Abstract
Parkin is an E3 ubiquitin ligase that mediates the ubiquitination of protein substrates. The mutations in the parkin gene can lead to a loss of function of parkin and cause autosomal recessive juvenile onset parkinsonism. Recently, parkin was reported to be involved in the regulation of mitophagy. Here, we identify the Bcl-2, an anti-apoptotic and autophagy inhibitory protein, as a substrate for parkin. Parkin directly binds to Bcl-2 via its C terminus and mediates the mono-ubiquitination of Bcl-2, which increases the steady-state levels of Bcl-2. Overexpression of parkin, but not its ligase-deficient forms, decreases autophagy marker LC3 conversion, whereas knockdown of parkin increases LC3 II levels. In HeLa cells, a parkin-deficient cell line, knockdown of parkin does not change LC3 conversion. Moreover, overexpression of parkin enhances the interactions between Bcl-2 and Beclin 1. Our results provide evidence that parkin mono-ubiquitinates Bcl-2 and regulates autophagy via Bcl-2.
Keywords: Autophagy, E3 Ubiquitin Ligase, Mitochondria, Parkinson Disease, Ubiquitination, Bcl-2, Parkin
Introduction
Parkinson disease (PD)2 is the second most common neurodegenerative disorder after Alzheimer disease (1) and is characterized by a distinct set of motor symptoms including tremor, muscle rigidity, postural instability, and bradykinesia (2). Although the cause of PD is poorly understood, there is evidence that both environmental factors and genetic factors contribute to its development. Recently, several genes have been reported to be associated with the pathogenesis of familial forms of PD. Mutations in the parkin gene (PARK2; OMIM600116) cause autosomal recessive juvenile onset parkinsonism (3). It has been shown that mutations in parkin account for nearly 50% of patients with the early onset familial PD cases (3–6) and more than 15% of sporadic PD cases with early onset (7).
Parkin is a 465-amino acid protein that contains an ubiquitin-like domain at its N terminus and two RING finger domains separated by an in-between-ring domain at its C terminus. Similar to other RING finger-containing proteins, parkin is an E3 ubiquitin ligase. Parkin ubiquitinates several target proteins and enhances their degradation via the ubiquitin-proteasome system (8, 9). Ubiquitination of a substrate is usually processed by a multi-step involving the sequential activity of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin-protein ligase (10). It was reported that parkin can selectively interact with the E2 enzymes, UbcH7 and UbcH8 (9, 11, 12). A number of protein substrates for parkin have been identified, including synphilin-1 (13, 14), CDCrel-1 and 2a (12, 15), Pael-R (16), synaptotagmin XI (17), α- and β-tubulin (18), RanBP2 (19), cyclin E (20), the aminoacyl-tRNA synthetase cofactor p38/AIMP2 (21, 22), Eps15 (23), and far upstream sequence element-binding protein 1 (24). Within these substrates, p38/AIMP2 and far upstream sequence element-binding protein 1 were reported to be accumulated in brains of parkin null mice, MPTP (1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine) treated mice, and sporadic PD cases (22, 24).
Parkin is dominantly located in cytosol (25), whereas it is also located in mitochondria (26). It regulates mitochondrial morphology by the PINK/Parkin pathway (27) and is involved in mitochondria biogenesis in proliferating cells (28). It was recently reported that parkin promotes mitophagy by its recruitment to impaired mitochondria (29). However, the mechanism by which parkin functions in mitochondria or its substrates that are associated with mitochondria is still not clear.
Here, we identity the anti-apoptotic protein, Bcl-2, as a novel substrate for parkin. Parkin increases the amount of Bcl-2 by mediating its mono-ubiquitination to enhance its stability. Overexpression of parkin decreases, whereas knockdown of parkin increases, LC3 conversion under normal conditions or starvation treatment. Moreover, overexpression of parkin increases the binding of Bcl-2 and Beclin 1. This study reveals that parkin is involved in the regulation of autophagy via mono-ubiquitinating Bcl-2.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
For creating constructs expressing FLAG-parkin and EGFP-parkin, we cut out the full-length parkin cDNA at BamHI/SalI sites from pGEX-5x-1-parkin and subcloned this fragment into p3×FLAG-myc-CMV-24 vector (Sigma) or pEGFP-C2 vector (BD Biosciences Clontech). The deletion mutants of parkin were generated by PCR with different primers using pGEX-5X-1-parkin as the template. pGEX-5X-1-parkin (N terminus, amino acids 1–219) was created by subcloning the PCR product, amplified with primers 5′-CGGGATCCATATGATAGTGTTTGTCAGG-3′ and 5′-ACGCGTCGACTCAGTCAGAGGTGGGGTGTGC-3′, into pGEX-5X-1 (Amersham Biosciences); pGEX-5X-1-parkin (C terminus, amino acids 220–465), with primers 5′-CGGGATCCATAAGGAAACACCAGTAGTC-3′ and 5′-ACGCGTCGACCTACACGTCGAACCAGTG-3′; pGEX-5X-1-parkin (R1, amino acids 220–318), with primers 5′-CGGGATCCATAAGGAAACACCAGTAGCT-3′ and 5′-ACGCGTCGACTCAATACTGCTGGTACCGGTTGTA-3′; and pGEX-5X-1-parkin (R2, amino acids 385–465), with primers 5′-CGGGATCCATGGAACAACTACTCAGGCCTA-3′ and 5′-ACGCGTCGACCTACACGTCGAACCAGTG-3′, respectively. For creating constructs expressing EGFP-parkin-N terminus, EGFP-parkin-C terminus, EGFP-parkin-R1, and EGFP-parkin-R2, we cut out these deletion mutant parkin cDNAs at BamHI/SalI sites from pGEX-5X-1-parkin-N terminus, pGEX-5X-1-parkin-C terminus, pGEX-5X-1-parkin-R1, and pGEX-5X-1-parkin-R2 and then subcloned these fragments into pEGFP-C2 vector (BD Biosciences Clontech). Mutants of parkin (K161N, T240R, C431F, and P437L) were generated by site-directed mutagenesis using a MutantBEST kit (Takara), with primers 5′-TCTCAGGGTACAGTGCAG-3′ and 5′-TTTCCCGGCTGCACTCTTT-3′ for K161N; 5′-GGTGCACAGACGTCAGG-3′ and 5′-TAATGCAAGTGATGTTCCGAC-3′ for T240R; 5′-TCATGCACATGAAGTGTCCGCAG-3′ and 5′-AGCCTCCATTTTTTTCCACTGG-3′ for C431F; and 5′-TGCAGCCCCAGTGCAGGCTCGAGTGG-3′ and 5′-GACACTTCATGTGCATGCAGCCTCC-3′ for P437L, respectively.
