Abstract
Inclusion bodies of aggregated mutant huntingtin (htt) fragments are a neuropathological hallmark of Huntington disease (HD). The molecular chaperones Hsp70 and Hsp40 colocalize to inclusion bodies and are neuroprotective in HD animal models. How these chaperones suppress mutant htt toxicity is unclear but might involve direct effects on mutant htt misfolding and aggregation. Using size exclusion chromatography and atomic force microscopy, we found that mutant htt fragments assemble into soluble oligomeric species with a broad size distribution, some of which reacted with the conformation-specific antibody A11. Hsp70 associated with A11-reactive oligomers in an Hsp40- and ATP-dependent manner and inhibited their formation coincident with suppression of caspase 3 activity in PC12 cells. Thus, Hsp70 and Hsp40 (DNAJB1) dynamically target specific subsets of soluble oligomers in a classic ATP-dependent reaction cycle, supporting a pathogenic role for these structures in HD.
Keywords: Amyloid, Chaperone Chaperonin, Heat Shock Protein, Neurodegeneration, Polyglutamine Disease, Protein Folding, Oligomer
Introduction
The accumulation of aggregated mutant htt in cytoplasmic and intranuclear inclusion bodies in neurons is a pathological hallmark of HD.4 Recent studies suggest that mutant htt is a highly dynamic protein that adopts multiple misfolded monomeric and higher order conformations that exist in a dynamic equilibrium (1, 2). Which of these structures, if any, mediate toxic gain-of-function protein interactions that cause neuronal dysfunction and cell death in HD is not well understood.
The assembly of mutant htt into amyloid-like fibrils was initially thought to be causally associated with the pathogenesis of HD (3, 4). More recent studies indicate that mutant htt and other disease-associated proteins with polyglutamine (polyQ) expansions also exist as smaller oligomers that have spherical, annular, and protofibrillar morphologies (5–7) that might be less energetically stable and more chemically reactive (and toxic) than fibrils. Consistent with this scenario, in cell and mouse models of HD, misfolded species of mutant htt in a diffuse fraction impair cellular machinery before fibrils accumulate (8–10). In a cellular model of HD, the survival rate of cells is greater in the presence of inclusion bodies that contain fibrils than in their absence (11). Importantly, mutant htt fragments and synthetic polyQ peptides form oligomers in a polyQ length-dependent manner in vitro and in vivo, consistent with a causative role for these structures in HD pathogenesis (2). However, oligomers are also highly heterogeneous in size, stability, and solubility (2). Therefore, only subsets of such structures might be most biologically active.
Many disease-causing proteins form toxic oligomers that react in a conformation-dependent manner with A11, an antibody that neutralizes their toxicity in cell-based assays (12). Thus, the misfolding of disease proteins to a similar oligomeric conformation might contribute to their toxicity. The molecular mechanism for this misfolding is not clear, but A11 seems to recognize a structural motif that is exposed only in oligomers and not in the native structure, random coil monomers, or mature fibrils (13).
Molecular chaperones, which are sequestered in inclusion bodies, suppress neurodegeneration in several animal models of protein misfolding diseases, including HD (14, 15). Chaperones are thought to suppress the toxicity of disease-associated proteins, at least in part, through direct effects on their misfolding, but the mechanisms are not well understood. Hsp70 and Hsp40 are major cytosolic molecular chaperones that, if overexpressed, suppress neurodegeneration in animal models of polyQ diseases (16–19). In contrast, deletion of Hsp70 markedly worsens pathogenesis in a mouse model of HD (20).
The molecular mechanism invoked by Hsp70 and Hsp40 in protein folding is well established. These chaperones regulate folding in a cooperative manner driven by Hsp70-dependent ATP hydrolysis, thereby promoting substrate refolding and preventing unspecific protein interactions that otherwise would lead to aggregation (21, 22). However, the mechanism by which these chaperones influence protein misfolding in HD and other neurodegenerative diseases is not completely clear. For example, it is not known whether Hsp70 and Hsp40 interact with soluble oligomeric forms of misfolded proteins or whether they use a similar ATP-dependent mechanism to refold reactive sequences exposed on oligomers. In earlier studies, Hsp70 and Hsp40 inhibited the formation of SDS-insoluble aggregates of mutant htt fragments by presumably acting on small, misfolded species that were structurally uncharacterized (15, 23). A subsequent study showed that Hsp70 and Hsp40 inhibited the formation of spherical and annular mutant htt oligomers, as shown by atomic force microscopy (AFM), but presumably by partitioning of mutant htt monomers (7). In animal models of polyQ expansion disorders, overexpression of Hsp70 and Hsp40 reduces pathogenesis without altering the accumulation or size of inclusion bodies, but increases the detergent solubility of the misfolded polyQ proteins (16, 18). Conversely, increased pathogenesis in the absence of Hsp70 in a mouse model of HD is associated with larger inclusion bodies without changes in the level of detergent-insoluble, fibrillar aggregates (7). Thus, chaperones such as Hsp70 and Hsp40 may mediate neuroprotection by acting on soluble monomeric and oligomeric species rather than insoluble fibrillar species.
In this study, we show that mutant htt forms soluble oligomers that are highly heterogeneous in size, some of which react with the conformation-specific antibody A11. We also found that Hsp70 suppresses the formation of A11-reactive species by interacting with them in a classic ATP-dependent reaction cycle that requires Hsp40 (DNAJB1). These results support the hypothesis that subsets of soluble mutant htt oligomers mediate pathogenesis in HD.
EXPERIMENTAL PROCEDURES
Protein Purification
GST-HD20Q and GST-HD53Q were purified as described (7, 15, 24). Immediately after SEC, fresh, unfrozen HD53Q and HD20Q were used for all experiments. Hsp40 (Dj1/DNAJB1/hdj-1) was expressed from plasmid pQE9 in Escherichia coli and purified as described (7). Bovine Hsc70 (Hsp70) from liver was purified as described (25, 26). Chaperones were frozen in liquid nitrogen and stored at −80 °C until ready for use in biochemical assays.
Antibodies
The following antibodies were used: anti-htt (MAB5374, clone EM48) and anti-GFP (MAB3580) from Chemicon; c-Myc (9E10/sc40) from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-Hsc70 (Spa815), inducible Hsp70 (Spa810), and Hsp40 (Spa400) from Stressgen Biotechnologies (Ann Arbor, MI). Antibody A11 (12) was provided by Charles Glabe (University of California, Irvine). Anti-htt antibodies MW1 and MW8 were kindly provided by Paul Patterson (California Institute of Technology).
