Abstract
Autotransporters are bacterial virulence factors that share a common mechanism by which they are transported to the cell surface. They consist of an N-terminal passenger domain and a C-terminal β-barrel, which has been implicated in translocation of the passenger across the outer membrane (OM). The mechanism of passenger translocation and folding is still unclear but involves a conserved region at the C terminus of the passenger domain, the so-called autochaperone domain. This domain functions in the stepwise translocation process and in the folding of the passenger domain after translocation. In the autotransporter hemoglobin protease (Hbp), the autochaperone domain consists of the last rung of the β-helix and a capping domain. To examine the role of this region, we have mutated several conserved aromatic residues that are oriented toward the core of the β-helix. We found that non-conservative mutations affected secretion with Trp1015 in the cap region as the most critical residue. Substitution at this position yielded a DegP-sensitive intermediate that is located at the periplasmic side of the OM. Further analysis revealed that Trp1015 is most likely required for initiation of processive folding of the β-helix at the cell surface, which drives sequential translocation of the Hbp passenger across the OM.
Keywords: Bacteria, Membrane, Protein Domains, Protein Secretion, Protein Translocation, Autotransport, E. coli, Hbp, SPATE, Type V Secretion
Introduction
Many large virulence factors that are secreted to the extracellular milieu of pathogenic Gram-negative bacteria are autotransporters (ATs)2 (1). The mechanism of autotransport appears to be more complicated and less autonomous than the name suggests (2, 3). Each AT has an N-terminal signal peptide to direct translocation across the inner membrane (IM) via the Sec translocon, a central passenger domain that contains the virulence function and a conserved C-terminal domain (the β-domain) that forms a β-barrel pore in the outer membrane (OM), as implied by the original model for secretion (4, 5). Although it is clear that the β-domain is required for translocation of the passenger across the OM, it is debated whether it functions unassisted as the actual translocation pore. For example, recent structural analysis of the β-domain of several ATs reveals a barrel pore with a very narrow inner diameter of ∼10 Å (6, 7). This is incompatible with the observed translocation of AT passengers that include partly folded domains and AT passengers that contain intramolecular disulfide bonds formed prior to secretion (6, 8–10).
Structural analysis of the secreted AT passengers pertactin (11), VacA (12), hemoglobin protease (Hbp) (13), and IgA protease (14) reveals a common overall topology: a long stem or stalk consisting of a right-handed β-helix to which other functional domains are appended. A β-helix is an extremely stable fold of β-strands, in this case of triangular rungs comprising three strands, that are interrupted by loops with variable length and are stacked to form a cross β-structure (15, 16). Other examples of this repetitive fold include P22 tailspike protein, pectate lyase, and a domain of the HET-s prion protein (17–19). Importantly, most of the hundreds of ATs analyzed are predicted to include extensive β-helical structure, suggesting a generic role in biogenesis and transport (20, 21). The Hbp structure includes 24 rungs forming a 125 Å long scaffold that is slightly kinked in the middle at the position where a domain of unknown function protrudes from the main stem (see Fig. 1A) (13). At the N terminus, the β-helix is connected with a juxtaposed globular domain that contains a serine protease triad. As seen in other β-helix proteins, the core of the helix is hydrophobic and largely filled with partly stacked aliphatic and aromatic side chains (13, 22).
FIGURE 1.
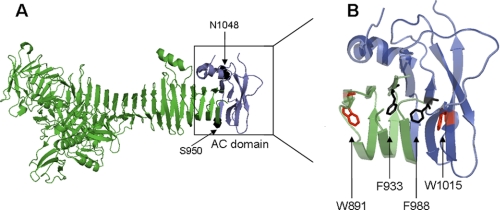
Structure of the E. coli Hbp passenger and its AC domain. A, ribbon diagram representing the crystal structure of the Hbp passenger (PDB ID: 1WXR); the region constituting the AC domain (residues 950–1048) is colored dark blue. B, close-up of the AC domain and adjacent β-helix rungs with several aromatic residues oriented inward into core of the β-helix. Residues targeted for substitution are indicated: Trp891, Phe933, Phe988, and Trp1015. The β-domain is not included in the figure.