Full-length Bcl-2 cDNA was first amplified by PCR using primers 5′-CGGAATTCATGECTCAGAGCAAC-3′ and 5′-GTCGACTCATTTCCGACTGAAG-3′ from a human fetal brain cDNA library (Clontech). The PCR product was then inserted into pET-21a (Novagen) or pEGFP-C2 (BD Biosciences Clontech) vectors via EcoRI/SalI sites.
In Vitro Binding Assays
For GST pulldown assays, an aliquot containing 20 μl of glutathione-agarose beads (GE Healthcare Life Sciences) was incubated with 20 μg of protein from the soluble fraction of Escherichia coli cell lysates containing GST or GST-parkin or GST-Bcl-xl for 30 min on ice. After washing with 1× PBS for three times to remove unbound materials, the beads were incubated with 50 μg of protein from the supernatants of E. coli crude extract containing Bcl-2 or Bcl-xl or Bax, which was expressed by pET-21a-Bcl-2, pET-21a-Bcl-xl, or pET-21a-Bax in 0.25 ml of HNTG buffer (20 mm Hepes-KOH, pH 7.5, 100 mm NaCl, 0.1% Triton X-100, and 10% glycerol) for 2 h at 4 °C. After incubation, the beads were washed seven times with 1 ml of 1× HNTG buffer. The proteins were eluted with 20 μl of SDS sample buffer and subjected to immunoblot analysis.
Cell Culture and Transfection
293 cells, SH-SY5Y cells, or HeLa cells were cultured in DMEM (GIBCO) containing 10% fetal bovine serum (Hyclone). The Lipofectamine 2000 reagent (Invitrogen) was used to transfect the cells with expressing plasmids according to the manufacturer's instructions. Forty-eight hours after transfection, the transfected cells were observed using an inverted system microscope IX71 (Olympus) or harvested for immunoblot or immunoprecipitation analyses. Dissociated HF cultures were prepared from postnatal 1-day-old Sprague-Dawley rats. The cells were gently dissociated with a plastic pipette after digestion with 0.5% trypsin (Invitrogen) at 37 °C. The dissociated cells were plated at a final density of 5 × 105/cm2 on polyethyleneimine-coated six-well plates (Corning) and cultured in Neurobasal medium (Invitrogen) containing 1×B27 supplement (Invitrogen) and 3 μg/ml glutamine (Sigma). Three days after culture, 5-fluoro-2′-deoxyuridine and uridine were added to a final concentration of 10 μm (Sigma) to repress the growth of glial cells.
Antibody Preparation
Polyclonal antisera against parkin were raised by immunizing New Zealand White rabbits with purified GST-parkin protein as an antigen.
RNA Interference
Double-stranded oligonucleotides targeting human parkin as described elsewhere (30). Double-stranded oligonucleotides targeting 5′-AACATCGCCCTGTGGATGACT-3′ of human Bcl-2 mRNA, or targeting 5′-TGCAAGGAAGCATACCAT-3′ of rat parkin mRNA were synthesized by Shanghai GenePharma (Shanghai, China), and an irrelevant oligonucleotide served as a negative control. The transfection was performed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Briefly, siRNA and Lipofectamine 2000 reagent (Invitrogen) were mixed in Opti-MEM medium (Invitrogen) and incubated for 30 min at room temperature to allow the complex formation. Then the cells were washed with Opti-MEM medium (Invitrogen), and the mixtures were added. Twelve hours after transfection, the culture medium was replaced by fresh complete medium. The cells were harvested 72 h after transfection, followed by further analysis.
Immunoblot Analysis
The proteins were separated by 12% SDS-PAGE and then transferred to polyvinylidene difluoride membrane (Millipore). The following primary antibodies were used. Monoclonal anti-Bcl-2 antibody was from Santa Cruz Biotechnology or Abcam. Monoclonal anti-FLAG antibody was from Sigma. Monoclonal anti-GAPDH antibody was from Chemicon. Monoclonal anti-GFP antibody was from Santa Cruz Biotechnology or Roche Applied Science. Monoclonal anti-HA, anti-P62, and anti-parkin antibodies were from Santa Cruz Biotechnology. Monoclonal anti-LC3 antibody was from Novus. Polyclonal anti-Beclin 1 antibodies were from Santa Cruz Biotechnology. The secondary antibodies, sheep anti-mouse IgG-HRP antibody or anti-rabbit IgG-HRP antibody, were from Amersham Biosciences. The proteins were visualized using an ECL detection kit (Amersham Biosciences).
To reprobe the membrane for another primary antibody, the membrane was first incubated with stripping buffer (50 mm Tris-HCl, pH 6.8, 0.1 mm β-mercaptoethanol, 20 mm SDS) for 30 min at 50 °C. Then the membrane was subjected to immunoblot analysis following the standard protocol.
Immunoprecipitation
293 cells co-expressing FLAG-parkin along with EGFP or EGFP-Bcl-2 were collected 48 h after transfection and sonicated in TSPI buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 1 mm EDTA, 1% Nonidet P-40 supplemented with complete mini protease inhibitor mixture (Roche Applied Science). Cellular debris was removed by centrifugation at 12,000 × g for 30 min at 4 °C. The supernatants were incubated with monoclonal anti-GFP antibody (Roche Applied Science) for 1 h at 4 °C. After incubation, protein G-Sepharose was used for precipitation. The beads were washed with TSPI buffer six times and then eluted with SDS sample buffer for immunoblot analysis.
Degradation Assay
Eighteen hours after transfection, 293 cells co-expressing FLAG-parkin along with EGFP or EGFP-Bcl-2 were treated with 100 μm cycloheximide (CHX) (Sigma) to inhibit protein synthesis. The cells were harvested at 0, 4, 8, 12, and 16 h after CHX treatment. The same volume of lysates was analyzed by immunoblot using anti-FLAG antibody (Sigma) and anti-GFP antibody (Santa Cruz).
Long-lived Protein Degradation Assay
HeLa cells expressing EGFP or EGFP-parkin were cultured in 6-well plates and labeled for 24 h at 37 °C with 65 μm concentrations of unlabeled leucine and 3H-labeled leucine (1 μCi/ml) (PerkinElmer Life Sciences) in DMEM containing 10% fetal bovine serum. The cells were then washed and incubated in medium containing excess cold leucine (2 mm) for 24 h to allow for degradation of short-lived proteins. These labeled cells were then incubated at 37 °C with either Essential Medium with Earle's Balanced Salts (Hyclone) or DMEM containing 10% fetal bovine serum. At 2, 4, and 6 h after starvation, 0.5 ml of the medium were taken out from each sample and mixed with 56 μl of 100% TCA. The samples were then vortexed and centrifuged at 12,000 × g for 5 min. The acid soluble radioactivity was determined by liquid scintillation counting. The cells were washed with PBS, and 1 ml of cold 10% TCA was added to fix the cellular proteins. The fixed cell monolayers were washed with 10% TCA and dissolved in 0.5 ml of 1 n NaOH at 37 °C. The radioactivity in each aliquot of 1 n NaOH was counted by liquid scintillation counting. The percentage of long-lived protein degradation at each time point was determined by the formula: % degradation = (3H counts at time point/sum of 3H counts at each time point + total cell-associated radioactivity) × 100.
