Abstract
During co-translational folding, the nascent polypeptide chain is extruded sequentially from the ribosome exit tunnel and, under severe conformational constraints, is dictated by its one-dimensional geometry. How do such vectorial constraints impact the folding pathway? Here, we combine single-molecule atomic force spectroscopy and steered molecular dynamics simulations to examine protein folding in the presence of one-dimensional constraints that are similar to those imposed on the nascent polypeptide chain. The simulations exquisitely reproduced the experimental unfolding and refolding force extension relationships and led to the full reconstruction of the vectorial folding pathway of a large polypeptide, the 253-residue consensus ankyrin repeat protein, NI6C. We show that fully stretched and then relaxed NI6C starts folding by the formation of local secondary structures, followed by the nucleation of three N-terminal repeats. This rate-limiting step is then followed by the vectorial and sequential folding of the remaining repeats. However, after partial unfolding, when allowed to refold, the C-terminal repeats successively regain structures without any nucleation step by using the intact N-terminal repeats as a template. These results suggest a pathway for the co-translational folding of repeat proteins and have implications for mechanotransduction.
Keywords: Atomic Force Microscopy, Computation, Computer Modeling, Protein Folding, Single-molecule Biophysics
Introduction
Significant progress has been made toward understanding the protein folding problem (1–5) through in vitro experiments on individual proteins (6–21) and their ensembles (22–24) and through computer simulations (25–36). However, much less is known about in vivo folding (37–39). Following protein synthesis, the nascent polypeptide chain (NPC)4 is extruded through the long (∼80 Å) and narrow (10–20 Å) ribosome exit tunnel in which the NPC starts its folding process (40). This co-translational folding has a strong vectorial character (39, 41). Recent studies using single-molecule techniques and NMR spectroscopy have captured interesting structural features in NPC synthesis and folding on the ribosome (40–47). As recently suggested by Cabrita et al. (41), the vectorial character of co-translational folding is in a way mimicked by force-induced unfolding experiments. Such mechanical experiments can be carried out, for example, in an atomic force microscope (AFM) (48–54), with optical tweezers (10,11), or by translocating proteins through a pore (55–57). These processes have been extensively modeled in computer simulations (30–36, 58–62). During mechanical unfolding and refolding in AFM, the N and C termini of the polypeptide chain are constrained to the pulling axis, which limits the conformational space of the chain in a vectorial fashion. Here, we used a combination of AFM-based single-molecule force spectroscopy (48–54) and steered molecular dynamics (SMD) (63–65) simulations to examine in detail the folding of NI6C, a consensus ankyrin repeat (AR) protein, under such vectorial constraints.
NI6C, which is composed of 253 amino acids, is organized into six identical internal repeats and two capping repeats (66). It was chosen as our model system because of (i) the fact that ARs are very common and have been identified in over 4700 proteins (25); (ii) its extended “vectorial” structure; (iii) its composition consisting of tandem repeats of nearly identical sequences, which should simplify the analysis of force spectroscopy data; (iv) expected robust refolding forces that can be captured by AFM (67–69); and (v) its extreme thermodynamic stability (66) that makes mechanical stretching and relaxing the only practical experimental approach to induce and follow the repeats' unfolding and refolding.
MATERIALS AND METHODS
DNA Cloning and Protein Expression
The gene sequence of NI6C (66) was synthesized by GenScript (Piscataway, NJ). The NI6C gene was inserted into the poly(I27) pRSETa vector (a kind gift from Jane Clarke, Ref. 70) using KpnI and NheI restriction sites, and the stop codon was added before the MluI restriction site. The engineered plasmids were transformed into Escherichia coli C41(DE3) and expressed using isopropyl β-d-thiogalactopyranoside induction. The expressed proteins were purified by a nickel affinity column, followed by size exclusion HPLC.
AFM-based Single-molecule Force Spectroscopy
All AFM stretching measurements were carried out on custom-built AFM instruments (67). The spring constant (kc) of each cantilever was calibrated in solution using the energy equipartition theorem (71). All force extension measurements were performed using BioLever AFM tips (kc ≈ 6 piconewtons/nm; Veeco) at pulling speeds between 5 and 100 nm/s at room temperature. The force peaks in the force extension curves were fitted to the WLC (worm-like chain) model (72).
