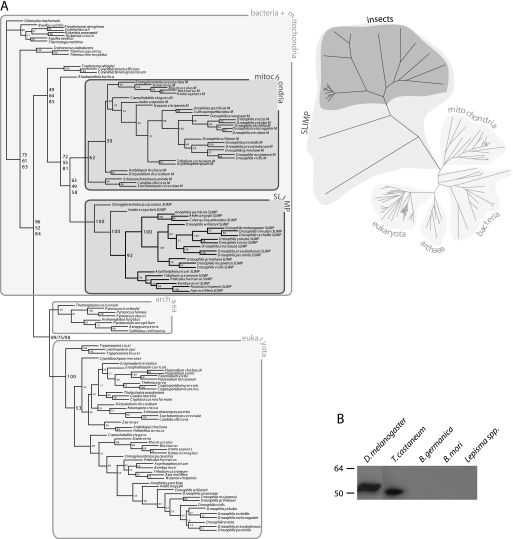
Analysis of the distribution of genes coding for SLIMP homologues reveals that they are present in all available insect genomes, as well as echinoderms and arachnids. The lack of genomic information prevents us from studying the distribution of the protein among arthropods, but it is apparent that SLIMP is universally distributed in insects. Phylogenetic analyses of SRS sequences show that SLIMP forms a fast evolving clade linked to mitochondrial SRS (Fig. 1A). This suggests that SLIMP is a descendant of a duplicated mitochondrial SRS gene that appeared early in the evolution of Eumetazoa and has persisted in all available insect genomes and in at least one species of sea urchins and one species of ticks.
FIGURE 1.
Phylogenetic analysis of SLIMP and its presence in different insect species. A, SLIMP and SRS phylogenetic trees. The left panel shows a maximum likelihood tree of SRS and SLIMP sequences. Bootstrap values from distance, parsimony, and maximum likelihood trees are shown in this order. All SLIMP proteins cluster together, sharing the same ancestor as mitochondrial SRS. A distance tree shown in the right panel indicates that the SLIMP clade evolved faster than SRS. B, the presence of the protein SLIMP in insect species detected by immunoblotting with an antibody against the N-terminal peptide of D. melanogaster SLIMP. The antibody detected the full-length protein in D. melanogaster (52.92 kDa) and was able to cross-react with a protein of similar size from T. castaneum (49.88 kDa).
To investigate whether SLIMP is produced in insects, we obtained crude extracts from adults of Lepisma spp. (silverfish, Thysanura), B. germanica (cockroach, Dictyoptera), B. mori (silkworm, Lepidoptera), T. castaneum (red flour beetle, Coleoptera), and D. melanogaster (fruit fly, Diptera). Fig. 1B shows that a positive signal, consistent with the predicted molecular weight of the protein in these species, could be detected in extracts of D. melanogaster and T. castaneum, indicating that genes coding for SLIMP are likely translated into proteins. No signal was observed in the rest of species tested. However, some of these species possess transcribable genes coding for SLIMP homologues (vide supra).
SLIMP Is a tRNA-binding Protein without tRNA Aminoacylation Activity
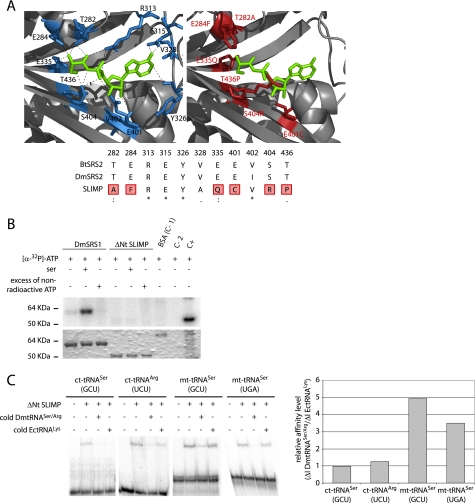
To determine whether SLIMP is a functional SRS, we analyzed it in silico and biochemically. First, we constructed a three-dimensional model of SLIMP to study whether the residues that shape the catalytic cavity in bona fide SRS were conserved in SLIMP. Fig. 2A shows that 6 of the 11 amino acids responsible for the recognition of the seryl-adenylate in B. taurus mitochondrial SRS are not conserved in SLIMP. Moreover, the mutated positions contain residues that are physically incompatible with the interactions established between serine, ATP, and SRS. In contrast, these positions are perfectly conserved in the canonical mitochondrial SRS from D. melanogaster (DmSRS2). Thus, computational models predict that SLIMP does not possess a typical SRS active site.
FIGURE 2.