At the C terminus of the passenger, approximately from residues 950 to 1048, a domain is located that has been named the autochaperone (AC) domain (13). It contains the last three rungs of the β-helical stem and a cap region that is partly disordered and connects the passenger to the β-domain. This AC domain is conserved among many ATs, including members of the SPATE (for serine protease autotransporters of Enterobacteriaceae) subfamily to which Hbp belongs (22). Deletion and mutagenesis experiments have shown that this region is crucial for efficient folding of several passenger domains (23–26). Recent evidence indicates that translocation of the passenger across the OM occurs in an C-to-N direction, possibly following the formation of a hairpin structure by the C terminus of the passenger in the β-domain channel (27, 28). In this scenario, the AC domain might function as a template for stacking of the β-helix to initiate folding (22). Folding may provide a pulling force to transfer the passenger across the OM, where other energy sources such as ATP or a proton motive force are absent. On the other hand, deletions in this region in the ATs Serratia marcescens serine protease (SSP) and BrkA appear to have little effect on secretion. Instead, the translocated passenger missing the AC region remains unfolded at the cell surface, where it is susceptible to OM proteases and externally added trypsin (23, 29).
Recent cross-linking experiments, using AT passengers that were stalled in the process of OM translocation, revealed contacts with subunits of the Bam complex (30, 31). The Bam complex functions in the general assembly of OM proteins (32), and one of its subunits, BamA, is an integral OM protein that has been shown to possess pore activity (33). It has been suggested that BamA may act directly as the protein-conducting channel for the AT passenger or indirectly by coordinating the assembly of the β-domain and translocation of the passenger in a concerted fashion (30, 31). In the latter model, the β-domain may still function as the actual channel for passenger translocation, assuming that it is accommodated in or near the Bam complex in an extended translocation-competent conformation that transiently forms during transport of the passenger. Following transport, the passenger domain of several ATs is proteolytically cleaved from the β-domain and released from the membrane.
In this study, we have analyzed the role of the Hbp AC domain in folding and secretion by mutating conserved residues in the AC domain. We find that Trp1015 and to a lesser extent Phe988 and Phe933 play a crucial role in translocation across the OM. Non-conserved substitutions of this residue stall translocation at an early stage and provoke degradation by the periplasmic protease DegP. The data are consistent with a role for the C-terminal region of the β-helix in the initiation of processive folding of the β-helix at the cell surface to drive sequential translocation of the Hbp passenger across the OM.
EXPERIMENTAL PROCEDURES
Strains and Medium
The Escherichia coli strains used in this study are MC1061 (34) and MC1061 degP::S210A (35). Cells were generally grown in M9 medium (36) containing 0.1% casamino acids (Difco), 0.2% glucose, and chloramphenicol (30 μg ml−1) at 30 °C.
Plasmid Construction
Hbp and its derivatives were expressed from vector pEH3 (37). Plasmids that contain wild-type Hbp and the Hbp derivatives Δβ-cleavage and L110C/G348C have been described previously (8). To create the substitutions by Ala, the hbp gene in plasmid pACYC-Hbp (38) was mutated by site-directed mutagenesis. The amplified product was subcloned into pEH3 containing wild-type hbp (pEH3-Hbp) by replacing a fragment using the KpnI and EcoRI restriction sites. This resulted in plasmids pEH3-HbpF988A and pEH3-HbpW1015A. A similar strategy was used to create the mutants W891A, F933A, W1015K, W1015F, and W1015Y. Plasmid pEH3-Hbp was mutated using nested PCR, essentially as described previously (39). The primers used for mutagenesis are listed in Table 1. Nucleotide sequences were confirmed by semi-automated DNA sequencing.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′)a |
|---|---|
| N-W988A-f | ctgataatctggcaagggcatcaac |
| N-W988A-b | gttgatgcccttgccagattatcag |
| N-W1015A-f | gacgggaaaaaagaggcggtcctcgatggttac |
| N-W1015A-b | gtaaccatcgaggaccgcctcttttttcccgtc |
| N-W891A-f | catgacagacacccaggcgtcgatgaacggaaactcc |
| N-W891A-b | gagtttccgttcatcgacgcctgggtgtctgtcatgc |
| N-W933A-f | cggttcagtcagcagctgtcatgcgtacagacc |
| N-W933A-b | gtctgtacgcatgacagctgctgactgaaccgc |
| N-W1015K-f | gacgggaaaaaagagaaagtcctcgatggttacc |
| N-W1015K-b | gtaaccatcgaggactttctcttttttcccgtcc |
| N-W1015F-f | gacgggaaaaaagagtttgtcctcgatggttacc |
| N-W1015F-b | gtaaccatcgaggacaaactcttttttcccgtcc |
| N-W1015Y-f | gacgggaaaaaagagtatgtcctcgatggttacc |
| N-W1015Y-b | gtaaccatcgaggacatactcttttttcccgtcc |
a The mutated residues are underlined.