CG-SMD Simulations
The initial geometry of NI6C was built based on Protein Data Bank code 2QYJ, coarse-grained to Cα representation, and the molecule was simulated using the structure-based CG force field (73), which has been successfully employed in studies of the folding of similar proteins (25, 34). The SMD simulations were performed with kc = 6 piconewtons/nm and Langevin friction coefficient γ = 1 amu/ps. The simulations approached quasi-equilibrium conditions with converged conformational samplings of the pulling and relaxing processes. The moving speed of the SMD point was 0.1 nm/ns, and the total sampling time for each simulation was 800 ns. Further details are provided in the supplemental “Materials and Methods.”
RESULTS
Mechanical Folding of NI6C under One-dimensional Constraints
Our approach is illustrated in Fig. 1A. The NI6C protein was flanked on each side by three I27 domains of titin serving as pulling handles and as a force spectroscopy reference for identifying single-molecule recordings (49, 50, 69, 70, 74–78). We first stretched the chimeric protein to unfold the NI6C portion, either fully or partially, and then allowed it to relax and refold while measuring the extension and tension (see details under supplemental “Materials and Methods”). To verify the convergence of the SMD simulations, we compared the force extension curves obtained at different pulling speeds within the range of 0.1–10 nm/ns. Furthermore, we also performed multiple simulations with the same SMD speed (v = 0.1 nm/ns) but different initial particle velocities. The multiple simulations generated almost identical force extension curves, suggesting that the conformational samplings in the SMD simulations converged very well. This comparison is included in supplemental Fig. S5.
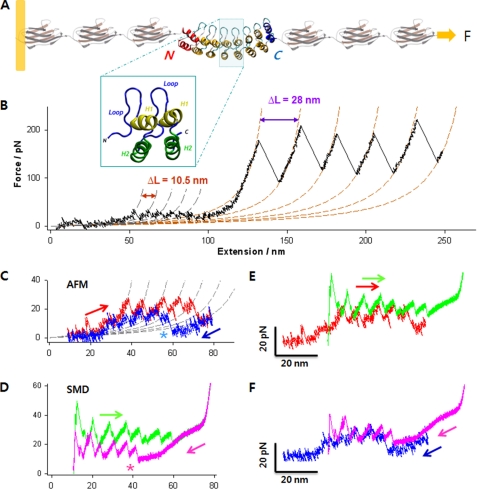
FIGURE 1.
Complete mechanical unfolding and refolding of the NI6C-I27 chimeric protein. A, structure of the chimeric polyprotein designed for the AFM experiments. NI6C contains eight ARs (66). Two internal repeats are shown in the blue box, each composed of two α-helices (H1 and H2) and a loop. The full sequence is shown in supplemental Fig. S1D. B, representative unfolding trace of NI6C-I27 obtained at a stretching speed of 100 nm/s. The AFM data are fitted to two families of WLC curves, one with contour length increment ΔL = 10.5 nm and persistence length p = 0.78 nm (gray dashed lines) and the other with ΔL = 28 nm and p = 0.36 nm (orange dashed lines). These values of ΔL are consistent with the fully stretched lengths of one AR and one I27 domain, respectively; hence, the two families of peaks correspond to the unfolding of individual ARs of NI6C and of I27 domains, respectively. pN, piconewtons. C, measured unfolding (red) and refolding (blue) force extension traces of NI6C at 30 nm/s, fitted to a family of WLC curves (gray dashed lines) with ΔL = 10.5 nm and p = 0.86 nm. Note that following the complete unfolding of all ARs, the first refolding force peak appeared only after the molecule was partially relaxed (asterisk). D, simulated unfolding (green) and refolding (pink) force extension traces of NI6C. Similar to the AFM data, the first SMD refolding force peak occurred only after the molecule has been significantly relaxed (asterisk). E, comparison of the unfolding force extension traces by SMD (green) and AFM (red). The SMD trace was shifted to the right by 20 nm to compensate for the initial length of I27 modules that contribute to the extension in the AFM measurements but are absent in the SMD simulations. F, comparison of the refolding force extension traces by SMD (pink) and AFM (blue) following the unfolding of NI6C. The same 20-nm shift was applied to the SMD trace.
Fig. 1B shows a typical AFM force extension trace of the NI6C-I27 construct obtained at a pulling speed of 100 nm/s. Evenly spaced by 10.5 nm, small unfolding force peaks at protein extensions below 100 nm (blue-shaded area) strongly suggest that they correspond to the stepwise unfolding of individual NI6C repeats (supplemental Figs. S1 and S2) (67, 79). These small force peaks are then followed by the characteristic sawtooth pattern of large force peaks corresponding to the mechanical unfolding of five of six I27 domains, providing direct evidence that the whole measurement was obtained on a single molecule containing all eight ARs. We used this recording as the reference force spectrogram to identify unfolding events of ARs in other measurements.