SLIMP structural analyses. A, active site structural analysis. B. taurus mitochondrial SRS structure (1WLE, left) is shown with the seryl-adenylate in green. Residues that contact the intermediate are depicted in blue. The SLIMP catalytic core three-dimensional model is shown to the right. The residues in red correspond to non-conserved residues that would disrupt the interaction between SRS substrates and SLIMP. The alignment shows that all the interacting residues from B. taurus are conserved in DmSRS2, whereas 6 over 11 of these are not conserved in SLIMP. B, ATP photocross-linking assay. ATP binding ability was assessed for DmSRS1 and ΔNt SLIMP. The upper panel shows the signal from [α-32P]ATP bound to the tested proteins, and the lower panel shows the Coomassie Blue-stained SDS-PAGE. Serine was added in some reactions to test whether it would increase the affinity of SLIMP for ATP. An excess of non-radioactive ATP was used to ensure the specificity of the interactions. DmSRS1 binds ATP specifically, but ΔNt SLIMP does not. Controls are BSA (negative control (C-1)), protein extracts (positive control (C+)), and buffer (negative control (C-2)). C, tRNA binding by ΔNt SLIMP measured by EMSA. The two mitochondrial tRNASer isoacceptors and two cytosolic tRNAs (tRNASer (GCU) and tRNAArg (UCU)) from D. melanogaster were tested. Competition assays were performed adding non-radiolabeled DmtRNASer/Arg or a heterologous tRNA (tRNALys from E. coli). Affinity values are represented as ΔI DmtRNASer/Arg/ΔI EctRNALys, where ΔI DmtRNASer/Arg is (signal intensity when adding radiolabeled DmtRNASer/Arg − signal intensity when adding 10× non-radiolabeled DmtRNASer/Arg), and ΔI EctRNALys is (signal intensity when adding radiolabeled DmtRNASer/Arg − intensity when adding 10× non-radiolabeled EctRNALys). Ratios ≤1 indicate nonspecific interactions. The values obtained for the two mitochondrial DmtRNASer (GCU and UGA) tested are 4.95 and 3.50, respectively, whereas the values for the cytosolic DmtRNASer (GCU) and DmtRNAArg (UCU) are similar to 1 (0.98 and 1.26, respectively).
We then tested whether SLIMP can aminoacylate tRNASer with serine. Pure SLIMP neither had detectable tRNASer aminoacylation activity, nor was it capable of catalyzing the first step of the aminoacylation reaction using a mixture of the 20 amino acids from the canonical genetic code as a substrate (supplemental Experimental Procedures and Fig. S1). We then performed ATP binding assays, which were also negative for SLIMP (Fig. 2B). SLIMP is not able to bind any of the initial substrates required for the serylation of tRNA by SRS.
SLIMP has a predicted N-terminal coiled-coil as canonical SRS. We analyzed whether the protein was able to bind tRNAs by performing electrophoretic gel mobility shift assays. To discard unspecific interactions, competition assays with unlabeled tRNAs were also done. Surprisingly, SLIMP is able to specifically bind the two mitochondrial tRNASer isoacceptors (UGA and GCU) with high affinity (Fig. 2C), whereas it exhibits low affinity for the cytosolic tRNASer (GCU) and tRNAArg (UCU).
SLIMP Is a Mitochondrial Protein Whose Translation Is Developmentally Regulated
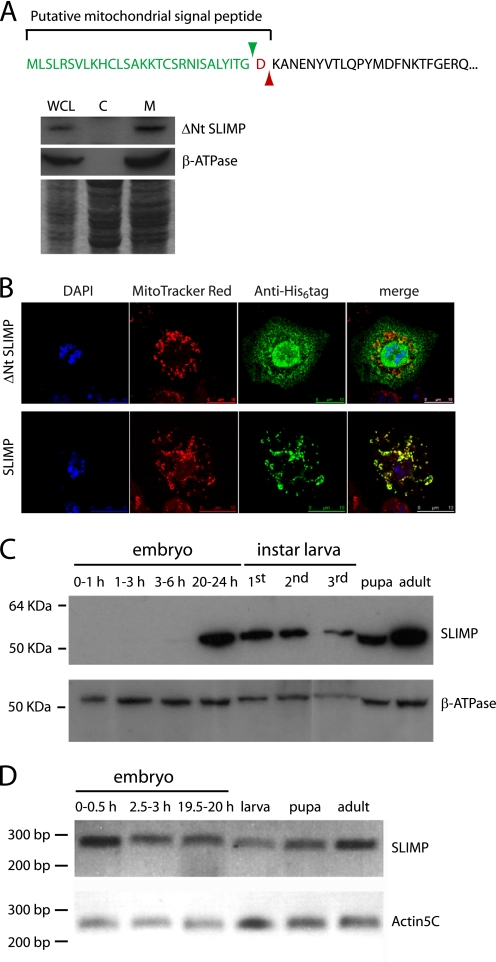
We determined the subcellular localization of SLIMP and its synthesis throughout the D. melanogaster life cycle. Prediction algorithms assign a high probability of mitochondrial localization to the protein (93.96% by MitoProt), a fact consistent with its phylogenetic relationship with bona fide mitochondrial SRS. The MitoProt and iPSORT software programs predict the presence of a 30–31-amino acid N-terminal signal peptide (Fig. 3A). We performed S2 cellular fractionation experiments to determine the presence of SLIMP in mitochondrial or cytosolic fractions. As expected, only the mitochondrial fractions contain mature SLIMP as detected by immunoblotting (Fig. 3A). Moreover, when immunostainings were performed on S2 cells expressing the full-length SLIMP, which is led by the mitochondrial signal peptide, the protein co-localized with the mitochondrial marker. In contrast, when S2 cells expressed SLIMP lacking the mitochondrial putative signal peptide (ΔNt SLIMP), the protein was not imported to the organelle. Therefore, SLIMP is a protein with a mitochondrial localization that depends on a 30-amino acid-long signal peptide (Fig. 3B).