RESULTS
Interactions of Parkin and Bcl-2
Because parkin is partially localized to mitochondria and is functionally associated with mitochondria (26, 28, 29), we questioned whether parkin interacts with Bcl-2 or its related proteins. To test this possibility, we performed GST pulldown assays to evaluate the binding of parkin with Bcl-2, Bcl-xl, or Bax using GST-parkin, which was coupled to glutathione-agarose beads to pull down Bcl-2 or Bcl-xl or Bax expressed by E. coli transformed with pET-21a-Bcl-2 or pET-21a-Bcl-xl or pET-21a-Bax. GST-parkin, but not GST alone, pulled down Bcl-2 (Fig. 1A) and Bcl-xl (Fig. 1B). However, it failed to pull down Bax (supplemental Fig. S1). To further confirm the interactions between parkin and Bcl-2, we performed co-immunoprecipitation assays using 293 cells expressing a combination of FLAG-parkin and EGFP-Bcl-2 or EGFP. FLAG-parkin was co-immunoprecipitated with EGFP-Bcl-2 using anti-GFP antibody but was not with EGFP (Fig. 1C). In similar co-immunoprecipitation experiments, Bcl-xl failed to be co-immunoprecipitated with EGFP-parkin (data not shown). These data suggest that parkin may interact only with Bcl-2, but not Bcl-xl, in mammalian cells, although parkin interacts with both Bcl-2 and Bcl-xl using recombinant proteins expressed by E. coli. To further examine the interactions between parkin and Bcl-2, we performed co-immunoprecipitation assays using mouse brain lysates with anti-parkin polyclonal antisera. Endogenous Bcl-2 was co-immunoprecipitated with endogenous parkin (Fig. 1D).
FIGURE 1.
Interactions of parkin and Bcl-2. A, GST-parkin interacts with Bcl-2 in vitro. The supernatant of E. coli crude extract containing recombinant Bcl-2 expressed by pET-21a-Bcl-2 was incubated with glutathione-agarose beads bound with GST or GST-parkin. After incubation, the beads were washed with HNTG buffer, and the bound proteins were probed with anti-Bcl-2 antibody. The lower panels show the inputs of GST, GST-parkin. B, GST-parkin interacts with Bcl-xl in vitro. Similar experiments as in A were performed using GST-parkin to pull down Bcl-xl, which is expressed by pET-21a-Bcl-xl. The lower panels show the inputs of GST, GST-parkin. C, FLAG-parkin is co-immunoprecipitated with EGFP-Bcl-2. 293 cells expressing a combination of EGFP-Bcl-2 and FLAG-parkin or FLAG were subjected to immunoprecipitation with anti-GFP antibody. Immunoprecipitants and inputs were detected with antibodies as indicated. GAPDH served as a loading control. D, endogenous parkin is co-immunoprecipitated with Bcl-2. Mouse brain lysates were subjected to immunoprecipitation with polyclonal antisera against parkin or empty rabbit sera. Immunoprecipitants and inputs were detected with antibodies to parkin and Bcl-2. The parkin and heavy chain bands are overlapped. ▴ indicates the nonspecific bands. E, similar experiments as in A were performed using GST-tagged parkin or its deletion mutants to pull down Bcl-2. Meanwhile, 293 cells expressing a combination of FLAG-Bcl-2 and EGFP-parkin or its deletion mutants were subjected to immunoprecipitation with anti-GFP antibody to further identify the binding domain of parkin with Bcl-2. IP, immunoprecipitation; IB, immunoblot.
To further determine which domain(s) of parkin are responsible for its interaction with Bcl-2, we generated different deletion mutants of parkin (Fig. 1E) and examined their interactions with Bcl-2 using GST pulldown assays. Our results showed that the full-length parkin, both the RING1 (R1) and RING2 (R2) domains and the C terminus containing two RINGs and an in-between-ring domain, pulled down Bcl-2, whereas the N terminus did not (Fig. 1E). Then we performed co-immunoprecipitation assays using 293 cells expressing a combination of FLAG-Bcl-2 and EGFP-parkin or its deletion mutants or EGFP. Our results showed that only full-length parkin and its RING1 domain interacted with Bcl-2, whereas the RING2 domain only had a very weak binding with Bcl-2 in cells (Fig. 1E), suggesting that RING domains, at least RING1, is important for their interactions.
Mono-ubiquitination of Bcl-2 and Increase of Its Stability by Parkin
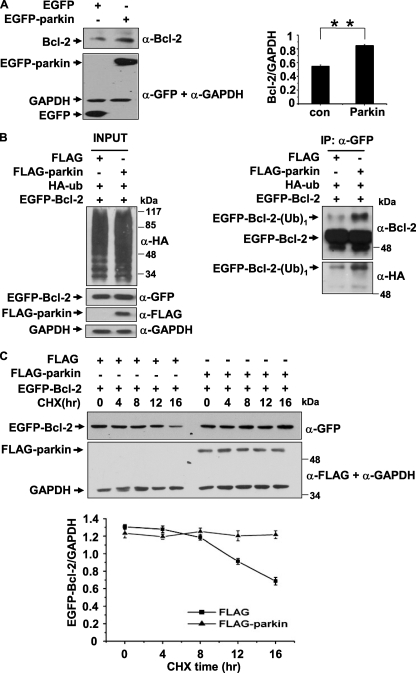
Because parkin is a well known E3 ligase (9) and we have shown that parkin interacts with Bcl-2, we questioned whether Bcl-2 is a substrate of parkin. First, we transfected 293 cells with EGFP or EGFP-parkin and examined the endogenous Bcl-2 levels using immunoblot analysis. In the presence of parkin, the endogenous Bcl-2 level was increased (Fig. 2A). Next, we co-transfected 293 cells with a combination of FLAG-parkin, EGFP-Bcl-2, and HA-ubiquitin and then performed immunoprecipitation and immunoblot assays to examine ubiquitination of Bcl-2. In the immunoprecipitants, a molecular mass corresponding to EGFP-Bcl-2 with one ubiquitin was detected much more strongly in the presence of parkin than in the absence of it using anti-GFP and anti-HA antibodies (Fig. 2B, right two panels), suggesting that parkin mono-ubiquitinates Bcl-2. Because mono-ubiquitination is a nondegradation signal for the substrates, we performed CHX chase assays to test whether parkin-mediated mono-ubiquitination of Bcl-2 could change its turnover. 293 cells co-expressing EGFP-Bcl-2 and FLAG or FLAG-parkin were treated with CHX at 48 h post-transfection to inhibit new protein syntheses. The total cell lysates were collected at different time points over the following 16 h and subjected to immunoblot analysis with anti-GFP, anti-FLAG, or anti-GAPDH antibodies. Bcl-2 was much more stable in the presence of parkin than in the absence of it (Fig. 2C). However, the K161N mutant failed to stabilize Bcl-2 (supplemental Fig. S2).