Reagents, Enzymes, and Sera
Dithiobis(succinimidyl) propionate (DSP) was purchased from Pierce. Restriction enzymes, T4-DNA ligase, proteinase K, Complete mini EDTA-free protease inhibitor, and Lumi-Light immunoblotting substrate were purchased from Roche Applied Science. The Phusion site-directed mutagenesis kit was from Finnzymes. The Cy3-conjugated donkey anti-rabbit antiserum was from Jackson ImmunoResearch. All other reagents and enzymes were from Sigma-Aldrich. The phenylmethylsulfonyl fluoride (PMSF) was from Roche Applied Science. Polyclonal antiserum against the Hbp passenger domain was from our own collection. Polyclonal antisera against BamA, BamB, SurA, and TolC were gifts from J. Tommassen (Utrecht University), T. Silhavy (Princeton University), R. Kolter (Harvard University), and V. Koronakis (University of Cambridge). The antisera against Leader peptidase (Lep) and OmpA were gifts from J.-W. de Gier (Stockholm University).
Preparation and Analyses of Membrane Fractions
All procedures were essentially carried out as described (40). Briefly, cells were collected and resuspended in 7 ml of buffer K (50 mm triethanolamine, 1 mm EDTA, 10% sucrose, Complete Mini EDTA-free protease inhibitor, 0.5 mm DNase, and 0.5 mm RNase). Cells were disrupted by two passages through a One Shot cell disrupter (Constant Systems). After ultracentrifugation (345,700 × g, 4 °C, 60 min), the membranes were resuspended in 450 μl of buffer K containing 55% (w/w) sucrose and placed at the bottom of a centrifuge tube. A sucrose gradient was created by layering on top of the crude membrane fraction consecutive volumes (500 μl) of buffer K containing 51, 48, 45, 42, 39, 36, 33, and 30% (w/w) sucrose, respectively. After centrifugation (230,000 × g, 40 h, 4 °C), 20 fractions (220 μl) were collected from the top of the gradients and analyzed by SDS-PAGE and immunoblotting.
Protease Treatment of Whole Cells
Cells were grown to an OD660 of ∼0.3 and induced with 1 mm of IPTG. One hour after induction, cells were harvested by centrifugation (9,300 × g, 1 min, room temperature). Cells were harvested and resuspended in 50 mm Tris-HCl, pH 7.4, and 20 mm CaCl2. Half of the sample was kept on ice, whereas the other half was subjected to sonication on ice using a tip sonicator (Branson Sonifier 250, output control 5, duty cycle 40%, 1 min). To remove intact cells and debris, sonicated samples were centrifuged (9,300 × g, 2 min, 4 °C). Whole cells and sonicated samples were incubated with proteinase K (100 μg ml−1) at 37 °C for 20 min. The reaction was stopped by the addition of 0.1 mm PMSF. All samples were TCA-precipitated and analyzed by SDS-PAGE and immunoblotting.
Indirect Immunofluorescence Microscopy
All procedures were carried out as described previously (41) except that for non-permeabilized cells, treatment with Triton X-100, EDTA, and lysozyme were omitted. Furthermore, cells were incubated with either a polyclonal primary antibody against the Hbp passenger or a polyclonal antibody against OmpA and subsequently a Cy3-conjugated donkey anti-rabbit secondary antibody.
Cross-linking in Whole Cells and Immunopurification
All procedures were carried out as described previously (30) except that expression of Hbp in the cells was induced using 1 mm IPTG. Furthermore, cells were disrupted using a One Shot cell disrupter (Constant Systems), and DTT was used to dissociate cross-linked products instead of β-mercaptoethanol.
RESULTS
Mutations in the Core of the Hbp AC Domain
To study the relationship between the folding of ATs and their translocation across the OM, we have made several mutations within the conserved AC domain of Hbp that extends from Ser950 to Asn1048 (13) and adjacent three rungs upstream from the AC domain. The AC domain consists of the last rung in the β-helix followed by a break in the β-helix structure at the conserved Ala979/Pro980 and a cap-like domain of three antiparallel β-strands at the C terminus. Notably, several loops that connect the β-strands in this region were not resolved in the crystal structure, indicating that they are flexible (13).