FIGURE 3.
SLIMP subcellular localization and expression pattern. A, cellular fractionation. Immunoblot of S2 cellular fractions (WCL, whole cell lysate; C, cytoplasmic fraction; M, mitochondrial fraction) was performed using an anti-SLIMP antibody. Mature SLIMP is found exclusively in the mitochondrial fraction. The predicted mitochondrial targeting sequence and processing site are displayed. B, S2 cells immunostaining. Cells expressing the full-length or the ΔNt version of SLIMP with a His6 tag were detected with an anti-His antibody. ΔNt SLIMP does not localize to the mitochondria, whereas the full-length protein is transported to mitochondria by the signal peptide. MitoTracker Red was used to label mitochondria, and DAPI was used to mark nuclei. C, immunoblot of SLIMP in samples of different developmental stages. The full-length protein is expressed from late embryonic stages (20–24-h embryos) onward. The antibody against β-ATPase was used as mitochondrial loading control. D, SLIMP mRNA levels during the life cycle. SLIMP mRNA levels, determined by semiquantitative RT-PCR, are maintained throughout the life cycle. The actin5C mRNA was used to normalize the mRNA levels.
The presence of the protein in different developmental stages was determined by immunoblot analysis of protein extracts. Fig. 3C shows that SLIMP is synthesized at late embryonic stages and remains present throughout the rest of the cycle. However, SLIMP mRNA can be detected at all stages of development (Fig. 3D), including early embryo phases. Thus, SLIMP expression seems to be temporally regulated at the translational level, a common process during the Drosophila life cycle (41). This suggests again that SLIMP is not functioning as a canonical aminoacyl-tRNA synthetase.
SLIMP Is an Essential Protein
We tested the functional importance of SLIMP in flies silencing its expression by means of RNA interference (RNAi) using the UAS-GAL4 system. The RNAiSLIMP strain 8 was crossed with different GAL4 drivers to induce the RNAi in the progeny in a constitutive ubiquitous (actin5C-GAL4 driver), or more restricted (patched-GAL4 and nubbin-GAL4; UAS-dcr2 drivers) expression pattern. To intensify the RNAi effect, another strain containing an extra copy of the dicer-2 gene was used (RNAiSLIMP strain 8-dcr2). To account for potential heterogeneity in the experiments, we also used an available strain from the Vienna Drosophila RNAi Center (VDRC) (RNAiSLIMP strain 33774) that silences the expression of the same gene by targeting another region of the mRNA. The two different RNAi sequences were able to significantly decrease SLIMP mRNA levels (supplemental Experimental Procedures and Fig. S2).