FIGURE 2.
Increase of Bcl-2 steady-state levels by parkin mono-ubiquitinating Bcl-2. A, increase of endogenous Bcl-2 by parkin. 293 cells were transfected with EGFP or EGFP-parkin. Cell lysates were subjected to immunoblot analysis using antibodies as indicated. The right panel shows the band intensity of Bcl-2 relative to that of GAPDH. The values are the means ± S.E. from three independent experiments. **, p < 0.01, one-way ANOVA. B, mono-ubiquitination of Bcl-2 by parkin. 293 cells expressing a combination of EGFP-Bcl-2, HA-ubiquitin, FLAG, or FLAG-parkin were subjected to immunoprecipitation using anti-GFP antibody. Immunoprecipitants and inputs were detected with antibodies as indicated. C, parkin increases the stability of Bcl-2. Forty-eight hours after transfection, 293 cells expressing EGFP-Bcl-2 with or without FLAG-parkin were treated with CHX (100 μg/ml). The cells were then harvested at 0, 4, 8, 12, and 16 h and subjected to immunoblot analysis with antibodies to GFP or FLAG. The band density of EGFP-Bcl-2 relative to that of GAPDH was shown. The values are the means ± S.E. from three independent experiments. IP, immunoprecipitation.
Inhibition of Autophagy under Normal Conditions or Starvation, but Enhancement of Autophagy upon CCCP Treatment, by Parkin
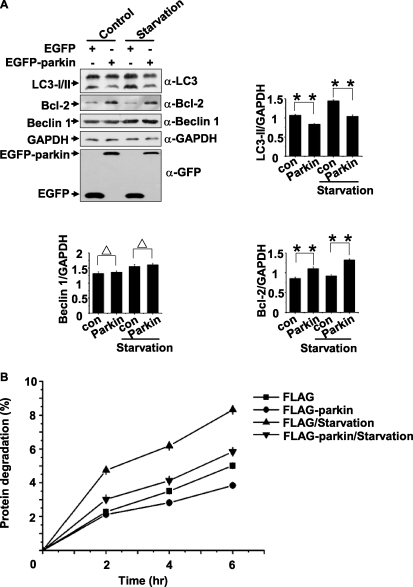
We have shown that parkin stabilizes Bcl-2, which is reported to be an inhibitory factor of autophagy (31). We questioned whether parkin regulates autophagy and whether its effects on autophagy are related with Bcl-2 levels. 293 cells expressing EGFP or EGFP-parkin were untreated (control) or treated with HBSS for 1 h (starvation). The autophagy protein LC3 is a widely used marker of mammalian autophagy (32), and the abundance of LC3-II or LC3-II/LC3-I ratio corresponds with the level of autophagosome formation (32, 33). In the presence of parkin, the endogenous Bcl-2 level was increased, and the LC3 conversion was decreased under normal conditions or starvation treatment (Fig. 3A), suggesting that parkin induces parallel changes with an increase of Bcl-2 and a decrease of autophagy activation. Beclin 1, an ATG gene product essential for autophagy (34), however, was not changed upon multiple treatments (Fig. 3A). Furthermore, the proteolysis rate of long-lived proteins in HeLa cells expressing EGFP-parkin was significantly decreased during normal conditions and starvation (Fig. 3B).
FIGURE 3.
Inhibition of autophagy by parkin under normal conditions or starvation. A, 293 cells transfected with EGFP or EGFP-parkin were incubated under normal conditions (control) or starvation (HBSS for 1 h). The cell lysates were subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II, Bcl-2, and Beclin 1 relative to that of GAPDH are shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01; Δ, p > 0.05, one-way ANOVA. B, Parkin inhibits autophagic degradation of long-lived proteins. HeLa cells expressing EGFP or EGFP-parkin were incubated for 24 h with 3H-labeled leucine (1 μCi/ml). The degradation of long-lived proteins was measured at the indicated time points in complete medium or Essential Medium with Earle's Balanced Salts. The values are the means ± S.E. from three independent experiments. con, control.
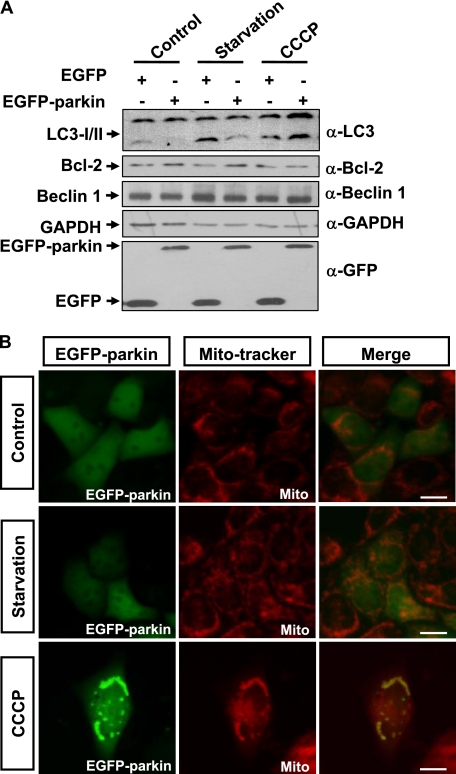
It was reported that parkin could mediate the engulfment of damaged mitochondria by autophagosomes after CCCP treatment (29). We therefore compared the effects of parkin on autophagy under normal conditions, starvation, and CCCP treatment. In 293 cells expressing EGFP-parkin, LC3-II level was decreased under normal conditions and starvation treatment; however, it was increased upon CCCP treatment (Fig. 4A).
FIGURE 4.
Enhancement of autophagy by parkin upon its mitochondrial localization. A, 293 cells transfected with EGFP or EGFP-parkin were incubated under various conditions, including complete medium (Control), HBSS for 1 h (Starvation), or 10 μm CCCP for 24 h (CCCP). The cell lysates were subjected to immunoblot analysis with antibodies as indicated. B, HeLa cells expressing EGFP-Parkin (green) were treated with Me2SO or HBSS for 1 h, or 10 μm CCCP for 24 h. The cells were then stained with 20 nm MitoTracker Red for 15 min and observed using an invert fluorescent microscope.
Parkin is recruited to mitochondria to induce autophagy under CCCP treatment (29). Because autophagy is decreased under normal conditions or starvation but increased under CCCP treatment, we examined the subcellular localization of EGFP-parkin in HeLa cells under normal conditions, starvation, and CCCP treatment. EGFP-parkin was predominately located in the cytosol under normal conditions and starvation treatment but apparently translocated to the mitochondria after CCCP treatment (Fig. 4B).