Closer examination of the structure of the AC domain and the three adjacent rungs revealed that 40% of the residues are hydrophobic. Of these hydrophobic residues, only a few are aromatic residues, and these are buried in the core of the β-helix. They all point inward at equivalent positions in the successive rungs to build a stack of side chains (Fig. 1B). The end of the stack is formed by the highly conserved Trp1015 residue. This residue is located in the cap and forms a hydrogen bond through its indole side chain with Asn985 and Phe988, which stabilizes a turn in the protein backbone. The positioning of stacked aromatic residues inside the core is quite common in β-helical structures and is thought to contribute to folding and stability (15, 42). To investigate the role of these residues in the biogenesis of Hbp, we selected four conserved aromatic residues for substitution by Ala: Trp891, Phe933, and Phe988 in the regular rungs and Trp1015 in the cap (Fig. 1B). We compared the biogenesis and secretion of these mutants with wild-type Hbp and the HbpΔβ-cleavage mutant, which carries a mutation in the autocatalytic cleavage site between the passenger and the β-domain. The passenger of this mutant is fully folded but remains covalently attached to the β-domain at the extracellular side of the OM (8).
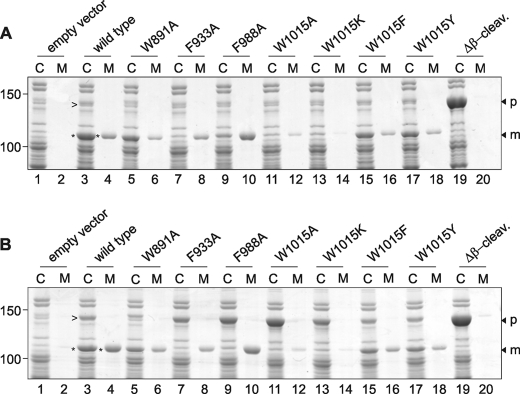
The wild-type and mutant genes were cloned under control of the lac promoter in the expression vector pEH3. To monitor expression and secretion, E. coli strain MC1061 harboring the Hbp plasmids was grown in minimal medium to early log phase and induced for Hbp expression with 1 mm IPTG. Production of Hbp and the mutants affected growth of the cultures somewhat, in particular the mutant W1015A (supplemental Fig. S1A). Culture samples were collected 2 h after induction and centrifuged to separate cells (C) and spent medium (M). Both fractions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 2A). Under these conditions, wild-type Hbp is expressed, processed, and secreted, as evident from the appearance of a 110-kDa product in the cell and medium fractions that represents the Hbp passenger (Fig. 2A, lanes 3 and 4). The identity was confirmed by immunoblotting using passenger-specific antibodies (not shown). It is known that upon translocation across the OM, the Hbp passenger is in part released into the medium and in part retained at the cell surface via an unknown mechanism (43, 44).
FIGURE 2.
Expression and secretion of Hbp AC domain mutants. A, expression and secretion of wild-type Hbp and mutants in E. coli MC1061. Expression of Hbp was induced by the addition of 1 mm IPTG when cultures reached early log phase. Samples were collected 2 h after induction, separated into cells (C) and spent medium (M) by centrifugation, and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. The relevant areas of the same gel are shown. Molecular mass (kDa) markers are indicated at the left side of the panels. Wild-type Hbp passenger and pro-form are indicated with an asterisk and arrowhead, respectively. The positions of the pro-form (p) and the mature passenger (m) are indicated on the right side of the panel. Δβ-cleav., Δβ-cleavage. B, expression and secretion of wild-type Hbp and the AC mutants in E. coli MC1061degP::S210A analyzed as described under A.
The mutations in the three rungs of the AC domain showed mild but variable effects on the detected levels of cell-associated and released Hbp. Levels for mutants W891A and F933A appeared lower than for wild type, whereas the total levels for F988A appeared comparable with wild type but with more of the mutant passenger released from the cells (Fig. 2A, lanes 5–10). Intriguingly, strongly reduced levels were observed for the Hbp W1015A mutant, suggesting that the biogenesis of this mutant was severely affected (Fig. 2A, lanes 11 and 12).
Previous studies have indicated a crucial role for the periplasmic chaperone and protease DegP in the quality control of Hbp biogenesis (8). Hbp derivatives impaired in translocation across the OM were degraded by DegP, resulting in lower detected levels. We further showed that expression of secretion-incompetent Hbp derivatives in the absence of DegP resulted in the accumulation of unprocessed pro-Hbp in the periplasm. Conceivably, the mutations introduced in the AC render the Hbp partly secretion-incompetent and prone to periplasmic degradation by DegP. To investigate this possibility, we analyzed the biogenesis of the Hbp mutants in E. coli strain MC1061degP::S210A. This mutant has an active site mutation rendering DegP defective in its protease function, whereas the chaperone activity is still intact (35). The induced expression of W1015A and to a lesser degree F988A and F933A had a negative effect on the culture growth in MC1061degP::S210A (supplemental Fig. S1B). It should be noted that production of wild-type and HbpΔβ-cleavage appeared less detrimental to culture growth under these conditions. This suggests that the presence of HbpW1015A induces a stress response that is deleterious to cells that lack the protease function of DegP. We have previously observed a similar growth arrest for secretion-incompetent Hbp mutants (8, 30), and it, therefore, appeared to indicate that Hbp secretion was severely affected for the W1015A mutant.