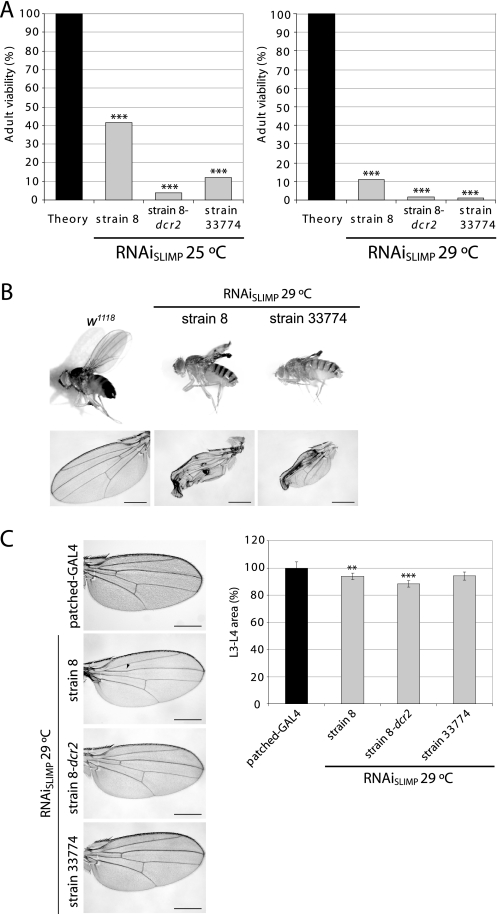
The constitutive ubiquitous expression of the RNAi of SLIMP was strongly deleterious to the development of the animals and significantly reduced their viability (Fig. 4A). The number of flies emerging at 25 °C ranged from 12% (RNAiSLIMP strain 33774) to 41.35% (RNAiSLIMP strain 8). The milder effect observed when using RNAiSLIMP strain 8 was intensified by the presence of dicer-2 gene (RNAiSLIMP strain 8-dcr2, 3.90% viability). Because the efficiency of the UAS-GAL4 system is temperature-dependent, the effect could be strengthened by growing the animals at 29 °C. In these conditions, adult viability was reduced for RNAiSLIMP strain 8 (41.35–11.25%), RNAiSLIMP strain 33774 (12–0.87%), and RNAiSLIMP strain 8-dcr2 (3.9–1.8%). Although variability between RNAi lines was observed, the lethality caused by the knockdown of SLIMP was significant in all cases.
FIGURE 4.
In vivo effect of SLIMP knockdown by RNA interference. A, constitutive and ubiquitous knockdown of SLIMP causes a decrease in adult viability at 25 °C and at 29 °C (***, p < 0.001). B, wing-restricted SLIMP depletion. RNAiSLIMP strains crossed at 29 °C with the nubbin-GAL4; UAS-dcr2 driver show severe tissue damage. Scale bars correspond to 500 μm. C, SLIMP silencing in the L3-L4 wing area. RNAiSLIMP strains are crossed with the patched-GAL4 driver that restricts the RNAi expression in the region flanked by longitudinal veins L3 and L4. Some individuals present a partial or total loss of the anterior cross vein (marked with an arrowhead). Graphs show the averages of all the measurements in percentage with S.E. when compared with the paternal line (**, p < 0.01; ***, p < 0.001). Scale bars correspond to 500 μm.
Tissue-restricted silencing showed that SLIMP is also essential for the correct development of adult structures. Fig. 4, B and C, show the effect of SLIMP depletion in the wing tissue by using two different GAL4 strains. In Fig. 4B, the RNAi was expressed in the larval wing disc zone that gives rise to the adult wing blade using the nubbin-GAL4 driver. Although the overall structure of the wing is not compromised, the tissue fails to unfold and collapses. This suggests that the damage caused by total or partial ablation of SLIMP is accumulative as an early induction of RNAi expression does not impair early stages of wing development.
Fig. 4C shows the effect of SLIMP silencing in the region flanked by longitudinal veins L3 and L4 in the adult wing, using a patched-GAL4 driver. The phenotype observed was characterized by a narrowing of this area in all the strains and a reduction or elimination of the anterior cross vein in some of the wings. This effect was cell-autonomous as adjacent regions to the silenced area were not affected (data not shown). The reduction in the L3-L4 area was caused by a decrease in the number of cells, and not by a reduction of cell size, as the cell density in the affected zone is not significantly different from the one in control wings (data not shown).
SLIMP Ablation Causes Oxidative Stress
We reasoned that the reduction in SLIMP could be affecting mitochondrial metabolic functions. It is known that mitochondrial dysfunction can cause an increase of reactive oxygen species and, as a result, oxidative stress. To test this hypothesis, we tested whether an increase in anti-oxidant molecules in the diet would be able to reduce the deleterious effect of SLIMP ablation by RNAi.