E3 Ligase Activity-dependent Autophagy Activation by Parkin
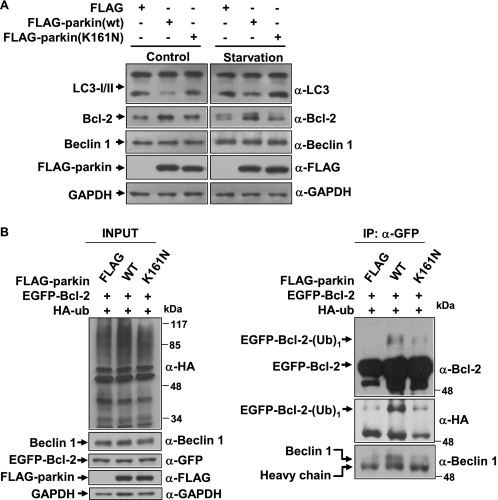
Parkin is a protein-ubiquitin E3 ligase (9), and the deficiency of its E3 ligase activity is associated with PD (3); we questioned whether the effects of parkin on autophagy and Bcl-2 are mediated by its E3 ligase function. To address this question, we created a FLAG-tagged disease-associated mutant of parkin (K161N), which selectively impaired its E3 ligase activity (35, 36), and transfected this mutant into 293 cells. The cells transfected with FLAG, FLAG-parkin (WT), or FLAG-parkin (K161N) were divided into two groups, in which one was without any treatment and another was treated with HBSS for 1 h (starvation). In both groups, parkin decreased LC3 conversion and increased Bcl-2 levels; however, the K161N mutant had no effects (Fig. 5A), suggesting that the E3 ligase activity is essential for parkin to regulate autophagy and Bcl-2.
FIGURE 5.
E3 ligase activity-dependent autophagy activation by parkin. A, E3 ligase activity deficient K161N mutant fails to regulate autophagy under normal conditions and starvation treatment (HBSS for 1 h). 293 cells expressing FLAG, FLAG-parkin, or FLAG-parkin (K161N) were subjected to immunoblot analysis with antibodies as indicated. B, Parkin, but not K161N mutant, mono-ubiquitinates Bcl-2 and increases its interactions with Beclin 1. 293 cells expressing a combination of EGFP-Bcl-2, HA-ubiquitin, and FLAG or FLAG-parkin (WT or K161N mutant) were subjected to immunoprecipitation using anti-GFP antibody. Immunoprecipitants and inputs were detected with antibodies as indicated. IP, immunoprecipitation.
We next co-transfected 293 cells with EGFP-Bcl-2 and HA-ubiquitin, together with FLAG-parkin or the K161N mutant. The cells were collected and subjected to immunoprecipitation assays using anti-GFP antibody. In the presence of parkin, a molecular mass corresponding to EGFP-Bcl-2 with one ubiquitin was detected, whereas it was not detected or only very weakly detected in the absence of parkin or in the presence of K161N mutant (Fig. 5B). Although both WT and mutant parkin had no effects on the protein level of endogenous Beclin 1, more Beclin 1 was co-immunoprecipitated in the presence of WT parkin than the K161N mutant (Fig. 5B), suggesting that WT parkin may increase the interactions of Bcl-2 and Beclin 1. These data suggest that parkin increased the amount of Bcl-2 by mediating the ubiquitination of Bcl-2 and changed its turnover. The increased Bcl-2 could bind more Beclin 1 to inhibit autophagy under normal conditions or starvation treatment.
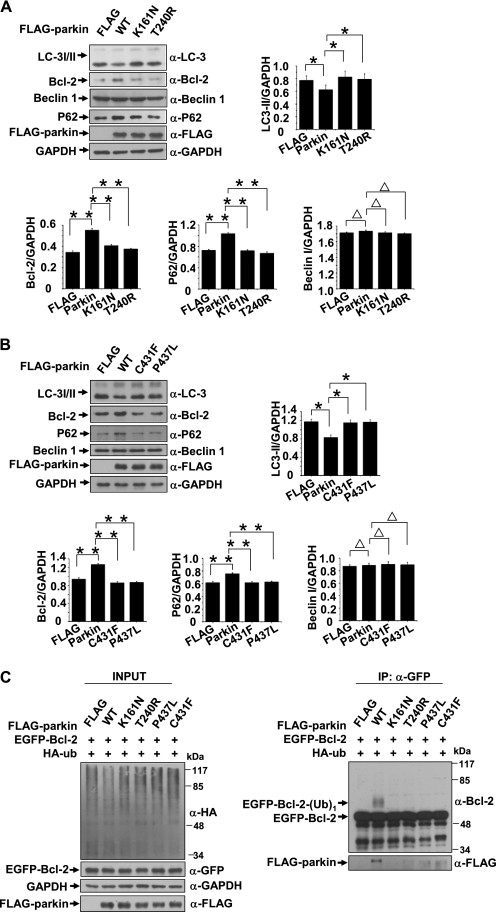
Failure to Regulate Autophagy by Disease-linked RING Domain Mutations
We have shown that the RING1 and RING2 domains of parkin interacted with Bcl-2 (Fig. 1E). The disease-linked mutations located in RING1 and RING2 domains of parkin impair its E3 ligase activity (35–38). We questioned whether these mutations could impair parkin-mediated autophagy activation and Bcl-2 ubiquitination. HeLa cells were transfected with FLAG, FLAG-parkin (WT), or its mutants (K161N, T240R, C431F, and P437L), and the cells were harvested 72 h after transfection. WT parkin decreased LC3 conversion and increased Bcl-2 level; however, all of the RING domain mutants had no effects (Fig. 6, A and B). p62, a substrate of autophagy, was also increased in WT parkin transfected cells but not in its mutant transfected cells (Fig. 6, A and B), further suggesting that the down-regulation of autophagy by parkin is associated with its E3 activity. We next co-transfected 293 cells with EGFP-Bcl-2 and HA-ubiquitin, together with FLAG-parkin or its mutant (K161N, T240R, C431F, or P437L). The cells were collected and subjected to immunoprecipitation assays using anti-GFP antibody. In the presence of parkin, a molecular mass corresponding to EGFP-Bcl-2 with one ubiquitin was detected, whereas it was not detected in the absence of parkin or in the presence of its RING domain mutants (Fig. 6C). We also found no or fewer interactions between these mutants and Bcl-2 (Fig. 6C, right, lower panel), suggesting that the impairment of parkin-mediated Bcl-2 ubiquitination conferred by disease-linked mutations located in RING1 and RING2 domains could be due to alterations of their interactions. Meanwhile, the N terminus of parkin failed to ubiquitinate Bcl-2 (supplemental Fig. S3), further suggesting that the RING domains at its C terminus are important for ubiquitinating Bcl-2.
FIGURE 6.