When analyzed by Coomassie Brilliant Blue staining upon SDS-PAGE, mutant W1015A accumulated almost exclusively as an ∼140-kDa product in the cell fraction (Fig. 2B, lane 11). This form co-migrates with the HbpΔβ-cleavage mutant that remains uncleaved (Fig. 2, A and B, lane 19). Immunodetection confirmed that this form represents the unprocessed Hbp (pro-Hbp) as it reacts with antibodies specific for the passenger and β-domain, respectively (not shown). Mutants F933A and F988A also showed an increase in accumulated pro-form, although less pronounced (Fig. 2B, lanes 7 and 9, respectively). These findings suggest that the reduced amounts of accumulated pro-form detected in wild-type strain MC1061 were not due to reduced initial expression levels but rather resulted from a defect in secretion and subsequent degradation. It indicates also that the mutant pro-forms accumulate in MC1061degP::S210A in a secretion-incompetent form and that this accumulation is responsible for the reduced cell growth.
Together, the data suggest that residue Trp1015 is crucial for efficient secretion of Hbp, whereas the role of Phe933 and Phe988 in secretion is less prominent. To investigate the role of Trp1015 in more detail, we made another non-conservative change to Lys at this position. This residue has a long aliphatic and charged side chain and could, like Trp, establish the hydrogen bond that stabilizes the turn in the peptide backbone. In addition, we introduced two conservative changes by introducing the aromatic residues Tyr and Phe. When expressed in MC1061, only small amounts of fully processed Hbp W1015K were detected in the cell fraction, whereas released W1015K in the culture medium was virtually undetectable (Fig. 2A, lanes 13 and 14). Expression of W1015K in the DegP mutant cells led to the accumulation of the Hbp pro-form, although less pronounced than W1015A, indicating a partial degradation by a protease other than DegP or lower initial expression levels of the protein (Fig. 2B, lanes 13 and 11, respectively). By contrast, when Trp was substituted for Tyr or Phe, i.e. conservative mutations, the expression and release of the passenger was comparable with wild-type Hbp (Fig. 2A, lanes 15–18). The lower amounts of the two mutants observed might be explained by the fact that neither Tyr nor Phe are able to establish the additional hydrogen bond, which apparently influences protein stability. In contrast, the wild-type Hbp and W1015Y and W1015F mutants accumulated similar amounts of pro-form in the DegP mutant cells (Fig. 2B, lanes 15–18) and had no additional effect on the growth of the cell culture (supplemental Fig. S1B). The combined data suggest that only conservative changes are allowed for Trp1015.
Localization and Topology of HbpW1015A
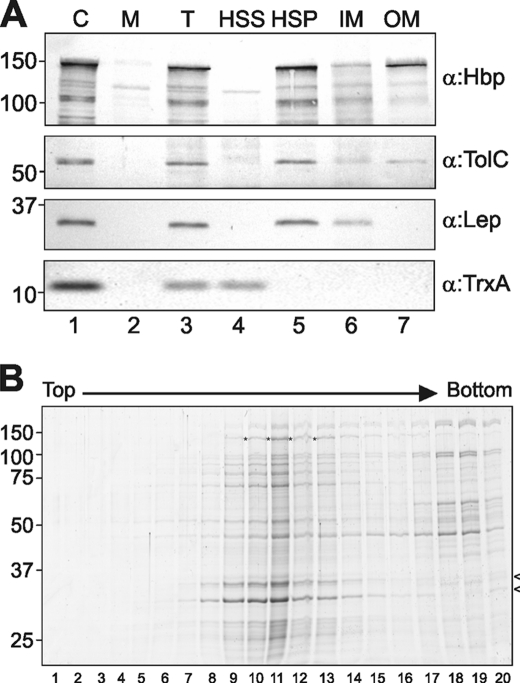
The data presented in Fig. 2B demonstrate that W1015A is poorly secreted and that the pro-form of W1015A is subject to degradation by DegP, suggesting that it accumulates in a protease-sensitive conformation that is, at least in part, exposed to the periplasm. To establish where the pro-form of the Hbp W1015A mutant accumulates, we expressed it in MC1061degP::S210A and examined its subcellular localization. The cells were lysed and subjected to high speed centrifugation to separate the membranes from the soluble material and analyzed by SDS-PAGE and immunoblotting (Fig. 3A). Clearly, the W1015A pro-Hbp co-fractionated with the membranes in the high speed pellet, together with the control OM protein TolC and control IM protein Lep but in contrast to the control cytosolic protein thioredoxin A (TrxA) (Fig. 3A, lane 5). To further distinguish between an association with the IM or OM, the crude membrane fraction was extracted with sodium lauroyl sarcosinate (sarkosyl) to solubilize the IMs. Using this procedure, W1015A was primarily detected in the insoluble OM fraction similar to the control