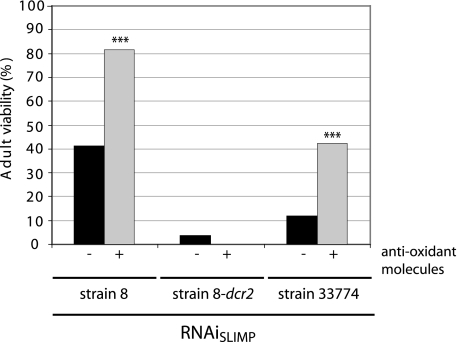
The anti-oxidant mix used contains, among others, well characterized scavengers such as selenium, α-lipoic acid, acetyl l-carnitine, and N-acetyl l-cysteine. As can be seen in Fig. 5, the addition of this anti-oxidant mix was capable of significantly improving the survival of flies despite depletion of SLIMP in those RNAi strains that already had the mildest effect (RNAiSLIMP strain 8 and strain 33774). This is reflected in a higher proportion of SLIMP-deprived flies reaching adulthood when compared with the proportion of viable flies grown without the anti-oxidant mix. For RNAiSLIMP strain 8, the viability increased from 41.35 to 81.52%, which indicates almost a complete recovery in the lethality produced by the RNAi, and for RNAiSLIMP strain 33774, it increased from 12 to 42.16%.
FIGURE 5.
Recovery of viability by anti-oxidant compounds. The addition of an anti-oxidant mixture (K-PAX) to the medium of the fly causes a partial recovery in adult viability. Actin5C-GAL4 driver and the RNAiSLIMP lines were crossed in parallel in the presence or absence of the anti-oxidant mixture at 25 °C. The proportion of viable adults carrying the RNAiSLIMP with regard to the total progeny increased for RNAiSLIMP strain 8 and RNAiSLIMP strain 33774 (***, p < 0.001).
SLIMP Silencing Affects Mitochondrial Morphology and Function
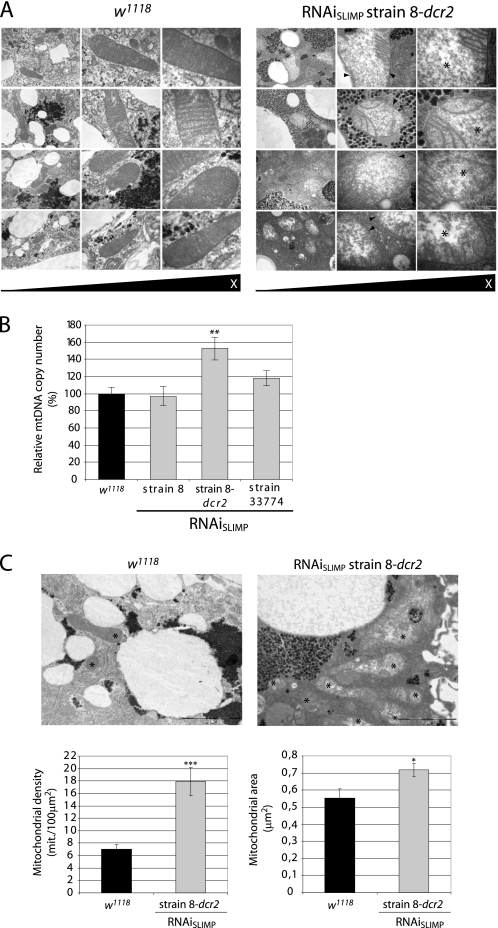
Larval fat bodies from RNAiSLIMP strain 8-dcr2 crossed at 29 °C with the actin5C-GAL4 were analyzed by electron microscopy. Mitochondrial morphology was strongly affected by SLIMP depletion (Fig. 6A). Mitochondrial inner membranes showed local or complete loss of cristae structures, and mitochondrial matrixes appeared swollen and less electron-dense than control samples.
FIGURE 6.
SLIMP knockdown affects mitochondrial morphology. A, electron microscopy images taken from fat bodies. RNAiSLIMP mitochondria show swollen matrixes with low electron density (marked with asterisks) and a partial or total loss of cristae structures (marked with arrowheads). Scale bars correspond to 1 μm, 500 nm, and 200 nm, respectively. B, relative mtDNA copy number determination. Relative mtDNA copy numbers in control larvae are expressed as 100%. Larvae from strains RNAiSLIMP 8-dcr2 and 33774 at 29 °C show an increase in the mtDNA copy number, indicative of an increase in mitochondria number. Columns represent the average of five replicates with S.E. (**, p < 0.01). C, mitochondrial density and area. EM images from larval fat bodies taken at 20,000× were used to analyze the mitochondrial density (mitochondria/100 μm2) and the mitochondrial area (μm2). Both measurements were taken from control and RNAiSLIMP strain 8-dcr2 at 29 °C (*, p < 0.05; ***, p < 0.001). Consistent with the relative mtDNA copy number, the density of mitochondria was higher in RNAi-treated tissues.