Failure to mono-ubiquitinating Bcl-2 and regulating autophagy by disease-linked RING1 and RING2 domain mutants. A and B, disease-linked RING1 and RING2 domain mutants (K161N, T240R, C431F, and P437L) fail to repress autophagy. HeLa cells expressing FLAG, FLAG-parkin, or disease-linked parkin mutants were subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II, Bcl-2, p62, or Beclin 1 relative to that of GAPDH were shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01; *, p < 0.05; Δ, p > 0.05, one-way ANOVA. C, disease-linked parkin mutants fail to mono-ubiquitinate Bcl-2. 293 cells expressing a combination of EGFP-Bcl-2 and HA-ubiquitin together with FLAG-parkin or its RING mutants (K161N, T240R, C431F, and P437L) were subjected to immunoprecipitation using anti-GFP antibody. Immunoprecipitants and inputs were detected with antibodies as indicated. IP, immunoprecipitation.
Knockdown of Parkin Enhances Autophagy Level
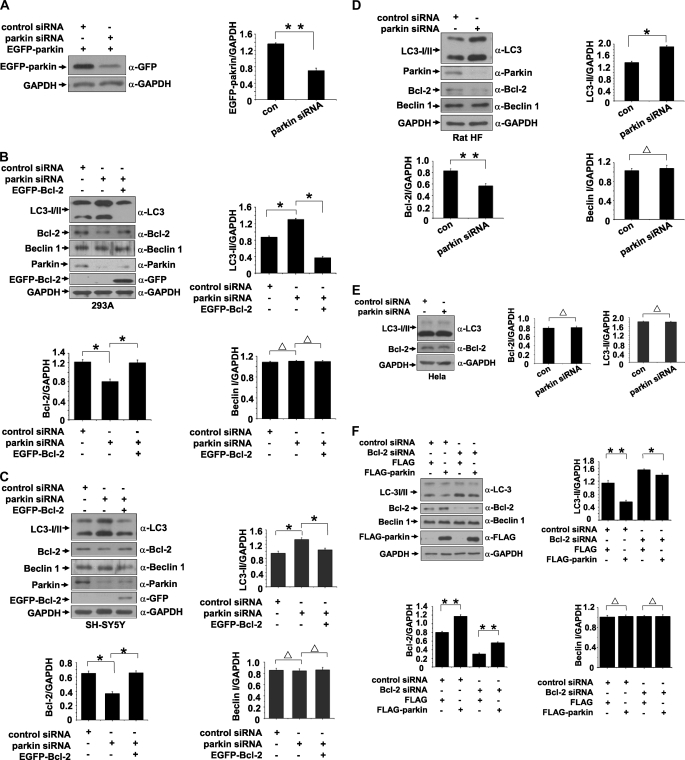
We further employed siRNA to knock parkin down to examine the effects of parkin. Transfection of the RNA oligonucleotides against parkin (parkin siRNA) in 293 cells successfully suppressed the protein level of EGFP-parkin as compared with the cells transfected with control siRNA (Fig. 7A), showing that parkin siRNA is effective.
FIGURE 7.
Increase of autophagy level by knockdown of parkin. A, RNA oligonucleotides against parkin (parkin siRNA) repress parkin expression effectively. The band density of EGFP-Bcl-2 relative to that of GAPDH was shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01, one-way ANOVA. B and C, knockdown of parkin enhances autophagy. 293 or SH-SY5Y cells were transfected with siRNAs against parkin or a combination of siRNAs against parkin and EGFP-Bcl-2. Negative control was also set by transfection of control siRNAs. Seventy-two hours after transfection, the total cell lysates were subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II, Bcl-2, and Beclin 1 relative to that of GAPDH were shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01; *, p < 0.05; Δ, p > 0.05, one-way ANOVA. D, knockdown of parkin enhances autophagy in primary cultured neuronal cells from rat HF. Cultured neuronal cells were transfected with siRNAs against parkin or negative control. Seventy-two hours after transfection, the total cell lysates were subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II, Bcl-2, and Beclin 1 relative to that of GAPDH were shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01; *, p < 0.05; Δ, p > 0.05, one-way ANOVA. E, HeLa cells were transfected with siRNAs against parkin or control siRNAs. The total cell lysates were collected 72 h after transfection and then subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II or Beclin 1 relative to that of GAPDH were shown. The values are the means ± S.E. from three independent experiments. Δ, p > 0.05, one-way ANOVA. F, knockdown of Bcl-2 attenuates parkin-induced down-regulation of autophagy. 293 cells were transfected with the RNA oligonucleotides against Bcl-2 (Bcl-2 siRNA) or control siRNA. The cells were then transfected with FLAG or FLAG-parkin. The total cell lysates were collected 72 h after transfection and then subjected to immunoblot analysis with antibodies as indicated. The band densities of LC3-II, Bcl-2, and Beclin 1 relative to that of GAPDH were shown. The values are the means ± S.E. from three independent experiments. **, p < 0.01; *, p < 0.05; Δ, p > 0.05, one-way ANOVA. con, control.
In 293 cells or SH-SY5Y cells that were transfected with parkin siRNA, the level of Bcl-2 was decreased, and the LC3-II conversion was increased; however, overexpression of EGFP-Bcl-2 greatly decreased LC3-II conversion that was induced by parkin siRNA (Fig. 7, B and C), suggesting that parkin functions in the upstream of Bcl-2. Similar results were obtained in primary cultured neuronal cells from rat HF that were transfected with parkin siRNA (Fig. 7D).
We next knocked parkin down in HeLa cells, in which parkin is not expressed because of deletions of parkin exons (39, 40). Knockdown of parkin failed to change the amounts of LC3-II and Bcl-2 in HeLa cells (Fig. 7E).
To further investigate the role of Bcl-2 in parkin-mediated autophagy regulation, we employed siRNA to knock Bcl-2 down in 293 cells. The cells expressing FLAG or FLAG-parkin were transfected with the RNA oligonucleotides against Bcl-2 (Bcl-2 siRNA) or control siRNA. Knockdown of Bcl-2 increased LC3-II conversion (Fig. 7F), suggesting that Bcl-2 cells negatively regulates autophagy. Overexpression of parkin significantly decreased LC3-II conversion, but it had a much lesser effect in Bcl-2 knockdown cells (Fig. 7F), further suggesting that Bcl-2 is involved in parkin-mediated autophagy regulation.
DISCUSSION
In our present study, we identified that Bcl-2 is a novel substrate for the E3 ubiquitin ligase parkin. Parkin directly interacts with Bcl-2 specifically through its C terminus and mediates the mono-ubiquitination of Bcl-2. Our results show that the mono-ubiquitination of Bcl-2 by parkin alters Bcl-2 turnover to increase its protein level. The PD-linked mutant forms of parkin (K161N, T240R, C431F, and P437F) (35–38), whose E3 ligase activities are selectively impaired, fail to regulate the mono-ubiquitination of Bcl-2. Recently studies have indicated that the parkin-mediated ubiquitination of particular protein substrates can change to degradation-independent roles (14, 15, 41). These proteasomal-independent substrates of parkin and their ubiquitination-mediated regulation may be important for the molecular pathogenesis of parkin-linked PD (14, 41).