Analysis of Mutant htt Aggregation by Dot-blot, Filter Trap, and Western Blot Analyses
After purification with SEC, HD20Q, and HD53Q were centrifuged (20,000 × g) for 30 min at 4 °C before each experiment to remove aggregates. HD53Q proteins were incubated at 6 μm in 20 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm dithiothreitol at 37 °C with shaking at 800 rpm. PreScission protease (4 units/100 μg of fusion protein; Amersham Biosciences) was added at time 0 to initiate the aggregation. At time 0 or as indicated, Hsp70 and Hsp40 were added alone or in combination to the aggregation reactions at an equimolar ratio to the fusion protein (6 μm). Reactions with Hsp70 and Hsp40 were supplemented with 3 mm ATP, 5 mm MgCl2, and an ATP-regenerating system (200 μg/ml creatine kinase and 20 mm creatine phosphate).
For dot-blot analyses, equal amounts of protein (2 μg) were removed from 6 μm aggregation reactions at various times after GST cleavage and applied to a nitrocellulose membrane (0.2-μm pore size; Schleicher & Schuell). The membrane was incubated in blocking buffer and treated as described (7, 12).
For the filter trap assay, aggregation of HD53Q was initiated as described above. HD53Q (4 μg) was removed from the 6 μm reaction, boiled in SDS loading buffer, and applied to a nitrocellulose membrane (0.45-μm pore size; Schleicher & Schuell) through a slot-blot manifold. The membranes were washed with 3% SDS buffer by vacuum filtering, incubated in blocking buffer, and treated as described above for dot-blots. Representative blots of trapped HD53Q aggregates with anti-htt antibody (MW8) are shown in Fig. 1E.
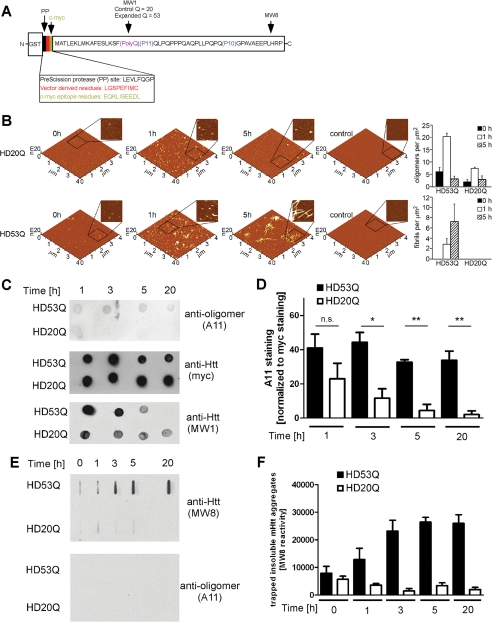
FIGURE 1.
The conformation-dependent anti-oligomer antibody A11 detects mutant htt oligomers but not monomers or fibrils. A, schematic of the GST-mutant htt exon 1 fusion protein with 53Q (HD53Q) showing a PreScission protease site between GST and the mutant htt fragment (not drawn to scale) and the locations of epitopes for MW8, MW1, and c-Myc antibodies. B, morphology of HD20Q and HD53Q aggregation reactions over time, analyzed by AFM. Representative AFM images of 6 μm aggregation reactions at 0, 1, and 5 h are shown. Zoomed in panels are 1 μm by 1 μm. Controls are denatured HD20Q and HD53Q in reaction buffer with 6 m guanidinium HCl. Analysis of oligomers and fibrils per unit area at 0, 1, and 5 h is presented. C, dot-blots of HD53Q and HD20Q after incubation with protease for various times, probed with antibodies as labeled. D, percentage of A11 reactivity relative to Myc reactivity (100%) in dot-blots of three independent experiments, quantified by densitometry using Image J. *, p < 0.05 versus HD20Q. ns, not significant at 1 h by t test. The values are the means ± S.E. E, filter trap assays of HD53Q and HD20Q after incubation with protease for various times. Insoluble HD53Q oligomers and fibrils were trapped and detected by MW8 but not by A11. F, quantification of filter trap assay of three independent experiments, quantified by densitometry using Image J. The values are the means ± S.E.
For Western blot analyses, proteins and aggregates were separated by SDS-PAGE and transferred to nitrocellulose membranes (0.45-μm pore size; Schleicher & Schuell) by standard Western transfer techniques and treated as described (7).
Analysis of Mutant htt Aggregation by AFM
All of the AFM images were collected in tapping mode with an MFP-3D AFM system (Asylum Research, Santa Barbara, CA). The samples were prepared as described for Western blot analysis or were sampled directly from the size exclusion column. Before imaging, a 10-μl sample was deposited onto freshly cleaved mica (Ted Pella, Redding, CA) and incubated for 1 min. The substrate was washed with 200 μl of ultrapure water and dried under a gentle steam of air. The images were taken with silicon cantilevers (Veeco Instruments, Santa Barbara, CA) with a nominal spring constant of 40 N/m and resonance frequency of ∼300 kHz. Typical imaging parameters were: drive amplitude, 150–500 kHz with set points of 0.7–0.8 V; scan frequencies, 2–4 Hz; image resolution, 512 × 512 points; and a scan size of 2–10 μm. The number of aggregates and aggregate size were determined with custom programs written with MATLAB (MathWorks, Natick, MA) equipped with the image processing toolbox. The programs locate individual objects in an AFM image and measure their volumes, heights, and other geometric characteristics. Geometrical simulations were used to compensate in part for contributions because of the finite shape and size of the tip, as described (27).
Analysis of Mutant htt Aggregation by SEC and Dot-blots
Supernatant from aggregation reactions (37 °C) obtained by centrifugation for 15 min at 20,000 × g were fractionated by SEC on a Superdex 200 10/30 column. To quench the kinetics of aggregation, all of the SEC experiments were performed at 4 °C and with a flow rate of 0.5 ml/min. The sample volume applied was 500 μl. The HR Suderdex200 10/30 column has an internal diameter of 10 mm. The height of the packed bed is 30–31 cm. The total bed volume is ∼24 ml. The sharp peaks with protein standards shows the resolving power of this column in the molecular weight range between 13 and 545 kDa (supplemental Fig. S1A). Fractions (500 μl volume/fraction) were collected and applied to a nitrocellulose membrane (0.1-μm pore size; Schleicher & Schuell) through a slot blot manifold (Hoefer PR 648) without any washing steps or detergent treatment. Both soluble monomeric and aggregated forms of HD53Q were retained in a nondenatured state on this charged, strong protein-binding membrane and analyzed with the antibodies as indicated. The amount of protein per fraction of HD53Q aggregates was quantified by densitometry of developed blots with NIH Image J. The effects of Hsp70/Hsp40 were analyzed in the presence of 3 mm ATP, 5 mm MgCl2, and an ATP-regenerating system (200 μg/ml creatine kinase and 20 mm creatine phosphate).