OM protein TolC but in contrast to the IM protein Lep that was completely extracted by the sarkosyl treatment (Fig. 3A, lanes 6 and 7, respectively). To exclude the presence of W1015A aggregates, which would co-fractionate with the OMs without actually being membrane-associated, we subjected the crude membrane fractions to flotation centrifugation through sucrose gradients. Fractions were withdrawn from the gradients and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 3B). The gradients showed that most of the W1015A pro-form had floated up with the membranes as it was found in fractions 10–13 and co-migrating with the porins (Fig. 3B, lanes 10–13), whereas (aggregated) membrane-free proteins are expected to remain in the bottom fraction with the highest sucrose concentration. Together, these data suggest that the majority of the W1015A pro-form is associated with the OM.
FIGURE 3.
Secretion-incompetent HbpW1015A is localized at the OM. A, MC1061degP::S210A cells expressing HbpW1015A were grown and induced as described in the legend for Fig. 2. One hour after induction, samples were collected and separated into cells (C) and spent medium (M) (lanes 1 and 2). After disruption of whole cells and removal of unlysed cells, the total cell lysate (T) was centrifuged at high speed to separate the membranes (high speed pellet, HSP) from the soluble fraction (high speed supernatant, HSS) (lanes 3–5). The membranes (HSP) were treated with 0.5% sarkosyl to solubilize the IMs and centrifuged again to collect the OMs as described previously (8). Samples of the fractions, derived from equal amounts of cell material, were analyzed by SDS-PAGE and immunoblotting using antiserum against Hbp (top panel). To verify the membrane separation, both the OM protein TolC and the IM protein Lep were detected in the fractions by immunoblotting. To verify the separation of high speed supernatant and HSP, the cytosolic protein thioredoxin A (TrxA) was detected. B, membranes (HSP) of cells expressing HbpW1015A were separated by flotation sucrose gradient centrifugation. Fractions were collected from the top of the gradient and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (lanes 1–20). The HbpW1015A pro-form is indicated by an asterisk; the identity of pro-Hbp was confirmed by immunoblotting (data not shown). Porins are indicated with an arrowhead on the right side of the panel.
HbpW1015A Is Peripherally Associated with the OM
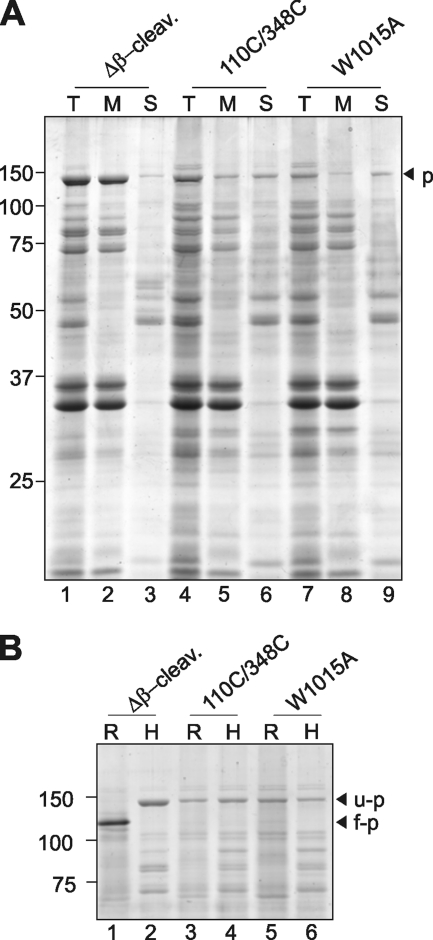
To investigate the nature of OM association of the unprocessed W1015A, the membrane fractions from the flotation experiment that contained the accumulated pro-form were pooled and subjected to urea extraction. This method enables separation of proteins that are fully inserted into the OM from proteins that are peripherally associated through protein-protein contacts. The pooled fractions containing the Hbp pro-form were incubated in a buffer containing 4 m urea and subjected to ultracentrifugation. Pellet and supernatant fractions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 4A). As a control, we analyzed the HbpΔβ-cleavage mutant of which the β-domain is fully assembled in the OM anchoring the uncleaved passenger to the cell surface (8, 30). As shown previously, the HbpΔβ-cleavage mutant was fully resistant to extraction similar to the porins in the ∼30-kDa region (Fig. 4A, lanes 1–3). As a further control, we analyzed the Hbp-L110C/G348C mutant that is known to be a pro-form intermediate that is stalled during translocation across the OM (8). It is to a large degree extractable with urea (Fig. 4A, lanes 4–6), confirming the earlier observations and consistent with the fact that its β-domain is not properly folded (30). Likewise, the Hbp W1015A mutant was efficiently extracted by urea, even more so than Hbp-L110C/G348C, suggesting that its β-domain is not integrated in the protein-protein interactions.