In Fig. 1 (C–F), we compare the results of AFM measurements and SMD simulations. The unfolding and refolding force extension traces obtained by SMD matched the AFM data remarkably well. This indicates that the computer simulations indeed reproduced the main events occurring in NI6C under the AFM control. Moderate differences between AFM and SMD data were observed primarily at small extensions (∼10 nm) due to the absence of I27 domains in the simulations. The presence of I27 domains in AFM measurements resulted in the initial length (and extension) of the construct being greater than that of NI6C alone, used in the simulations. It also affected the slope of force extension curves especially in low extension regions. This effect in high extension regions was less significant because the long unraveled polypeptide chain dominated the overall elasticity.
Folding Pathway and Time Evolution of Native Contacts of NI6C
To analyze the entire unfolding and refolding trajectories of NI6C, we built its native contact map (see details supplemental “Materials and Methods”) and monitored its time evolution (Fig. 2). It can be seen that the unfolding process occurred in a vectorial fashion from the C to the N terminus, and the refolding process followed the reverse order. A folding sequence that starts at the N terminus, as revealed here by the SMD simulations, would allow co-translational folding to achieve maximal speed. The analysis of native contacts at both termini indicated that the structure of NI6C is slightly asymmetric. As shown in Table 1, helices H1 and H2 in the N-terminal repeats are longer than those in the internal repeats, and the corresponding helices in the C-terminal repeat are the shortest. This asymmetry means that the N-terminal repeat has more stabilizing interactions than the C-terminal repeat; correspondingly, the latter consistently unfolded first. Unfolding is unlikely to start from an internal repeat because that would require breaking roughly twice as many native contacts with neighboring repeats when comparing with starting from a terminal repeat. The unfolding of some native AR proteins was found to proceed in the opposite direction to that of NI6C (80, 81). It would be interesting to see whether that unfolding order can be similarly explained by an analysis of native contacts.
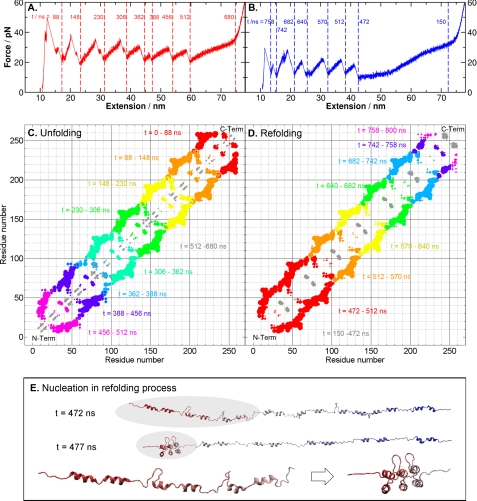
FIGURE 2.
Unfolding and refolding sequences of NI6C. A, simulated complete unfolding force extension traces of NI6C, with timestamps marked between each major force peak, indicating the breaking of a tertiary structure. pN, piconewtons. B, simulated complete refolding force extension traces of NI6C, with timestamps. C, changes in the distance between native contacts during the complete unfolding process, with color code from red to gray representing different time spans defined in A. For each contact, a varying size of dot is used. The size (S) of a dot is proportional to the change in the distance between the residues of an original native contact within a certain time span, i.e. S = c(r(t2) − r(t1)), where r(t) is the residual distance at time t, and c is a scaling constant. This change in contact distance clearly shows that the breakup of native contacts during the mechanical unfolding proceeded from the C to N terminus. The gray regions represent the unfolding of local α-helices (H1 and H2), corresponding to the plateau between 512 and 680 ns in A. D, changes in native contact distance during the complete refolding process, with color-coded time spans. From t = 150 to 472 ns, local structures were formed (shown by the gray regions), followed by the simultaneous folding of three repeats at the N terminus. This nucleation event produced the first force peak that appeared between t = 472 and 512 ns in B. E, snapshots of the NI6C structures before and after the nucleation event with timestamps. The zoomed inset shows the conformational change in the nucleation region (shaded) at t = 472 and 477 ns. Three N-terminal repeats folded within 5 ns and formed a nucleation core, which facilitated the folding of the rest of the polypeptide chain.
TABLE 1.