Hsp70/Hsp40 Mutant htt Oligomer Interaction Assay
To analyze the interaction between Hsp70/Hsp40 and soluble mutant htt oligomers, HD53Q proteins were incubated at 6 μm in 20 mm Tris-HCl, pH 7.4, 20 mm KCl, 5 mm MgCl2 at 37 °C with shaking at 800 rpm for 3 h. Chaperones with 3 mm ATP and an ATP-regenerating system were added, and the reaction was incubated for an additional 45 min. Protein cofractionation was determined by SEC as described (28, 29). Fractions (500 μl each) were collected, and the proteins were precipitated with methanol/chloroform (30).
Luciferase Refolding Assay
Firefly luciferase (L1792; Sigma) was diluted in unfolding buffer (40 mm Hepes-KOH, pH 7.5, 6 m guanidinium HCl) to a concentration of 1 mm and denatured by incubation at 25 °C for 30 min. Denatured luciferase was diluted (final concentration, 100 nm) into refolding buffer (25 mm HEPES-KOH, pH 7.5, 150 mm KCl, 20 mm MgAc, 5% glycerol, 2 mm ATP, and ATP-regenerating system) containing 3 μm Hsp70 and/or 3 μm Hsp40. The reaction mixtures were incubated for various times at 30 °C, and luciferase activity was measured in a luminometer (Monolight 2010; Pegasus Scientific, Rockville, MD) for 10 s after the addition of substrate (Promega).
For the competition assay, denatured luciferase (100 nm) was diluted into aggregation reactions of 6 μm HD53Q at 3 h (conditions where oligomers predominate) or 5 h (conditions where fibrils predominate) or bovine serum albumin (6 μm) that also forms dimers, trimers, and higher oligomers. Hsp70 and Hsp40 were added to the aggregation reaction 20 min before denatured luciferase was added. Refolding activity was measured after 30 min of incubation.
Analysis of Mutant htt Aggregation in PC12 Cells
The 14A2.5 PC12 line was generated and propagated as described (31). This cell line expresses a truncated htt peptide containing the first 17 amino acids of htt exon 1 and 103Qs fused in-frame to EGFP (31). The cells were maintained in complete medium with continued selection: Dulbecco's modified Eagle's medium (5% glucose) with 10% horse serum, 5% fetal bovine serum, 1% penicillin/streptomycin, and 200 mg/ml G418. Mutant htt expression was induced with ponasterone A (5 μm) for the indicated times. Plasmid pCMV40 was used to express Hsp40, and plasmid pCMV70 was used to express human inducible Hsp70 (32). The cells were transfected with 2 μg of pCMV40 and pCMV70 by the standard Lipofectamine method. One day after transfection, the cells were treated with ponasterone A. After the indicated time points, the cells were lysed in passive lysis buffer (E194A; Promega, Madison, WI) with protease inhibitor mixture (11697498001; Roche Applied Science). Cell lysate (60 μg) was analyzed by SEC and Western blotting as described above.
Cell Toxicity Assay
PC12 cells were harvested from 24-well plates and washed with PBS. Caspase 3 assays were performed in duplicate using EnZCheck caspase 3 assay kit no. 1 (Invitrogen/Molecular Probes) as described by the manufacturer. PC12 cells were not used if above ∼70% confluency, and caspase activity was normalized to total protein concentration.
Statistical Analysis
The samples were analyzed by Student's t test or by analysis of variance. Statistically significant differences (*, p < 0.05; **, p < 0.01; and ***, p < 0.001) are marked with asterisks.
RESULTS
Antibody A11 Detects Mutant htt Oligomers but Not Monomers or SDS-insoluble Aggregates
To identify specific oligomers of mutant htt fragments, we tested the ability of the conformation-dependent antibody A11 to detect oligomers, monomers, or higher ordered aggregates of mutant htt in vitro. We purified proteins from E. coli encoding the first exon of htt with polyQ repeats in the normal (HD20Q) and pathogenic (HD53Q) range as fusions to GST (Fig. 1A). After purification, uncleaved GST-HD53Q eluted as an apparent dimer (supplemental Fig. S1B), was detergent-soluble and appeared to be unaggregated by AFM analysis (Fig. 1B; t = 0). Incubation of the fusion proteins with a site-specific protease at 37 °C resulted in the removal of the GST moiety, release of the htt fragments, and their rapid aggregation into several oligomers with a broad size distribution in the SEC profile (5–600 kDa) (supplemental Fig. S1C) and AFM images (Fig. 1B).
To characterize oligomer formation by HD53Q fragments, we used dot-blot, filter trap, and SDS-PAGE/Western blotting assays. In the dot-blot assay, reaction products were applied to a nitrocellulose membrane at specific times after release of HD53Q from GST and probed with A11 as well as an antibody that recognizes a c-Myc epitope at the N terminus and MW1 that recognizes the polyQ tract of HD20Q and HD53Q (Fig. 1, A and C). Similar levels of HD20Q and HD53Q were detected by anti-c-Myc at each time point (Fig. 1C). However, higher levels of A11 reactivity were observed with HD53Q relative to HD20Q (Fig. 1, C and D). Interestingly, MW1 detects soluble HD20Q and HD53Q species at early time points but not insoluble forms at 20 h. In a filter trap assay (an assay that detects large (>0.45 μm), SDS-insoluble aggregates like fibrils), A11 and MW1 did not detect SDS-insoluble HD53Q aggregates that were readily detected by MW8 (Fig. 1, E and F), an antibody that detects insoluble mutant htt aggregates and inclusion bodies (1). A11 did not detect HD53Q monomers by SDS-PAGE/Western blot analysis (data not shown).
A Subset of Soluble Mutant htt Oligomers Reacts with A11
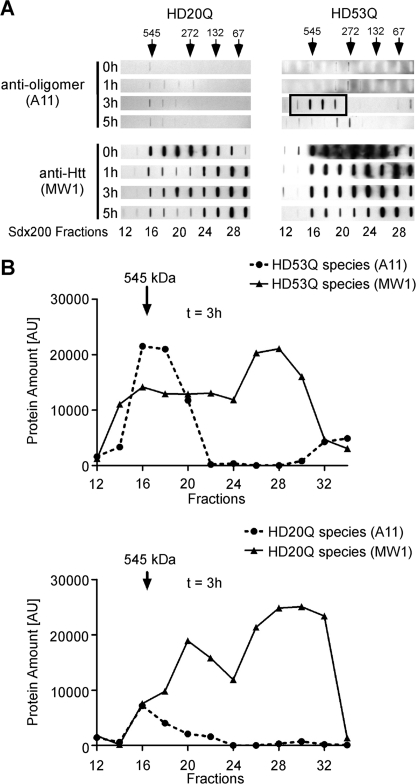
To determine whether A11 recognizes soluble oligomers formed by HD53Q, we fractionated soluble mutant htt oligomers by SEC. 500-μl HD20Q and HD53Q aggregation reactions were centrifuged at 20,000 × g, 500 μl of supernatant was loaded onto an HR Superdex 200 10/30 column at different time points of aggregation, and 500-μl aliquots collected from column fractions were directly applied to a nitrocellulose membrane (100-nm pore size) in the absence of detergent or heat denaturation. The membranes were probed with antibodies A11 and MW1. After cleavage, MW1 recognized a broad range (67–545 kDa) of oligomers formed by HD20Q and HD53Q at all time points of aggregation (Fig. 2 and supplemental Fig. S1C). However, A11 detected only HD53Q oligomers of ∼272–545 kDa that formed at 3 h (Fig. 2 and supplemental Fig. S1C). A11 also detected low levels of HD20Q at 3 h, but not at later time points (Fig. 2).