FIGURE 4.
The β-domain of HbpW1015A mutant is not fully inserted and folded in the OM. A, Coomassie Brilliant Blue-stained SDS-PAGE of urea-treated outer OM preparations from cells expressing the indicated Hbp derivatives. Fractions containing the outer membranes, as described in the legend for Fig. 3 (HSP), were treated with 4 m urea for 30 min on ice. Solubilized material (S) was separated from the integral membrane fraction (M) by centrifugation at 183,800 × g for 30 min and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. Input material was included for reference (T). Δβ-cleav., Δβ-cleavage; p, pro-form. B, Coomassie Brilliant Blue-stained PAGE of semi-native and heat-treated OM preparations from cells expressing the indicated Hbp derivatives. Fractions containing the outer membranes were either solubilized in sample buffer containing 0.2% SDS and incubated at room temperature (R) or solubilized in sample buffer containing 2% SDS at 95 °C for 10 min (H). Unfolded (u-p) and folded (f-p) forms of pro-Hbp are indicated on the right side of the panels; the identity of pro-Hbp was confirmed by immunoblotting (data not shown).
Next, we analyzed whether the β-domain of the W1015A mutant had attained its fully folded β-barrel conformation, which is a prerequisite for integral membrane insertion (45). The β-domain of Hbp attains a β-barrel conformation, which is so stable that it can only be completely denatured upon heating in SDS-containing sample buffer. This can be monitored using semi-native SDS-PAGE, in which the heat-denatured β-barrel protein runs at a different position than its non-heated native form (7). We assessed the heat modifiability of the Hbp pro-form as a measure of proper folding of its β-domain. The pooled membrane fractions containing the pro-form were solubilized in sample buffer and incubated at room temperature (R) or heated at 95 °C (H) for complete denaturation. The samples were subsequently analyzed by semi-native PAGE and Coomassie Brilliant Blue staining (Fig. 4B). Clearly, the incubation temperature did not alter the electrophoretic mobility of the W1015A pro-form, indicating that its β-domain is not properly folded in a barrel structure (Fig. 4B, lanes 5 and 6). Furthermore, and consistent with earlier findings (30), the HbpΔβ-cleavage mutant, which has its β-domain inserted into the OM, displayed heat modifiability, whereas the Hbp-L110C/G348C (like W1015A) did not show any shift (Fig. 4B, lanes 1–4).
Taken together, the data indicate that although the W1015A mutant associates with the OM, its β-domain is not properly folded into a β-barrel conformation and unable to form an integral anchor for the passenger in the OM. Consequently, the W1015A pro-form is only peripherally associated with the OM, rendering it sensitive to degradation by DegP.