To determine whether mitochondrial density was also altered by SLIMP silencing, relative genomic mtDNA copy numbers were calculated by quantitative PCR. The relative amount of mtDNA in larvae with silenced SLIMP increased proportionally to the degree of lethality caused by the silencing (Fig. 6B), suggesting that silencing triggered a gradual compensatory mechanism that increased mitochondrial number. This result was confirmed by comparing mitochondrial density in EM images of control and treated larval fat bodies. Mitochondrial density is 7.01 mitochondria/100 μm2 in control larvae, and 17.89 mitochondria/100 μm2 in treated larvae, an increase of a 155.2% (Fig. 6C). The average mitochondrial size was also significantly higher in SLIMP-silenced fat bodies (Fig. 6C).
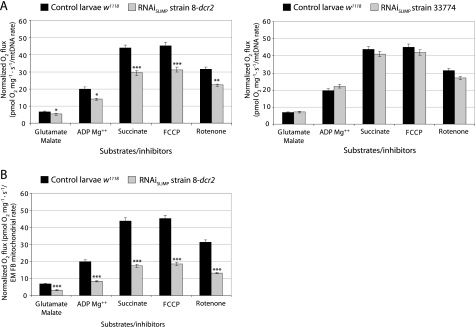
We then tested whether mitochondrial function was compromised in SLIMP-silenced flies. We analyzed respiratory activity by monitoring oxygen consumption of larval tissues. The respiratory control ratio (oxygen uptake in the presence of ADP divided by that in the absence of ADP) in knockdown larvae does not show a significant decrease, indicating that the respiratory chain is correctly coupled to oxidative phosphorylation (Fig. 7). Respiratory values of SLIMP-silenced larvae normalized by mitochondrial density (inferred from the relative mtDNA copy numbers (Fig. 7A) and from electron micrograph analyses (Fig. 7B)) were considerably reduced in animals with the highest silencing defects (RNAiSLIMP strain 8-dcr2) but were normal in larvae with a milder silencing effect (RNAiSLIMP strain 33774).
FIGURE 7.
SLIMP RNAi compromises mitochondrial respiration. The mitochondrial oxygen consumption profile from larvae under constitutive induction of RNAiSLIMP is displayed. Oxygen consumption values normalized by mitochondrial density show a decrease in the mitochondrial respiration for RNAiSLIMP strain 8-dcr2. All graphs give the average of consumed oxygen with S.E. (*, p < 0.05; **, p < 0.01, ***, p < 0.001). A, graphs normalized according to the mtDNA relative copy number. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. B, graph normalized by number of mitochondria in fat bodies (FB) determined by EM.
To summarize, SLIMP depletion severely affects mitochondrial structure, biogenesis, and function and probably triggers a compensatory response to maintain mitochondrial function by means of increasing the number and size of existing mitochondria.
DISCUSSION
The extended evolutionary history of ARS has generated a large number of related proteins that perform other cellular functions. Most of the known ARS paralogues have wide phylogenetic distributions that indicate an ancient origin, and most are relatively small polypeptides that only share homology with one domain of their related ARS. SLIMP is unusual in that it is the result of a relatively recent gene duplication (at the base of the metazoan branches) of an SRS gene and retains significant sequence identity with the complete sequence of its paralogous SRS.
The fast evolutionary rate experienced by SLIMP genes (Fig. 1A) is in contrast to the conservation displayed by SRS and suggests that the function of SLIMP in insects is not under the same functional constraints experienced by ARS. The conservation of the gene across the class Insecta, however, indicates that the function of the protein is important in these animals.
The sequence identity that SLIMP shares with SRS allows us to predict that this protein has an identical fold to a typical seryl-tRNA synthetase. Indeed, computer algorithms strongly predict the presence of a coiled-coil structure in the N terminus of SLIMP, as found in most cytosolic and mitochondrial SRS. However, the function of SLIMP is unrelated to SRS, as suggested by its active site architecture and its inability to charge tRNASer or activate serine. The lack of binding affinity of the protein for ATP prevents this protein from catalyzing the aminoacylation of tRNA and, presumably, activating any standard or non-standard amino acid. On the other hand, the coiled-coil structure that SLIMP shares with SRS is also present in DNA-binding proteins, raising the possibility that the function of SLIMP may involve the binding of nucleic acids.
The significance of the specific binding of mitochondrial tRNASer by SLIMP is unclear at the moment. A possibility is that despite its functional evolution, SLIMP has maintained an affinity for the peculiar conformation of mitochondrial tRNASer. Supporting this idea is the fact that the serylated amounts of mitochondrial tRNASer of SLIMP-deprived larvae did not show a significant deviation from control animals (supplemental Experimental Procedures and Fig. S3). Gel filtration chromatography data also indicate that SLIMP retains the dimeric structure characteristic of SRS (supplemental Experimental Procedures and Fig. S4). Thus, the affinity of SLIMP by tRNASer may simply be a reflection of the evolutionary origin of the protein.