FIGURE 2.
A subset of soluble mutant htt oligomers are detected by the A11 antibody. Supernatant obtained after aggregation of HD53Q and HD20Q (6 μm) for the indicated times was analyzed by SEC on a Superdex 200 column. A, nondenatured fractions were vacuumed onto a nitrocellulose membrane using a slot-blot manifold and probed with anti-htt (MW1) and A11 antibodies. Arrows indicate positions of molecular mass markers (kDa). The black box indicates A11 detection of HD53Q aggregates at 3 h. B, antibody detection, quantified by densitometry with Image J. Soluble htt species were quantified with MW1 and compared with htt oligomers detected by A11.
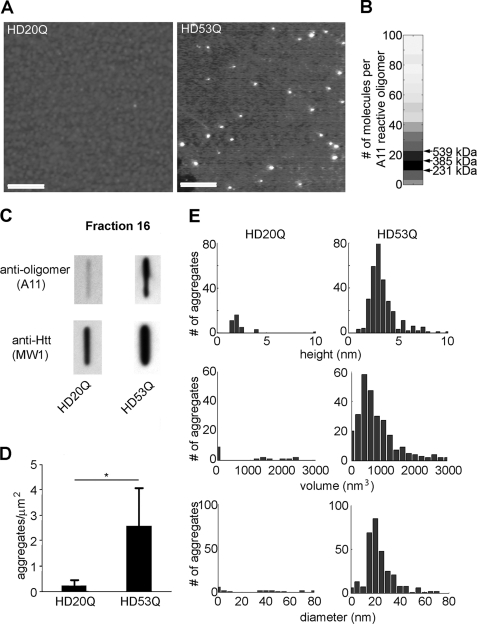
To analyze the morphology of this subset of soluble HD53Q oligomers, we used AFM to examine the A11-reactive fraction 16 (∼500 kDa) from the aggregation reaction at 3 h (Fig. 3 and supplemental Fig. S2). Oligomeric aggregates were readily detected in this fraction but were rarely observed in the equivalent fraction from the HD20Q sample (Fig. 3A). To estimate the number of protein molecules per A11-reactive HD53Q oligomer, we used the corrected volume distributions of oligomeric species in fractions that showed A11 reactivity. The volume of an individual HD53Q protein was estimated based on its molecular weight and the average density of protein (33, 34). By dividing the corrected volume of each individual aggregate by the volume of a single monomer, the number of molecules per each oligomer was estimated with the assumption of perfect packing of individual monomers within the oligomer. Using these analyses, we estimated that 10–25 molecules per HD53Q oligomer formed in a size range of 230–530 kDa (Fig. 3B). Quantification of multiple independent AFM images showed ∼10-fold more oligomers in the HD53Q sample than in the HD20Q sample (p < 0.01; Fig. 3D). HD53Q oligomers had globular structures with average heights of 3–5 nm, diameters of 20–25 nm, and volumes of 400–800 nm3 (Fig. 3, A and E), consistent with past measurements of HD53Q oligomer size (2, 7).
FIGURE 3.
A11 detects soluble mutant htt oligomers with a globular structure. A, AFM images of SEC fraction 16 from HD20Q and HD53Q aggregation reactions (3 h) (see Fig. 2). Scale bars, 400 nm. B, subunit composition of A11-reactive HD53Q oligomers. Based on corrected volume measurement and the molecular mass of HD53Q, the numbers of molecules/oligomers in A11-reactive fractions were calculated for each fraction from AFM images. Darker shades represent a greater abundance of oligomers composed of that number of molecules. Arrows indicate the size range where ∼200–500-kDa A11 reactive oligomers would be observed. C, fraction 16 was applied to nitrocellulose (100-nm pore size) through a slot-blot manifold and analyzed with A11 and MW1 antibodies. D, number of aggregates/μm2 in AFM images. The values are the means ± S.E. E, height, volume, and diameter histograms of oligomers from fraction 16 observed by AFM.
A11-reactive Mutant htt Oligomers Form in a polyQ Length-dependent Manner
If A11-reactive oligomers are pathogenic in HD, their formation, abundance, and/or persistence should correlate to polyQ length. To determine the effects of polyQ length on the formation of A11-reactive oligomers, we purified mutant htt fragments with increasing polyQ lengths (HD20Q, HD35Q, HD46Q, and HD53Q) and used SEC followed by direct application of the fractions to nitrocellulose membrane to measure A11 reactivity to mutant htt oligomers after 3 h of aggregation at 6 μm. Indeed, A11 reacted with oligomers formed by HD46Q and HD53Q but not with those formed by mutant htt fragments with nonpathogenic polyQ expansions (HD20Q or HD35Q) (supplemental Fig. S3). Thus, the formation of A11-reactive oligomers by mutant htt correlates tightly with polyQ length.
Anti-oligomer Antibody A11 Does Not React with Full-length Hsp70
Previously, we showed by AFM that Hsp70 and Hsp40 suppress formation of mutant htt oligomers but do not destabilize preformed oligomers (7), suggesting that these chaperones act on mutant htt monomers or possibly early soluble oligomers. Therefore, we wanted to determine whether Hsp70 and Hsp40 modulate A11-reactive mutant htt oligomers. However, we first wanted to ensure that A11 does not react directly with Hsp70 or Hsp40. In a dot-blot assay, 2 μg of recombinant Hsp40 or of Hsp70 purified from bovine liver (that was fully active in luciferase refolding assays) did not react with A11 (supplemental Fig. S4A), in contrast to the results of a previous study (35). Interestingly, the purified substrate-binding domain of Hsp70 alone was strongly A11-reactive (supplemental Fig. S4A). SEC analysis and small angle x-ray scattering revealed that the substrate-binding domain of Hsp70 readily oligomerizes but that of full-length Hsp70 does not (supplemental Fig. S4B and data not shown).
A11 reportedly has chaperone-like refolding activity (35). In a classical refolding assay for molecular chaperones, Hsp70 and Hsp40 robustly refolded denatured luciferase in the presence of ATP, but A11 did not (supplemental Fig. S4C). Moreover, stoichiometric concentrations of A11 did not impair luciferase refolding by Hsp70 and Hsp40.