Count of the native contacts in the α-helical domains (H1 and H2) in the N terminus, C terminus, and internal repeats of NI6C
| Domain | Sequencea | No. of residues | No. of all contactsb | No. of inter-repeat contactsb |
|---|---|---|---|---|
| N-terminal | Asp7–Gln30 | 24 | 85 | 49 |
| C-terminal | Thr241–Gln259 | 19 | 70 | 45 |
| Internal | Thr109–Ala129 | 21 | 129 | 101 |
a NI6C was built using the x-ray structure of NI3C (Protein Data Bank code 2QYJ); the residues were renumbered (see details in supplemental Fig. S4).
b The differences in the numbers of native and inter-repeat contacts lead to significant differences in stability between the repeats, as indicated by the energy function.
Nucleation of N-terminal α-Helices, the Rate-limiting Step for Folding
It is worth noting that the unfolding of the last repeat was followed by a plateau in the SMD force extension trace (Fig. 1D). On the basis of the contacts shown in gray in Fig. 2C (t = 512–680 ns) and the protein dynamics in the unfolding trajectory captured in supplemental Movie S1, we ascertained that the plateau corresponded to the unwinding of the helical structures that remained folded until this stage. Similar plateaus were observed in the refolding traces, where they preceded the occurrence of the refolding force peaks (Fig. 1D). According to the contact map in Fig. 2D and supplemental Movie S2, the folding of NI6C started with the formation of local α-helices (H1 and H2) within t = 150–472 ns. No tertiary structures were formed until t = 472 ns, at which point three repeats at the N terminus (Asp13–Gly103) folded simultaneously within 5 ns (t = 472–477 ns). This happened after several failed attempts to create a folded nucleation core in different regions of the fluctuating polypeptide chain (supplemental Movie S2). This observation indicates the importance of nucleation in the folding of AR proteins under one-dimensional constraints and is consistent with the prediction that the main folding barrier corresponds to the simultaneous folding of two to three consecutive repeats (25, 82). A similar nucleation process was proposed by Wetzel et al. (66) in the chemical and thermal folding of consensus AR proteins, but it has never before been observed directly. Once the nucleation event occurred and a stable folded stack was formed, the rest of the polypeptide chain folded in a vectorial fashion, repeat by repeat, using the stack as the folding template (supplemental Movie S2).
Refolding of NI6C from Partially Unraveled Structures
To examine in detail the late stage of AR folding, which may occur in the presence of folded N-terminal repeats, we designed AFM experiments and corresponding SMD simulations in which only part of NI6C was unraveled and then allowed to relax and refold. This was achieved by limiting the extension of the chimeric protein below the contour length of NI6C (<90 nm). This procedure allowed us to specify the number of repeats that unfolded or remained folded at the end of the stretching phase. During the subsequent relaxation phase, the refolding behavior of the repeats that already unraveled was followed. These short extension measurements could be performed at significantly lower pulling speeds, affording better force resolution. The force extension data obtained at a stretching speed of 5 nm/s are shown in Fig. 3A and are compared with the data obtained at 30 nm/s in supplemental Fig. S1C. Even though the spacing between major unfolding force peaks measured at low and high pulling speeds are comparable, the force traces obtained at slower stretching speeds resolve finer details of the unfolding and refolding processes. For example, at a pulling speed of 5 nm/s, the major peaks were split into pairs of smaller subpeaks with ΔL ≈ 3–5 nm as shown in Fig. 3A, suggesting that individual repeats unfold in two or more steps (supplemental Fig. S1).
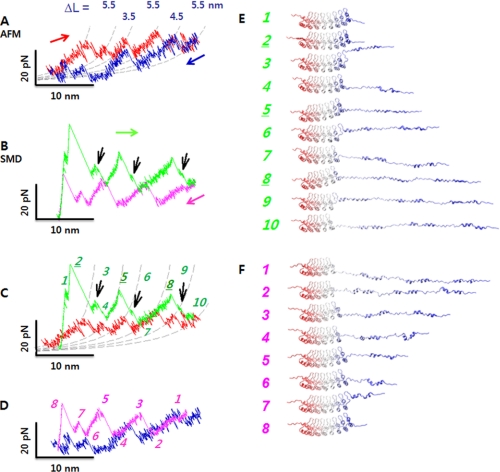
FIGURE 3.