It is also unclear whether SLIMP homologues found in echinoderms and arachnids are functionally related to SLIMP from insects. It is possible that a duplicated SRS acquired a new function related to idiosyncratic features of invertebrate mitochondria early in the evolution of these animals. Indeed, SLIMP may represent an alternative to a mitochondrial function that is performed by other molecules in other metazoans. Determining the evolutionary history of SLIMP and its exact function in invertebrates will probably provide new information on the function and evolution of metazoan mitochondria.
Although the actual role of SLIMP remains unknown, we have determined that it is essential in D. melanogaster. The depletion of the protein severely reduces the successful completion of tissue development and, locally, causes gross deformation of the organs affected. The fact that anti-oxidant molecules can alleviate the effect of SLIMP depletion is suggestive of a role of the protein in mitochondrial metabolism. Excessive generation of reactive oxygen species by mitochondria takes place when pathological conditions affect the components of the respiratory chain. These defects lead to the generation of reactive electrons that, combined with oxygen, result in oxidative stress. An increase in mtDNA copy number suggests that the mitochondrial dysfunction caused by SLIMP silencing may be compensated by a reactive oxygen species-mediated increase in mitochondrial biogenesis, as it has been reported previously in mammals (42, 43). SLIMP has no detectable sequence identity with components of the respiratory chain. Thus, although we cannot rule out a function of SLIMP in the electron transport chain, we favor the hypothesis that SLIMP may be important for mitochondrial DNA replication or mitochondria biogenesis. This situation would be comparable with the glycyl-tRNA synthetase (GRS) paralogue polymerase γ accessory subunit (44, 45).
Interestingly, deprivation of SLIMP has been recently identified in a genetic mosaic screen as a compensator of the cell growth arrest phenotype caused in Drosophila cells by the inactivation of transcription factor dE2F1 (46). Although the significance of this observation is still unclear, it suggests to us that the mitochondrial role of SLIMP, or a product of its function, may be acting as a signal between mitochondria and nuclear transcription factors that regulate cell proliferation.
In summary, SLIMP represents a new type of ARS-like protein that has acquired an essential function in insects despite a relatively modest divergence from a canonical SRS structure. This represents a new demonstration of the ability of SRS-related polypeptides to rapidly adopt other biological roles and joins the instances of mammalian ARS and ARS-related domains that carry out secondary regulatory roles in these species.
Acknowledgments
We are grateful to Dr. Andreu Casali (IBMB-CSIC/IRB) for providing fly stocks, to Dr. Xavier Bellés (CSIC-UPF) for supplying individuals of several insect species, and to Dr. Magali Frugier (Centre National de la Recherche Scientifique (CNRS)) for the help provided with the EMSA techniques. We thank Dr. Antonio Zorzano (IRB) for helpful discussions regarding respirometry assays. We are grateful to Dr. Jon D. Kaiser (K-PAX Inc.) for providing the anti-oxidant mixture used in this work.
This work was supported by Grant BIO2006-01558 from the Spanish Ministry of Science and Education.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S4.
- ARS
- aminoacyl-tRNA synthetase(s)
- SRS
- seryl-tRNA synthetase(s)
- SLIMP
- seryl-tRNA synthetase-like insect mitochondrial protein
- Nt
- N terminus.