Hsp70 and Hsp40 Inhibit Mutant htt Aggregation in an ATP-dependent Manner
Although we showed that Hsp70 and Hsp40 inhibit oligomer formation by a mutant htt fragment in an ATP-dependent manner (7), we did not know whether they functionally interact with soluble mutant htt oligomers. To confirm the ATP dependence of the inhibition, we added Hsp70, Hsp40 (DNAJB1), or both, along with protease, to mutant htt aggregation reactions at time 0. Coincubation of 6 μm HD53Q with an equimolar concentration of Hsp70 and Hsp40 in the presence of ATP and an ATP-regenerating system markedly reduced the formation of insoluble aggregates, shown by Western blotting with MW8 antibody (supplemental Fig. S5A). Hsp40 or Hsp70 alone inhibited mutant htt aggregation less efficiently, and ATP was necessary for chaperone-mediated inhibition of mutant htt aggregation (supplemental Fig. S5A).
Hsp70 and Hsp40 Interact Dynamically with Soluble Mutant htt Oligomers in an ATP-dependent Manner Mediated by Hsp40
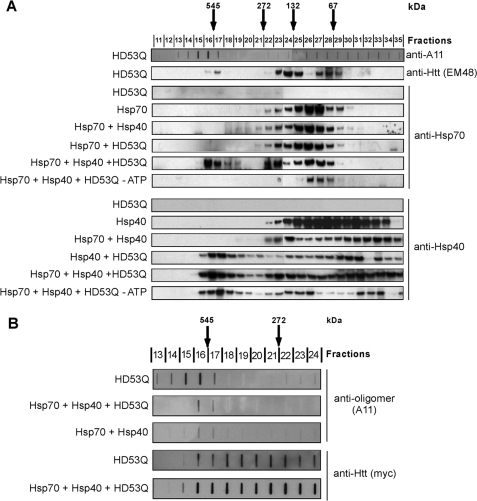
Previous studies by us (7, 15) and others (17, 23, 36) used indirect approaches to suggest that Hsp70 and Hsp40 interact directly with monomeric or early oligomeric forms of mutant htt. Here we wanted to determine whether Hsp70 and Hsp40 interact directly with soluble oligomers. After allowing HD53Q to aggregate for 3 h (under conditions in which HD53Q predominantly forms oligomers), Hsp70 alone or in combination with Hsp40, and with or without ATP, was added to the reaction and incubated for 45 min. Aggregation reactions were centrifuged at 20,000 × g for 30 min, and the supernatants were separated on a Superdex 200 column. Analysis by SDS-PAGE and Western blotting with mutant htt antibody (EM48) detected oligomeric HD53Q species in several fractions, including species that eluted at an apparent molecular mass of ∼500 kDa (Fig. 4A) and overlapped partially with oligomers in fractions detected by A11. When a mixture of HD53Q oligomers and Hsp40 was run on SEC, a significant amount of Hsp40 coeluted with mutant htt oligomers (Fig. 4A), consistent with the formation of a stable complex of the two proteins. This binding was independent of ATP. When Hsp70 alone was incubated with HD53Q oligomers and examined by SEC, no comigration was observed; however, when added together, Hsp70 and Hsp40 coeluted with high molecular mass mutant htt oligomers (∼500 kDa) in a trimeric complex that was not formed in the absence of ATP (Fig. 4A and supplemental Fig. S5B). Thus, Hsp40 recruits Hsp70 to mutant htt oligomers, an interaction that is likely to be functional, given its strict dependence on ATP.
FIGURE 4.
Hsp70 and Hsp40 associate specifically with soluble mutant htt oligomers and attenuate their formation in an ATP-dependent manner. A, analysis of the Hsp70/Hsp40 interaction with soluble HD53Q oligomers. Supernatant of HD53Q aggregation reaction (3 h) alone or a mixture of HD53Q with Hsp70, Hsp40, or both, was loaded onto a Superdex 200 column, and the fractions were analyzed by SDS-PAGE/Western blot with the indicated antibodies. Molecular chaperones were added after 3 h of HD53Q aggregation (when oligomers predominate) and incubated for 45 min before fractionation by SEC. Arrows indicate positions of molecular mass standards (kDa). B, effect of Hsp70/Hsp40 on formation of A11-reactive oligomers. Supernatants from 3 h HD53Q aggregation reactions in the absence or presence of Hsp70 and Hsp40 (chaperones added at 0 h) were loaded onto a Superdex 200 column. Fractions were placed on a nitrocellulose membrane (100-nm pore size) and probed with A11 or anti-htt (Myc) antibodies.
Hsp70 and Hsp40 Inhibit Formation of A11-reactive Mutant htt Oligomers in Vitro
We next used SEC to analyze the effects of Hsp70 and Hsp40 on the formation of A11-reactive mutant htt oligomers when added prior to initiating aggregation reactions. Interestingly, these chaperones inhibited only the formation of the A11-reactive subset of oligomers (Fig. 4B). Surprisingly, the levels of other oligomeric species detected with an anti-htt antibody (c-Myc) were unchanged (Fig. 4B). These results suggest that Hsp70 and Hsp40 either suppress the formation of A11-reactive mutant htt oligomers or refold them to a conformation that is not recognized by A11.
Mutant htt Oligomers Inhibit Hsp70 and Hsp40 Refolding Activity in a Dose-dependent Manner
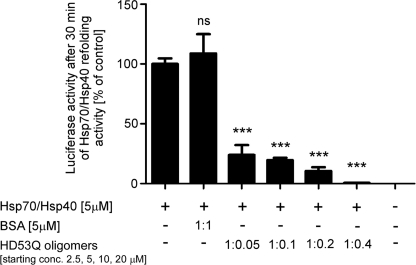
We then determined whether mutant htt oligomers influence the refolding activity of the chaperones by competing with luciferase as a substrate. Although BSA (nondenatured) did not affect Hsp70/Hsp40-dependent luciferase refolding, mutant htt oligomers (formed after 3 h of aggregation) potently inhibited chaperone-mediated refolding activity in a dose-dependent manner (Fig. 5). This inhibition is probably due to direct interaction with the chaperones because mutant htt oligomers did not affect intrinsic luciferase refolding activity, nor did they affect the native luciferase activity (supplemental Fig. S5, C and D). We calculated the oligomer concentration by estimating that A11 oligomers have a subunit composition of ∼10 molecules/oligomer after 3 h of aggregation (Fig. 3B). A starting concentration of 20 μm HD53Q monomers for the aggregation reaction (chaperone concentration in the refolding reaction was 5 μm) results in an oligomer:chaperone molar ratio of 1:2.5. Therefore, a mutant htt oligomer concentration of less than half of the chaperone concentration results in complete inhibition of refolding activity (Fig. 5), suggesting that more than one chaperone complex binds to each mutant htt oligomer. Fibrils also inhibited chaperone-dependent luciferase refolding, but to a lesser extent than oligomers (data not shown).
FIGURE 5.