Partial unfolding followed by refolding of NI6C. A, force extension curves corresponding to partial unfolding (red) and refolding (blue) of NI6C at a pulling velocity of 5 nm/s. Dashed curves show the WLC fits with p = 0.9 nm. pN, piconewtons. B, partial unfolding (green) and refolding (pink) force extension traces obtained from the SMD simulations (supplemental Figs. S4–S6). Black arrows indicate small unfolding force peaks separated by major unfolding force peaks. C, comparison of SMD (green) and AFM (red) unfolding force extension traces. D, comparison of SMD (pink) and AFM (blue) refolding traces. E, series of snapshots along the SMD partial unfolding trajectory. Numbers correspond to the peaks in C. Three repeats unfolded sequentially from the C to N terminus; five N-terminal repeats remained intact (see snapshot 10). F, series of snapshots in the SMD refolding trajectory. Numbers correspond to the peaks in D. The refolding process followed the reverse order of the unfolding process.
The relaxation traces obtained on a partially unfolded NI6C protein exhibit robust well separated refolding force peaks that coincide with the unfolding force peaks. This suggests that upon relaxation, the polypeptide chain contracts in a stepwise manner while refolding the unraveled repeats (Fig. 3A and supplemental Fig. S2, A and B). Transient unfolding and refolding events were also frequently observed at low stretching speeds, indicating that these transitions occur at near equilibrium (51).
To gain insight into the events captured by AFM during partial unfolding/refolding, we performed SMD simulations in which three of the eight repeats were unraveled and then relaxed under mechanical control. In Fig. 3 (B–D), we show the SMD results and their comparison with the AFM data. As before, the agreement between the experiment and simulation is very satisfactory.
In Fig. 3 (E and F), we show two series of snapshots from the SMD trajectories; each snapshot corresponds to a numbered peak in Fig. 3 (C or D). The snapshots illustrate the vectorial unfolding and refolding of the C-terminal three repeats. In the unfolding trace, successive unfolding events of the three repeats are captured by conformational changes in snapshots 2–4, 5–7, and 8–10. During the unfolding process, the hairpin loops (Fig. 1A, inset) tended to remain folded after the detachment of the two α-helices, H1 and H2, from the stack. Loop unfolding produced small subpeaks in the force extension trace labeled as subpeaks 3, 6, and 9 in Fig. 3C. Accordingly, the unfolding of one repeat consisted of two steps: the detachment and straightening of α-helices H1 and H2 (peaks 2, 5, and 8), followed by the opening of the hairpin loop (subpeaks 3, 6, and 9). The refolding of these repeats took a similar pathway but in the reverse direction; however, helices H1 and H2 and the loop in one repeat typically folded in a single step.
DISCUSSION
We used AFM-based single-molecule force spectroscopy to unravel the polypeptide chain and to follow its relaxation under one-dimensional geometrical constraints. We also simulated these stretch/relax processes by structure-based coarse-grained SMD calculations. The remarkable hitherto unattained accuracy with which SMD simulations recreated the experimental force spectrograms allowed us to confidently reconstruct the vectorial folding pathway of NI6C. Thus, this combination of AFM measurements with structure-based computer simulations presents a powerful tool for advancing our understanding of protein folding under geometrical constraints.
First, our results indicate that the folding of NI6C under geometrical constraints is hierarchical and does not follow a simple two-state process indicated in bulk folding measurements (83). The folding of NI6C starts with the formation of local α-helices. The formation of these local secondary structures greatly reduces the dimension of the conformational space and accelerates the formation of tertiary structure elements. A similar hierarchy has also been observed in other systems (35, 42, 43, 84–87).
Second, the results from multiple SMD simulations clearly demonstrate that the rate-limiting step in the vectorial folding process of the entire NI6C protein involves the nucleation of three N-terminal repeats. These observations together strongly suggest that the co-translational folding of ARs may proceed by forming α-helical segments still in the ribosome exit tunnel (43), and after extruding their length corresponding to at least three repeats, they nucleate. Further folding may involve a simple rapid addition of the remaining C-terminal repeats, without any nucleation step, using the existing nucleated structure as the folding template. We hypothesize that such a nucleation-free fast mechanical folding of terminal repeats may be advantageous for putative biological functions of repeat proteins in mechanotransduction (32, 88, 89) should they be challenged with a partial mechanical unfolding.
Third, we note that during the co-translational folding, the NPC is most likely not subjected to significant external mechanical forces, although it may be under some tension as a result of the interaction between the extruded part of the NPC with the ribosome-bound chaperone trigger factor that assists the folding (38, 90). We believe that the application of a small external force to a folding polypeptide chain during AFM refolding measurements, which provides a practical means to restrain the protein in a vectorial space, by itself does not significantly alter the folding pathway of NI6C. The results in this work suggest that the refolding processes of NI6C occur spontaneously from the N to C terminus without the assistance or hindrance of an external force. The fact that the end of the polypeptide chain is still tethered (through the I27 handle) to the AFM tip should have minimal effect except perhaps keeping the folding along one direction, which mimics the effect of the wall of the ribosome exit tunnel.