REFERENCES
- 1.Ibba M., Soll D. (2000) Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2.Park S. G., Schimmel P., Kim S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. (1990) Nature 347, 249–255 [DOI] [PubMed] [Google Scholar]
- 4.Pabo C. O., Sauer R. T. (1984) Ann. Rev. Biochem. 53, 293–321 [DOI] [PubMed] [Google Scholar]
- 5.Amsterdam A., Nissen R. M., Sun Z., Swindell E. C., Farrington S., Hopkins N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzog W., Müller K., Huisken J., Stainier D. Y. (2009) Circ. Res. 104, 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukui H., Hanaoka R., Kawahara A. (2009) Circ. Res. 104, 1253–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilokapic S., Korencic D., Söll D., Weygand-Durasevic I. (2004) Eur. J. Biochem. 271, 694–702 [DOI] [PubMed] [Google Scholar]
- 9.Korencic D., Polycarpo C., Weygand-Durasevic I., Söll D. (2004) J. Biol. Chem. 279, 48780–48786 [DOI] [PubMed] [Google Scholar]
- 10.Jaric J., Bilokapic S., Lesjak S., Crnkovic A., Ban N., Weygand-Durasevic I. (2009) J. Biol. Chem. 284, 30643–30651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chimnaronk S., Gravers Jeppesen M., Suzuki T., Nyborg J., Watanabe K. (2005) EMBO J. 24, 3369–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-Pesini E., Lott M. T., Procaccio V., Poole J. C., Brandon M. C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D. C. (2007) Nucleic Acids Res. 35, D823–D828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheper G. C., van der Klok T., van Andel R. J., van Berkel C. G., Sissler M., Smet J., Muravina T. I., Serkov S. V., Uziel G., Bugiani M., Schiffmann R., Krägeloh-Mann I., Smeitink J. A., Florentz C., Van Coster R., Pronk J. C., van der Knaap M. S. (2007) Nat. Genet. 39, 534–539 [DOI] [PubMed] [Google Scholar]
- 14.Edvardson S., Shaag A., Kolesnikova O., Gomori J. M., Tarassov I., Einbinder T., Saada A., Elpeleg O. (2007) Am. J. Hum. Genet. 81, 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley L. G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S. C., Thorburn D., Ryan M. T., Giegé R., Bahlo M., Christodoulou J. (2010) Am. J. Hum. Genet. 87, 52–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apwiler R., Martin M., O'Donovan C., Magrane M., Alam-Faruque Y., Antunes R., Barrell D., Bely B., Bingley M., Binns D., Bower L., Browne P., Chan W., Dimmer E., Eberhardt R., Fedotov A., Foulger R., Garavelli J., Huntley R., Jacobsen J., Kleen M., Laiho K., Leinonen R., Legge D., Lin Q., Liu W., Luo J. (2010) Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2009) Nucleic Acids Res. 37, D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt K. D., Tatusova T., Maglott D. R. (2007) Nucleic Acids Res. 35, D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 20.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. (1988) Annu. Rev. Genet. 22, 521–565 [DOI] [PubMed] [Google Scholar]
- 22.Guindon S., Gascuel O. (2003) Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 23.Jühling F., Mörl M., Hartmann R. K., Sprinzl M., Stadler P. F., Pütz J. (2009) Nucleic Acids Res. 37, D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 25.Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002) Bioinformatics 18, 298–305 [DOI] [PubMed] [Google Scholar]
- 26.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 28.Lee Y. S., Carthew R. W. (2003) Methods 30, 322–329 [DOI] [PubMed] [Google Scholar]
- 29.Rubin G. M., Spradling A. C. (1982) Science 218, 348–353 [DOI] [PubMed] [Google Scholar]
- 30.Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 31.Brand A. H., Perrimon N. (1993) Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 32.Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., Wang X. (1997) Science 275, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 33.Schneider I. (1972) J. Embryol. Exp. Morphol. 27, 353–365 [PubMed] [Google Scholar]
- 34.Di Nocera P. P., Dawid I. B. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7095–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Vol. 3, appendix A1.20, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Peña P., Garesse R. (1993) Biochem. Biophys. Res. Commun. 195, 785–791 [DOI] [PubMed] [Google Scholar]
- 37.Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 38.Ludlam A. V., McNatt M. W., Carr K. M., Kaguni J. M. (2001) J. Biol. Chem. 276, 27345–27353 [DOI] [PubMed] [Google Scholar]
- 39.Geslain R., Aeby E., Guitart T., Jones T. E., Castro de Moura M., Charrière F., Schneider A., Ribas de Pouplana L. (2006) J. Biol. Chem. 281, 38217–38225 [DOI] [PubMed] [Google Scholar]
- 40.Boushel R., Gnaiger E., Schjerling P., Skovbro M., Kraunsøe R., Dela F. (2007) Diabetologia 50, 790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm J. E., Smibert C. A. (2005) Biol. Cell 97, 235–252 [DOI] [PubMed] [Google Scholar]
- 42.Lee H. C., Wei Y. H. (2005) Int. J. Biochem. Cell Biol. 37, 822–834 [DOI] [PubMed] [Google Scholar]
- 43.Moreno-Loshuertos R., Acín-Pérez R., Fernández-Silva P., Movilla N., Pérez-Martos A., Rodriguez de Cordoba S., Gallardo M. E., Enríquez J. A. (2006) Nat. Genet. 38, 1261–1268 [DOI] [PubMed] [Google Scholar]
- 44.Carrodeguas J. A., Kobayashi R., Lim S. E., Copeland W. C., Bogenhagen D. F. (1999) Mol. Cell. Biol. 19, 4039–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrodeguas J. A., Theis K., Bogenhagen D. F., Kisker C. (2001) Mol. Cell 7, 43–54 [DOI] [PubMed] [Google Scholar]
- 46.Ambrus A. M., Rasheva V. I., Nicolay B. N., Frolov M. V. (2009) Genetics 183, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]