It has been demonstrated that the selective ablation of autophagy genes in the central nervous system leads to neurodegeneration and premature death (42, 43). An early report shows that autophagosome accumulates in the brains of patients with PD (44). These results suggest that autophagy is associated with PD and neurodegeneration. A recent study indicates that in the neuronal cultures of midbrain from parkin null mice, markers of autophagy, such as LC3 II/I, are increased (45), suggesting that knock-out of parkin may induce autophagy. In our study, we show that overexpression of parkin represses autophagy, whereas knockdown of parkin induces autophagy. Taken together, these data suggest that parkin may negatively regulate autophagy.
Beclin 1 is a component of complex with class III PI3K and other proteins, including UVRAG, Ambra-1, Bif-1 and anti-apoptotic Bcl-2 family members (46). Bcl-2 functions as an autophagy inhibitory protein via its interaction with Beclin 1, and the autophagy level maintained by Bcl-2 is compatible with cell survival, rather than cell death (31). In our present study, we demonstrate that parkin, but not its E3 ligase-deficient mutants, mono-ubiquitinates Bcl-2 and alters the Bcl-2 steady-state levels and turnover, thereby increasing the amount of Bcl-2 (Figs. 2, 5, and 6). The increased Bcl-2 by parkin can bind more Beclin 1. Knockdown of parkin decreases Bcl-2 levels and increases LC3-II conversion in different cell lines as well as in primary cultured neuronal cells (Fig. 7). Overexpression of Bcl-2 blocks LC3 conversion induced by knockdown of parkin, whereas knockdown of Bcl-2 attenuates parkin-mediated autophagy repression (Fig. 7). Taken together, these data suggest that parkin is involved in autophagy inhibition via Bcl-2/Beclin 1 pathway under normal conditions or starvation treatment.
It was reported that parkin is recruited to impaired mitochondria to induce mitophagy upon CCCP treatment (29). Consistent with this finding, our data show that parkin increases autophagy upon CCCP treatment. However, under normal conditions and starvation, parkin inhibits autophagy (Figs. 3 and 4). Parkin is diffusively distributed in the cytoplasm in normal conditions and starvation but apparently translocated to mitochondria under CCCP treatment. It is therefore possible that parkin regulates autophagy differentially by different pathways in cytosol and mitochondria. Cytosolic parkin may mono-ubiquitinate Bcl-2 to stabilize Bcl-2. Increases of Bcl-2 may regulate autophagy in an adequate level for cellular homeostasis or protect cells against the long term, cumulative damage resulting from the environment. Under certain conditions, such as mitochondria damage, parkin could be recruited to the damaged mitochondria and promote its degradation by autophagy to clear the damaged mitochondria.
Parkin has protective effects against various stimuli either in vivo or in cultured cells. For example, it protects cells against apoptosis induced by 6-hydroxydopamine or dopamine, which could produce reactive oxygen species to activate the caspase cascade (47). Parkin was also reported to inhibit the release of cytochrome c, thereby decreasing the downstream activation of caspases (26, 48). These data suggest that parkin may function in mitochondria to protect cells against oxidative stresses and to inhibit cytochrome c release. Bcl-2 is well known to function in mitochondria to inhibit cytochrome c release and the subsequent caspase activation (49). Interestingly, it was reported that Bcl-2, similar to parkin, has the same protective effects on 6-OHDA and dopamine-induced apoptosis that is mediated by reactive oxygen species (50, 51). In our study, we show that parkin, but not its E3-deficient mutants, increases the Bcl-2 protein level. It is therefore possible that the protective effects of parkin against various stimuli may be mediated by Bcl-2.
In summary, we have identified that parkin is involved in autophagy regulation, differentially in cytosol and mitochondria. Parkin, but not its E3 ligase-deficient mutants, mono-ubiquitinates Bcl-2 to stabilize it, resulting in an increase of its binding to Beclin 1, thereby repressing autophagy in normal conditions or starvation.
Supplementary Material
This work was supported in part by National Natural Sciences Foundation of China Grants 30770664 and 30970921, National High-Tech Research and Development Program of China Project 973 Grant 2006CB500703 and Project 863 Grant 2006AA02A408, and Chinese Academy of Sciences Knowledge Innovation Project Grant KSCX2-YW-R138.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PD
- Parkinson disease
- Bax
- Bcl-2-associated X protein
- Bcl-xl
- B cell lymphoma leukemia-xL
- EGFP
- enhanced green fluorescent protein
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone or carbonyl cyanide m-chlorophenylhydrazone.
REFERENCES
- 1.Huang Y., Cheung L., Rowe D., Halliday G. (2004) Brain Res. Brain Res. Rev. 46, 44–70 [DOI] [PubMed] [Google Scholar]
- 2.Olanow C. W., Tatton W. G. (1999) Annu. Rev. Neurosci. 22, 123–144 [DOI] [PubMed] [Google Scholar]
- 3.West A. B., Maidment N. T. (2004) Hum. Genet. 114, 327–336 [DOI] [PubMed] [Google Scholar]
- 4.Lücking C. B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B. S., Meco G., Denèfle P., Wood N. W., Agid Y., Brice A. (2000) N. Engl. J. Med. 342, 1560–1567 [DOI] [PubMed] [Google Scholar]
- 5.Scott W. K., Nance M. A., Watts R. L., Hubble J. P., Koller W. C., Lyons K., Pahwa R., Stern M. B., Colcher A., Hiner B. C., Jankovic J., Ondo W. G., Allen F. H., Jr., Goetz C. G., Small G. W., Masterman D., Mastaglia F., Laing N. G., Stajich J. M., Slotterbeck B., Booze M. W., Ribble R. C., Rampersaud E., West S. G., Gibson R. A., Middleton L. T., Roses A. D., Haines J. L., Scott B. L., Vance J. M., Pericak-Vance M. A. (2001) J. Am. Med. Assoc. 286, 2239–2244 [DOI] [PubMed] [Google Scholar]
- 6.West A., Periquet M., Lincoln S., Lucking C. B., Nicholl D., Bonifati V., Rawal N., Gasser T., Lohmann E., Deleuze J. F., Maraganore D., Levey A., Wood N., Durr A., Hardy J., Brice A., Farrer M. (2002) Am. J. Med. Genet. 114, 584–591 [DOI] [PubMed] [Google Scholar]