Therefore, we propose that the vectorial folding pathway reconstructed here for NI6C may be representative of the co-translational folding of thousands of AR domains and other repeat proteins that form similar extended structures composed of stacked α-helical repeats. This conclusion is supported by our recent observation that native ARs of ankyrin-R, armadillo repeats of β-catenin, and HEAT repeats of clathrin all unfold and refold under one-dimensional constraints in a stepwise manner similar to that displayed by the consensus ARs of NI6C (68).
Supplementary Material
Acknowledgment
We thank Jane Clarke for providing the plasmid with I27 domains.
This work was supported, in whole or in part, by National Institutes of Health Grants GM079663 (to P. E. M. and V. B.) and GM058187 (to H.-X. Z.). This work was also supported by National Science Foundation Grant MCB-0717770 (to P. E. M. and W. Y.) and a Duke University Center of Theoretical and Mathematical Sciences graduate fellowship (to X. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. S1–S6, Table S1, Movies S1 and S2, and additional references..
- NPC
- nascent polypeptide chain
- AFM
- atomic force microscope/microscopy
- SMD
- steered molecular dynamics
- AR
- ankyrin repeat.
REFERENCES
- 1.Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. (1995) Proteins Struct. Funct. Genet. 21, 167–195 [DOI] [PubMed] [Google Scholar]
- 2.Kim P. S., Baldwin R. L. (1990) Annu. Rev. Biochem. 59, 631–660 [DOI] [PubMed] [Google Scholar]
- 3.Englander S. W., Mayne L., Krishna M. M. G. (2007) Q. Rev. Biophys. 40, 287–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dill K. A., Ozkan S. B., Shell M. S., Weikl T. R. (2008) Annu. Rev. Biophys. 37, 289–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin R. L. (2007) J. Mol. Biol. 371, 283–301 [DOI] [PubMed] [Google Scholar]
- 6.Borgia A., Williams P. M., Clarke J. (2008) Annu. Rev. Biochem. 77, 101–125 [DOI] [PubMed] [Google Scholar]
- 7.Schuler B., Eaton W. A. (2008) Curr. Opin. Struct. Biol. 18, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang F., Ying L., Fersht A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16239–16244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez J. M., Li H. (2004) Science 303, 1674–1678 [DOI] [PubMed] [Google Scholar]
- 10.Cecconi C., Shank E. A., Bustamante C., Marqusee S. (2005) Science 309, 2057–2060 [DOI] [PubMed] [Google Scholar]
- 11.Shank E. A., Cecconi C., Dill J. W., Marqusee S., Bustamante C. (2010) Nature 465, 637–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler M., Gottschalk K. E., Janovjak H., Muller D. J., Gaub H. E. (2006) J. Mol. Biol. 357, 644–654 [DOI] [PubMed] [Google Scholar]
- 13.Puchner E. M., Gaub H. E. (2009) Curr. Opin. Struct. Biol. 19, 605–614 [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Manyes S., Dougan L., Badilla C. L., Brujic J., Fernández J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10534–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Manyes S., Dougan L., Fernández J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10540–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walther K. A., Gräter F., Dougan L., Badilla C. L., Berne B. J., Fernandez J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7916–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dougan L., Koti A. S., Genchev G., Lu H., Fernandez J. M. (2008) Chem. Phys. Chem. 9, 2836–2847 [DOI] [PubMed] [Google Scholar]
- 18.Berkovich R., Garcia-Manyes S., Urbakh M., Klafter J., Fernandez J. M. (2010) Biophys. J. 98, 2692–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma D., Perisic O., Peng Q., Cao Y., Lam C., Lu H., Li H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9278–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dougan L., Feng G., Lu H., Fernandez J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Movileanu L. (2009) Trends Biotechnol. 27, 333–341 [DOI] [PubMed] [Google Scholar]
- 22.Mayor U., Guydosh N. R., Johnson C. M., Grossmann J. G., Sato S., Jas G. S., Freund S. M., Alonso D. O., Daggett V., Fersht A. R. (2003) Nature 421, 863–867 [DOI] [PubMed] [Google Scholar]