Mutant htt oligomers inhibit Hsp70/Hsp40-dependent luciferase refolding. Denatured luciferase (100 nm) refolded by Hsp70/Hsp40 after 30 min in the presence and absence of HD53Q oligomers from a 3-h aggregation reaction or control protein. The molar ratios (chaperone:oligomer) were estimated based on predicted subunit composition of A11 oligomers after 3 h of aggregation (∼10 molecules/oligomer) in Fig. 3B. The values are the means ± S.E. of three independent experiments. ***, p < 0.001; ns, not significant (one-way analysis of variance).
Expression of Hsp70 and Hsp40 Inhibits the Formation of A11-reactive Mutant htt Oligomers and Toxicity in PC12 Cells
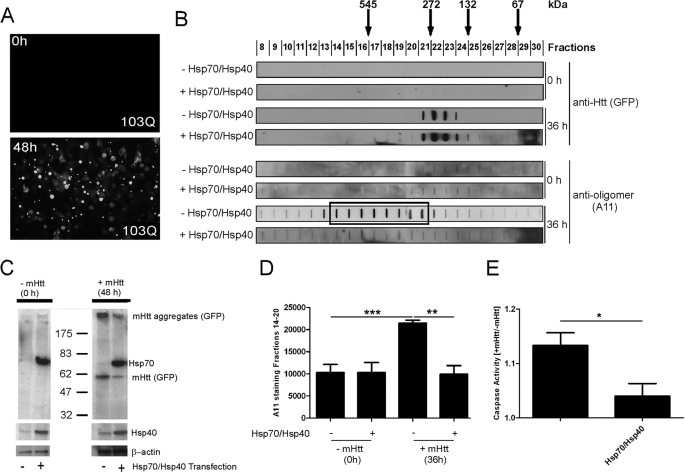
To determine whether mutant htt forms A11-reactive oligomers in mammalian cells, we examined soluble lysates from PC12 cells that stably and inducibly express a mutant htt fragment fused at the C terminus to enhanced GFP (HD103Q-EGFP) that leads to insoluble aggregate and inclusion body formation (31) (Fig. 6A and supplemental Fig. S6A). Lysates were analyzed by SEC before and 36 h after HD103Q-EGFP induction (SDS-insoluble aggregates first appear at 12 h and reach maximum at 48 h). Slot-blot analysis with an antibody to GFP showed that HD103Q-EGFP formed oligomers that fractionated by SEC at apparent molecular masses of ∼100–400 kDa (Fig. 6B and supplemental Fig. S6B). In contrast, A11 detected oligomers of ∼270–600 kDa (Fig. 6B, boxed area, and supplemental Fig. S6B), at similar apparent molecular masses to that observed with HD53Q under cell-free conditions (Fig. 2A). Because only a minority of the A11-reactive fractions were detected with a GFP antibody (supplemental Fig. S6B) but were readily detected with other htt-specific antibodies (data not shown), we presume that the GFP epitope is masked or partially occluded in the majority of A11-reactive oligomers in PC12 cells.
FIGURE 6.
Hsp70 and Hsp40 suppress formation of A11-reactive mutant htt oligomers and toxicity in PC12 cells. A, analysis of HD103Q-EGFP inclusion bodies by fluorescence microscopy. The addition of hormone (5 μm ponasterone A) to cell medium induced HD103Q-EGFP expression and caused the formation of GFP-positive inclusion bodies. B, the soluble supernatant of a homogenate from HD103Q-expressing PC12 cells (60 μg) in the absence and presence of Hsp70/Hsp40 overexpression was fractionated by SEC on a Superdex 200 column. Fractions were analyzed by slot-blot manifold with GFP and A11 antibodies. The boxed area indicates A11-reactive fractions. C, Western blot analysis of PC12 cell lysate (20 μg) with anti-GFP, anti-Hsp70 (inducible form), and anti-Hsp40 antibodies before and after HD103Q-EGFP expression. Overexpression of Hsp70 and Hsp40 increased the level of these chaperones and slightly reduced insoluble HD103Q-EGFP aggregates that remain in the well (anti-GFP). D, A11 reactivity in fractions 14–20, quantified by densitometry. The values are the means ± S.D. of three experiments. **, p < 0.01; ***, p < 0.001 (Student's t test). E, expression of a mutant htt fragment in PC12 cells causes an increase in caspase 3 activity that is suppressed when Hsp70/Hsp40 is overexpressed. Caspase 3 activity is shown as the ratio of values from induced relative to noninduced cells.
We next cotransfected PC12 cells with Hsp70 and Hsp40 plasmids driven by the CMV promoter to determine whether these chaperones influence the formation of HD103Q-EGFP oligomers. Overexpression of Hsp70 and Hsp40 reduced the total levels of SDS-insoluble mutant htt as observed by Western analysis (Fig. 6C), confirming findings in yeast and flies (15, 16). Interestingly, Hsp70/Hsp40 overexpression significantly decreased the levels of A11-reactive oligomers but not the levels of lower molecular mass oligomers detected with a GFP antibody (Fig. 6, B and D).
Next, we investigated the effect of molecular chaperones on mutant htt toxicity in these cells. Although mutant htt does not cause cytotoxicity per se in this cell line, caspase 3 activity increases progressively up to 4 days after induction relative to control cells (37). Overexpression of Hsp70 and Hsp40 suppressed caspase 3 activity to levels found in noninduced cells (Fig. 6E). Thus, soluble A11-reactive oligomers form in PC12 cells in a manner that correlates with toxic metabolic changes and levels of Hsp70 and Hsp40.
DISCUSSION
In this study, we show that a mutant htt fragment with an expanded polyQ repeat formed soluble oligomers of 60–600 kDa in vitro and in cells. These oligomers were fractionated readily by SEC, and a subset of them displayed an epitope recognized by the anti-oligomer antibody A11. In addition, the molecular chaperones Hsp70 and Hsp40 acted cooperatively in an ATP-dependent manner to interact with mutant htt oligomers and interfere with their formation.
Soluble mutant htt oligomers resolved by SEC displayed differential reactivity to a panel of monoclonal antibodies, suggesting that they are misfolded into discrete higher order structures in which specific epitopes are exposed, buried, or absent. Reactivity to some antibodies also changed with incubation time in vitro and in PC12 cells. Thus, soluble mutant htt oligomers appear to be highly dynamic and heterogeneous structures, consistent with recent AFM analyses (2). In our recent studies, monoclonal antibodies that recognize expanded polyQ repeats had diverse effects on the aggregation of monomeric mutant htt (1). This finding suggested that the polyQ repeat in mutant htt monomers also samples different conformations that can be discriminated readily by antibodies. Compelling support for this hypothesis came from a study in which x-ray crystallography showed that the polyQ repeat in a wild-type htt fragment stably adopts multiple independent secondary structures, including random coil and α-helix (38, 39). In line with these studies, one attractive hypothesis is that it is the chameleon-like nature of mutant htt that results in the numerous abnormal protein-interactions that mediate pathogenesis in HD. However, some subsets of misfolded mutant htt conformations might be more chemically reactive/toxic than others, depending on their secondary, tertiary, and quaternary structures. Consistent with this hypothesis, here we showed that soluble mutant htt oligomers of ∼272–545 kDa, but not smaller oligomers, react with the conformation-specific antibody A11, which neutralizes the toxicity of oligomers formed by synthetic polyQ peptides (12).