- 7.Periquet M., Latouche M., Lohmann E., Rawal N., De Michele G., Ricard S., Teive H., Fraix V., Vidailhet M., Nicholl D., Barone P., Wood N. W., Raskin S., Deleuze J. F., Agid Y., Dürr A., Brice A. (2003) Brain 126, 1271–1278 [DOI] [PubMed] [Google Scholar]
- 8.Sakata E., Yamaguchi Y., Kurimoto E., Kikuchi J., Yokoyama S., Yamada S., Kawahara H., Yokosawa H., Hattori N., Mizuno Y., Tanaka K., Kato K. (2003) EMBO Rep. 4, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 10.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 11.Imai Y., Soda M., Takahashi R. (2000) J. Biol. Chem. 275, 35661–35664 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung K. K., Zhang Y., Lim K. L., Tanaka Y., Huang H., Gao J., Ross C. A., Dawson V. L., Dawson T. M. (2001) Nat. Med. 7, 1144–1150 [DOI] [PubMed] [Google Scholar]
- 14.Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi P., Snyder H., Petrucelli L., Theisler C., Chong M., Zhang Y., Lim K., Chung K. K., Kehoe K., D'Adamio L., Lee J. M., Cochran E., Bowser R., Dawson T. M., Wolozin B. (2003) Brain Res. Mol. Brain Res. 117, 179–189 [DOI] [PubMed] [Google Scholar]
- 16.Imai Y., Soda M., Inoue H., Hattori N., Mizuno Y., Takahashi R. (2001) Cell 105, 891–902 [DOI] [PubMed] [Google Scholar]
- 17.Huynh D. P., Scoles D. R., Nguyen D., Pulst S. M. (2003) Hum. Mol. Genet. 12, 2587–2597 [DOI] [PubMed] [Google Scholar]
- 18.Ren Y., Zhao J., Feng J. (2003) J. Neurosci. 23, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Um J. W., Min D. S., Rhim H., Kim J., Paik S. R., Chung K. C. (2006) J. Biol. Chem. 281, 3595–3603 [DOI] [PubMed] [Google Scholar]
- 20.Staropoli J. F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. (2003) Neuron 37, 735–749 [DOI] [PubMed] [Google Scholar]
- 21.Corti O., Hampe C., Koutnikova H., Darios F., Jacquier S., Prigent A., Robinson J. C., Pradier L., Ruberg M., Mirande M., Hirsch E., Rooney T., Fournier A., Brice A. (2003) Hum. Mol. Genet. 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 22.Ko H. S., von Coelln R., Sriram S. R., Kim S. W., Chung K. K., Pletnikova O., Troncoso J., Johnson B., Saffary R., Goh E. L., Song H., Park B. J., Kim M. J., Kim S., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 7968–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallon L., Bélanger C. M., Corera A. T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., Brice A., van Bergen En Henegouwen P. M., Fon E. A. (2006) Nat. Cell Biol. 8, 834–842 [DOI] [PubMed] [Google Scholar]
- 24.Ko H. S., Kim S. W., Sriram S. R., Dawson V. L., Dawson T. M. (2006) J. Biol. Chem. 281, 16193–16196 [DOI] [PubMed] [Google Scholar]
- 25.Shimura H., Hattori N., Kubo S., Yoshikawa M., Kitada T., Matsumine H., Asakawa S., Minoshima S., Yamamura Y., Shimizu N., Mizuno Y. (1999) Ann. Neurol. 45, 668–672 [DOI] [PubMed] [Google Scholar]
- 26.Darios F., Corti O., Lücking C. B., Hampe C., Muriel M. P., Abbas N., Gu W. J., Hirsch E. C., Rooney T., Ruberg M., Brice A. (2003) Hum. Mol. Genet. 12, 517–526 [DOI] [PubMed] [Google Scholar]
- 27.Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., Matsumoto T. (2006) Hum. Mol. Genet. 15, 883–895 [DOI] [PubMed] [Google Scholar]
- 29.Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortiboys H., Thomas K. J., Koopman W. J., Klaffke S., Abou-Sleiman P., Olpin S., Wood N. W., Willems P. H., Smeitink J. A., Cookson M. R., Bandmann O. (2008) Ann. Neurol. 64, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 32.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 35.Abbas N., Lücking C. B., Ricard S., Dürr A., Bonifati V., De Michele G., Bouley S., Vaughan J. R., Gasser T., Marconi R., Broussolle E., Brefel-Courbon C., Harhangi B. S., Oostra B. A., Fabrizio E., Böhme G. A., Pradier L., Wood N. W., Filla A., Meco G., Denefle P., Agid Y., Brice A. (1999) Hum. Mol. Genet. 8, 567–574 [DOI] [PubMed] [Google Scholar]
- 36.Periquet M., Lücking C., Vaughan J., Bonifati V., Dürr A., De Michele G., Horstink M., Farrer M., Illarioshkin S. N., Pollak P., Borg M., Brefel-Courbon C., Denefle P., Meco G., Gasser T., Breteler M. M., Wood N., Agid Y., Brice A. (2001) Am. J. Hum. Genet. 68, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruyama M., Ikeuchi T., Saito M., Ishikawa A., Yuasa T., Tanaka H., Hayashi S., Wakabayashi K., Takahashi H., Tsuji S. (2000) Ann. Neurol. 48, 245–250 [PubMed] [Google Scholar]
- 38.Nichols W. C., Pankratz N., Uniacke S. K., Pauciulo M. W., Halter C., Rudolph A., Conneally P. M., Foroud T. (2002) J. Med. Genet. 39, 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denison S. R., Wang F., Becker N. A., Schüle B., Kock N., Phillips L. A., Klein C., Smith D. I. (2003) Oncogene 22, 8370–8378 [DOI] [PubMed] [Google Scholar]
- 40.Pawlyk A. C., Giasson B. I., Sampathu D. M., Perez F. A., Lim K. L., Dawson V. L., Dawson T. M., Palmiter R. D., Trojanowski J. Q., Lee V. M. (2003) J. Biol. Chem. 278, 48120–48128 [DOI] [PubMed] [Google Scholar]
- 41.Moore D. J., West A. B., Dikeman D. A., Dawson V. L., Dawson T. M. (2008) J. Neurochem. 105, 1806–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 43.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 44.Williams A., Jahreiss L., Sarkar S., Saiki S., Menzies F. M., Ravikumar B., Rubinsztein D. C. (2006) Curr. Top. Dev. Biol. 76, 89–101 [DOI] [PubMed] [Google Scholar]
- 45.Casarejos M. J., Solano R. M., Rodriguez-Navarro J. A., Gómez A., Perucho J., Castaño J. G., García de Yébenes J., Mena M. A. (2009) J. Neurochem. 110, 1523–1537 [DOI] [PubMed] [Google Scholar]
- 46.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H., Ren Y., Zhao J., Feng J. (2004) Hum. Mol. Genet. 13, 1745–1754 [DOI] [PubMed] [Google Scholar]
- 48.Berger A. K., Cortese G. P., Amodeo K. D., Weihofen A., Letai A., LaVoie M. J. (2009) Hum. Mol. Genet. 18, 4317–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., Wang X. (1997) Science 275, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 50.Offen D., Ziv I., Panet H., Wasserman L., Stein R., Melamed E., Barzilai A. (1997) Cell Mol. Neurobiol. 17, 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takai N., Nakanishi H., Tanabe K., Nishioku T., Sugiyama T., Fujiwara M., Yamamoto K. (1998) J. Neurosci. Res. 54, 214–222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.