- 23.Cellmer T., Henry E. R., Hofrichter J., Eaton W. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18320–18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werbeck N. D., Itzhaki L. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7863–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreiro D. U., Cho S. S., Komives E. A., Wolynes P. G. (2005) J. Mol. Biol. 354, 679–692 [DOI] [PubMed] [Google Scholar]
- 26.Best R. B., Hummer G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6732–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaeffer R. D., Fersht A., Daggett V. (2008) Curr. Opin. Struct. Biol. 18, 4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berezhkovskii A., Hummer G., Szabo A. (2009) J. Chem. Phys. 130, 205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freddolino P. L., Schulten K. (2009) Biophys. J. 97, 2338–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao M., Lu H., Schulten K. (2001) Biophys. J. 81, 2268–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Best R. B., Fowler S. B., Herrera J. L., Steward A., Paci E., Clarke J. (2003) J. Mol. Biol. 330, 867–877 [DOI] [PubMed] [Google Scholar]
- 32.Sotomayor M., Corey D. P., Schulten K. (2005) Structure 13, 669–682 [DOI] [PubMed] [Google Scholar]
- 33.Sotomayor M., Schulten K. (2007) Science 316, 1144–1148 [DOI] [PubMed] [Google Scholar]
- 34.Li M. S., Hu C. K., Klimov D. K., Thirumalai D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyeon C., Dima R. I., Thirumalai D. (2006) Structure 14, 1633–1645 [DOI] [PubMed] [Google Scholar]
- 36.Mickler M., Dima R. I., Dietz H., Hyeon C., Thirumalai D., Rief M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20268–20273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komar A. A. (2009) Trends Biochem. Sci. 34, 16–24 [DOI] [PubMed] [Google Scholar]
- 38.Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 39.Clark P. L. (2004) Trends Biochem. Sci. 29, 527–534 [DOI] [PubMed] [Google Scholar]
- 40.Cabrita L. D., Dobson C. M., Christodoulou J. (2010) Curr. Opin. Struct. Biol. 20, 33–45 [DOI] [PubMed] [Google Scholar]
- 41.Cabrita L. D., Hsu S. T., Launay H., Dobson C. M., Christodoulou J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22239–22244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 43.Bhushan S., Gartmann M., Halic M., Armache J. P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., Beckmann R. (2010) Nat. Struct. Mol. Biol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Qin H., Kudaravalli R. D., Kirillov S. V., Dempsey G. T., Pan D., Cooperman B. S., Goldman Y. E. (2007) Biochemistry 46, 10767–10775 [DOI] [PubMed] [Google Scholar]
- 45.Wen J. D., Lancaster L., Hodges C., Zeri A. C., Yoshimura S. H., Noller H. F., Bustamante C., Tinoco I. (2008) Nature 452, 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uemura S., Aitken C. E., Korlach J., Flusberg B. A., Turner S. W., Puglisi J. D. (2010) Nature 464, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tinoco I., Wen J. D. (2009) Phys. Biol. 6, 025006 [DOI] [PubMed] [Google Scholar]
- 48.Oberhauser A. F., Marszalek P. E., Erickson H. P., Fernandez J. M. (1998) Nature 393, 181–185 [DOI] [PubMed] [Google Scholar]
- 49.Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 50.Marszalek P. E., Lu H., Li H., Carrion-Vazquez M., Oberhauser A. F., Schulten K., Fernandez J. M. (1999) Nature 402, 100–103 [DOI] [PubMed] [Google Scholar]
- 51.Junker J. P., Ziegler F., Rief M. (2009) Science 323, 633–637 [DOI] [PubMed] [Google Scholar]
- 52.Li H., Linke W. A., Oberhauser A. F., Carrion-Vazquez M., Kerkvliet J. G., Lu H., Marszalek P. E., Fernandez J. M. (2002) Nature 418, 998–1002 [DOI] [PubMed] [Google Scholar]
- 53.Lv S., Dudek D. M., Cao Y., Balamurali M. M., Gosline J., Li H. (2010) Nature 465, 69–73 [DOI] [PubMed] [Google Scholar]
- 54.Oberhauser A. F., Carrión-Vázquez M. (2008) J. Biol. Chem. 283, 6617–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oukhaled G., Mathé J., Biance A. L., Bacri L., Betton J. M., Lairez D., Pelta J., Auvray L. (2007) Phys. Rev. Lett. 98, 158101. [DOI] [PubMed] [Google Scholar]
- 56.Mohammad M. M., Prakash S., Matouschek A., Movileanu L. (2008) J. Am. Chem. Soc. 130, 4081–4088 [DOI] [PubMed] [Google Scholar]