Molecular chaperones (e.g. Hsp70 and Hsp40) ensure that proteins fold properly and prevent them from misfolding and are thus potent suppressors of polyQ toxicity in vivo (16, 18). However, the molecular mechanisms by which they suppress the toxicity of mutant htt are not clear. Our previous studies suggested that Hsp70 and Hsp40 attenuate oligomer formation by partitioning monomers (7). Here we showed that these chaperones also associate directly and dynamically with soluble mutant htt oligomers. Importantly, these chaperones had variable affinities for different subsets of oligomers. Hsp70 and Hsp40 were completely absent from some SEC fractions that contained mutant htt oligomers and were most strongly associated with oligomers in fractions that also showed A11 reactivity. Moreover, when overexpressed in PC12 cells expressing a toxic mutant htt fragment, Hsp70 and Hsp40 suppressed the formation of A11-reactive oligomers but not other mutant htt oligomers coincident with suppression of caspase 3 activation. In yeast and mammalian cells, the chaperonin TRiC suppresses mutant htt aggregation and toxicity (40, 41). An independent study in yeast showed that TRiC interacts synergistically with Hsp70/Hsp40 to modulate the formation of A11-reactive oligomers by mutant htt (24, 41). Although confirmation of these studies in more physiologically relevant models of HD is needed, these results are consistent with the hypothesis that soluble A11-reactive mutant htt oligomers might be a pathogenic conformer that is recognized by these chaperones.
Hsp70 and Hsp40 suppressed mutant htt aggregation far more efficiently then either chaperone alone, as reported (15–17). However, we demonstrated that Hsp70 alone does not interact with soluble oligomers. Rather, the interaction of Hsp70 with mutant htt oligomers was mediated entirely by Hsp40 (DNAJB1), which is required for productive folding of model substrates by Hsp70 (26, 42–44). In these reactions, Hsp40 (the more abundant protein) stabilizes the substrate in a conformation suitable for interaction with Hsp70, which then transiently interacts with the substrate in a manner that allows it to reach a folding- or degradation-competent state (43, 44). Importantly, just as with model substrates, ATP was required for the interaction between Hsp70/Hsp40 and soluble mutant htt oligomers. Given the conformational flexibility exhibited in mutant htt monomers, it is tempting to speculate that Hsp40 recognizes an unfolded/misfolded motif in mutant htt exposed on the surface of soluble oligomers and that Hsp70 refolds this sequence in an ATP-dependent manner to a nontoxic fold, thus converting a toxic oligomer to one that is benign (Fig. 7). Consistent with our results, a recent study showed in mammalian cells that DNAJB members of the Hsp40 family might play a central role in early recognition and binding of polyQ protein aggregates (36).
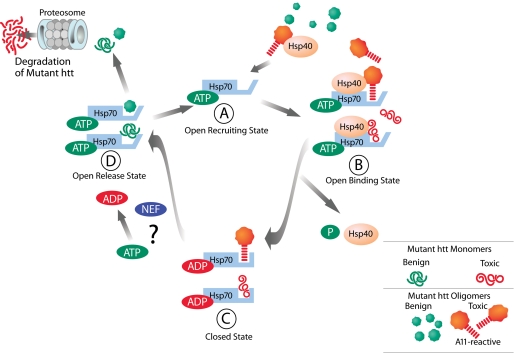
FIGURE 7.
Proposed model for Hsp70/Hsp40 function in mutant htt misfolding in HD. Expansion of the polyQ repeat of mutant htt causes misfolding and formation of discrete subsets of misfolded species (toxic and benign monomers and oligomers), with only toxic conformations recognized specifically by Hsp40 (DNAJB). A, Hsp40 transfers mutant htt substrates to ATP-bound Hsp70 (open conformation). B, in the open state, Hsp70, with Hsp40, forms a trimeric complex with misfolded mutant htt species, and Hsp40 stimulates the ATPase activity of Hsp70. Hydrolysis of ATP leads to ADP-Hsp70 bound state (closed state) and release of Hsp40 and Pi. C, in the ADP-bound state, Hsp70 shows a high affinity for misfolded mutant htt species. Nucleotide exchange factors (NEF) likely mediate the exchange of ADP for ATP, resulting in an open state of Hsp70 and conversion of toxic to benign conformers. D, the benign conformers are transferred to another chaperone system or are competent for degradation by the proteasome or by autophagy.
We propose a model of how Hsp70 and Hsp40 modulate mutant htt misfolding in HD (Fig. 7). Expansion of the polyQ repeat in mutant htt causes misfolding and formation of different subsets of intermediate species (monomers and oligomers), some of which adopt toxic conformations recognized by Hsp40 that are presented to Hsp70 (Fig. 7A). Hsp40 would then stimulate the ATPase activity of Hsp70, resulting in the formation of a high affinity ADP·Hsp70·mutant htt oligomer complex (Fig. 7, B and C). Although not addressed in this study, it is likely that nucleotide exchange factors are responsible for ATP replacement and release of benign mutant htt conformations that can either be degraded or transferred to other chaperone systems (Fig. 7D).
One perplexing feature of HD and other polyQ disorders is their relatively late age of onset. Molecular chaperones and other factors with important roles in proteostasis (45) appear to robustly cope with the burden of mutant htt misfolding for decades but clearly become overwhelmed as a function of age. Because the activities of most molecular chaperones and the proteasome are strictly ATP-dependent, the late onset of HD might reflect, in part, pronounced deficits in ATP production in HD patients (46). Therefore, in a symptomatic state in HD, decreased ATP levels would lead to a decrease in productive interactions between Hsp70 and substrates and would increase the concentration of ADP-bound Hsp70 (closed state), causing it to become trapped in complexes with misfolded mutant htt conformations or other Hsp70 substrates that would lead to Hsp70 aggregation/sequestration with mutant htt aggregates. Consistent with such a model, Hsp70 coaggregates with α-synuclein in the presence of ADP (47). Moreover, when combined with impairment of the protein degradation systems that is thought to occur in HD and related disorders, decreased chaperone function would lead to an overwhelming increase of toxic mutant htt species and cell death. In parallel, the accumulation of benign mutant htt aggregates (such as amorphous and fibrillar structures) under these conditions may initially represent a cellular coping response to prevent an increase of toxic oligomers. It is also important to note that under these conditions, all of the normal functions of chaperones in protein folding and other cellular processes are likely to become profoundly impaired (48).