- 57.Movileanu L., Schmittschmitt J. P., Scholtz J. M., Bayley H. (2005) Biophys. J. 89, 1030–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ammenti A., Cecconi F., Marini Bettolo Marconi U., Vulpiani A. (2009) J. Phys. Chem. B 113, 10348–10356 [DOI] [PubMed] [Google Scholar]
- 59.Kirmizialtin S., Ganesan V., Makarov D. E. (2004) J. Chem. Phys. 121, 10268–10277 [DOI] [PubMed] [Google Scholar]
- 60.Makarov D. E. (2009) Acc. Chem. Res. 42, 281–289 [DOI] [PubMed] [Google Scholar]
- 61.Tian P., Andricioaei I. (2005) J. Mol. Biol. 350, 1017–1034 [DOI] [PubMed] [Google Scholar]
- 62.West D. K., Brockwell D. J., Paci E. (2006) Biophys. J. 91, L51–L53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu H., Schulten K. (1999) Proteins Struct. Funct. Bioinformat. 35, 453–463 [PubMed] [Google Scholar]
- 64.Lu H., Schulten K. (2000) Biophys. J. 79, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isralewitz B., Baudry J., Gullingsrud J., Kosztin D., Schulten K. (2001) J. Mol. Graph. Model. 19, 13–25 [DOI] [PubMed] [Google Scholar]
- 66.Wetzel S. K., Settanni G., Kenig M., Binz H. K., Plückthun A. (2008) J. Mol. Biol. 376, 241–257 [DOI] [PubMed] [Google Scholar]
- 67.Lee G., Abdi K., Jiang Y., Michaely P., Bennett V., Marszalek P. E. (2006) Nature 440, 246–249 [DOI] [PubMed] [Google Scholar]
- 68.Kim M., Abdi K., Lee G., Rabbi M., Lee W., Yang M., Schofield C. J., Bennett V., Marszalek P. E. (2010) Biophys. J. 98, 3086–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serquera D., Lee W., Settanni G., Marszalek P. E., Paci E., Itzhaki L. S. (2010) Biophys. J. 98, 1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steward A., Toca-Herrera J. L., Clarke J. (2002) Protein Sci. 11, 2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Florin E. L., Rief M., Lehmann H., Ludwig M., Dornmair C., Moy V. T., Gaub H. E. (1995) Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 72.Bustamante C., Marko J. F., Siggia E. D., Smith S. (1994) Science 265, 1599–1600 [DOI] [PubMed] [Google Scholar]
- 73.Clementi C., Nymeyer H., Onuchic J. N. (2000) J. Mol. Biol. 298, 937–953 [DOI] [PubMed] [Google Scholar]
- 74.Perez-Jimenez R., Garcia-Manyes S., Ainavarapu S. R., Fernandez J. M. (2006) J. Biol. Chem. 281, 40010–40014 [DOI] [PubMed] [Google Scholar]
- 75.Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Oberhauser A. F., Fowler S. B., Clarke J., Fernandez J. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6527–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams P. M., Fowler S. B., Best R. B., Toca-Herrera J. L., Scott K. A., Steward A., Clarke J. (2003) Nature 422, 446–449 [DOI] [PubMed] [Google Scholar]
- 78.Dietz H., Rief M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16192–16197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L., Wetzel S., Plückthun A., Fernandez J. M. (2006) Biophys. J. 90, L30–L32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang K. S., Fersht A. R., Itzhaki L. S. (2003) Structure 11, 67–73 [DOI] [PubMed] [Google Scholar]
- 81.Werbeck N. D., Rowling P. J., Chellamuthu V. R., Itzhaki L. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9982–9987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferreiro D. U., Wolynes P. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9853–9854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mello C. C., Barrick D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14102–14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu Y., Alonso D. O., Maki K., Huang C. Y., Lahr S. J., Daggett V., Roder H., DeGrado W. F., Gai F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15486–15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weikl T. R., Palassini M., Dill K. A. (2004) Protein Sci. 13, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mukherjee S., Chowdhury P., Bunagan M. R., Gai F. (2008) J. Phys. Chem. B 112, 9146–9150 [DOI] [PubMed] [Google Scholar]
- 87.Du D., Gai F. (2006) Biochemistry 45, 13131–13139 [DOI] [PubMed] [Google Scholar]
- 88.Howard J., Bechstedt S. (2004) Curr. Biol. 14, R224–R226 [DOI] [PubMed] [Google Scholar]
- 89.Corey D. P., Sotomayor M. (2004) Nature 428, 901–903 [DOI] [PubMed] [Google Scholar]
- 90.Kramer G., Boehringer D., Ban N., Bukau B. (2009) Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.