In summary, we provide the first direct evidence that Hsp70 and Hsp40 functionally interact with subsets of soluble mutant htt oligomers that may play a pathogenic role in HD. Understanding the molecular mechanisms of chaperone functions in mutant htt misfolding may provide insights that will catalyze the development of novel strategies to combat this and other devastating neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank H. Kampinga for Hsp40 and Hsp70 mammalian expression plasmids, Kazutayo Terada for the Hsp40 (DNAJB1) construct for recombinant expression, Paul Patterson for MW antibodies, J. Miller for critically reading the manuscript, and G. Howard and S. Ordway for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS054753 and R01NS047237. This work was also supported by the HighQ Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- HD
- Huntington disease
- polyQ
- polyglutamine
- AFM
- atomic force microscopy
- SEC
- size exclusion chromatography
- EGFP
- enhanced GFP.
REFERENCES
- 1.Legleiter J., Lotz G. P., Miller J., Ko J., Ng C., Williams G. L., Finkbeiner S., Patterson P. H., Muchowski P. J. (2009) J. Biol. Chem. 284, 21647–21658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legleiter J., Mitchell E., Lotz G. P., Sapp E., Ng C., DiFiglia M., Thompson L. M., Muchowski P. J. (2010) J. Biol. Chem. 285, 14777–14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 4.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 5.Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M., Machida Y., Nishikawa Y., Akagi T., Hashikawa T., Fujisawa T., Nukina N. (2003) J. Biol. Chem. 278, 34717–34724 [DOI] [PubMed] [Google Scholar]
- 7.Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 8.Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 9.Mitra S., Tsvetkov A. S., Finkbeiner S. (2009) J. Biol. Chem. 284, 4398–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega Z., Díaz-Hernández M., Maynard C. J., Hernández F., Dantuma N. P., Lucas J. J. (2010) J. Neurosci. 30, 3675–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 12.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 13.Glabe C. G. (2008) J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muchowski P. J., Wacker J. L. (2005) Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 15.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warrick J. M., Chan H. Y., Gray-Board G. L., Chai Y., Paulson H. L., Bonini N. M. (1999) Nat. Genet. 23, 425–428 [DOI] [PubMed] [Google Scholar]
- 17.Jana N. R., Tanaka M., Wang G., Nukina N. (2000) Hum. Mol. Genet. 9, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 18.Cummings C. J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H. T., Dillmann W. H., Zoghbi H. Y. (2001) Hum. Mol. Genet. 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 19.Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M., Sobue G. (2003) J. Neurosci. 23, 2203–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wacker J. L., Huang S. Y., Steele A. D., Aron R., Lotz G. P., Nguyen Q., Giorgini F., Roberson E. D., Lindquist S., Masliah E., Muchowski P. J. (2009) J. Neurosci. 29, 9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erbse A., Mayer M. P., Bukau B. (2004) Biochem. Soc. Trans. 32, 617–621 [DOI] [PubMed] [Google Scholar]
- 22.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 23.Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F. U. (2004) Mol. Cell 15, 95–105 [DOI] [PubMed] [Google Scholar]
- 24.Behrends C., Langer C. A., Boteva R., Böttcher U. M., Stemp M. J., Schaffar G., Rao B. V., Giese A., Kretzschmar H., Siegers K., Hartl F. U. (2006) Mol. Cell 23, 887–897 [DOI] [PubMed] [Google Scholar]
- 25.Schlossman D. M., Schmid S. L., Braell W. A., Rothman J. E. (1984) J. Cell Biol. 99, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami Y., Höhfeld J., Ohtsuka K., Hartl F. U. (1996) J. Biol. Chem. 271, 19617–19624 [DOI] [PubMed] [Google Scholar]
- 27.Legleiter J., DeMattos R. B., Holtzman D. M., Kowalewski T. (2004) J. Colloid Interface Sci. 278, 96–106 [DOI] [PubMed] [Google Scholar]
- 28.Lotz G. P., Lin H., Harst A., Obermann W. M. (2003) J. Biol. Chem. 278, 17228–17235 [DOI] [PubMed] [Google Scholar]
- 29.Lotz G. P., Brychzy A., Heinz S., Obermann W. M. (2008) J. Cell Sci. 121, 717–723 [DOI] [PubMed] [Google Scholar]
- 30.Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 31.Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., Housman D. E., Marsh J. L., Thompson L. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5950–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michels A. A., Kanon B., Konings A. W., Ohtsuka K., Bensaude O., Kampinga H. H. (1997) J. Biol. Chem. 272, 33283–33289 [DOI] [PubMed] [Google Scholar]
- 33.Gekko K., Noguchi H. (1979) J. Phys. Chem. 83, [Google Scholar]
- 34.Squire P. G., Himmel M. E. (1979) Arch. Biochem. Biophys. 196, 165–177 [DOI] [PubMed] [Google Scholar]
- 35.Yoshiike Y., Minai R., Matsuo Y., Chen Y. R., Kimura T., Takashima A. (2008) PLoS ONE 3, e3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hageman J., Rujano M. A., van Waarde M. A., Kakkar V., Dirks R. P., Govorukhina N., Oosterveld-Hut H. M., Lubsen N. H., Kampinga H. H. (2010) Mol. Cell 37, 355–369 [DOI] [PubMed] [Google Scholar]
- 37.Apostol B. L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E. S., Olson J. M., Kazantsev A., Marsh J. L., Thompson L. M. (2006) Hum. Mol. Genet. 15, 273–285 [DOI] [PubMed] [Google Scholar]
- 38.Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J., Rutenber E., Muchowski P. J. (2009) Structure 17, 1151–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura A., Kubota H., Pack C. G., Matsumoto G., Hirayama S., Takahashi Y., Kimura H., Kinjo M., Morimoto R. I., Nagata K. (2006) Nat. Cell Biol. 8, 1163–1170 [DOI] [PubMed] [Google Scholar]
- 41.Tam S., Geller R., Spiess C., Frydman J. (2006) Nat. Cell Biol. 8, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman B. C., Morimoto R. I. (1996) EMBO J. 15, 2969–2979 [PMC free article] [PubMed] [Google Scholar]
- 43.Hartl F. U., Hayer-Hartl M. (2002) Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 44.Mayer M. P., Bukau B. (2005) Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 46.Browne S. E., Beal M. F. (2004) Neurochem. Res. 29, 531–546 [DOI] [PubMed] [Google Scholar]
- 47.Roodveldt C., Bertoncini C. W., Andersson A., van der Goot A. T., Hsu S. T., Fernández-Montesinos R., de Jong J., van Ham T. J., Nollen E. A., Pozo D., Christodoulou J., Dobson C. M. (2009) EMBO J. 28, 3758–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. (2006) Science 311, 1471–1474 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.