HbpW1015A Is Not Detected at the Cell Surface
HbpW1015A is associated with the OM and at least partially exposed to the periplasm. Previously, we had established that the secretion-stalled Hbp L110C/G348C mutant is not fully inserted in the OM. Nevertheless, parts of that mutant were exposed at the cell surface (8). To explore whether parts of the W1015A mutant were also surface-exposed, we examined its accessibility to proteinase K that was added externally to whole MC1061degP::S210A cells. The accessibility of wild-type Hbp expressed in the same strain served as positive control. Upon translocation and proteolytic release of wild-type Hbp passenger from the β-domain, large portions remain associated to the cell surface. Cell-associated passenger Hbp is sensitive to proteinase K and yields a specific cleavage pattern of products of ∼85 and ∼60 kDa that correspond to the stable β-helical core structure of Hbp (Fig. 5, lanes 1 and 2) (8). The integrity of the bacterial cells during the procedure was verified by the lack of degradation of the periplasmic chaperone SurA (Fig. 5, lower panel). In contrast to wild-type Hbp, the pro-form W1015A remained intact upon proteinase K treatment of whole cells, suggesting either that it is not exposed at the cell surface or that only a small portion is exposed that is shielded from the protease (Fig. 5, lanes 3 and 4). To exclude the possibility that the W1015A pro-form is intrinsically protease-resistant, sonicated cells were incubated with proteinase K, which resulted in the complete degradation of the W1015A pro-form (Fig. 5, lanes 7 and 8). Upon sonication, the periplasmic SurA protein was also degraded, as expected (Fig. 5, lower panel). Furthermore, wild-type Hbp is degraded more efficiently under these conditions (Fig. 5, lanes 5 and 6), possibly due to additional protease activity in the lysed sample. Overall, the experiments indicate that the W1015A mutant associates with the OM but is mainly located in the periplasm.
FIGURE 5.
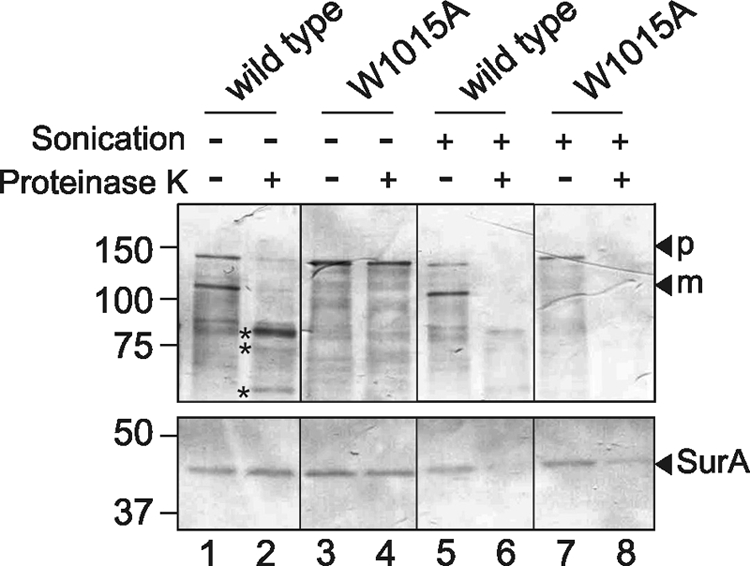
HbpW1015A is accessible to protease digestion in the periplasm. Expression of Hbp and Hbp derivatives in MC1061degP::S210A cells was induced as described in the legend for Fig. 3. One hour after induction, cells were subjected to treatment with proteinase K. Proteins separated by SDS-PAGE were probed by immunoblotting with an antiserum against mature Hbp passenger (top panels) and with an antibody against the periplasmic chaperone SurA (bottom panels). The HbpW1015A mature form (m) and pro-form (p) are indicated at the right side of the panels; the digestion fragments of wild-type Hbp passenger are indicated with an asterisk.
To corroborate these results, potential surface exposure was also monitored using whole cell immunofluorescence. MC1061degP::S210A cells expressing Hbp were fixed and subjected to indirect immunofluorescence using a polyclonal antiserum directed against the Hbp passenger (Fig. 6). As shown before, clear circumferential labeling was observed for cells expressing wild-type Hbp, the HbpΔβ-cleavage mutant (Fig. 6, A and B), and the Hbp-L110C/G348C intermediate, which are all known to expose at least part of their Hbp passenger at the cell surface (8). Cells that carried an empty vector did not show any labeling (data not shown). Similarly, and in line with the proteinase K experiments, cells that express the W1015A pro-form hardly showed any labeling (Fig. 6C), suggesting that the passenger is not surface-exposed. When cells were permeabilized with EDTA and lysozyme, efficient labeling by the serum was obtained, which showed that the mutant Hbp is expressed and recognized by the antiserum (Fig. 6F). Furthermore, antibodies against the periplasmic domain of OmpA were only able to label permeabilized and not untreated cells, confirming that labeling antibodies cannot cross the OM spontaneously (Fig. 6, G–L). Apparently, and different from what was found for the stalled Hbp-L110C/G348C mutant (8), the W1015A mutant fails to cross the OM and remains trapped within the periplasm. Together, the localization experiments indicate that the HbpW1015A pro-form remains in the periplasm in a conformation that is DegP-sensitive and loosely associated with the OM.
FIGURE 6.
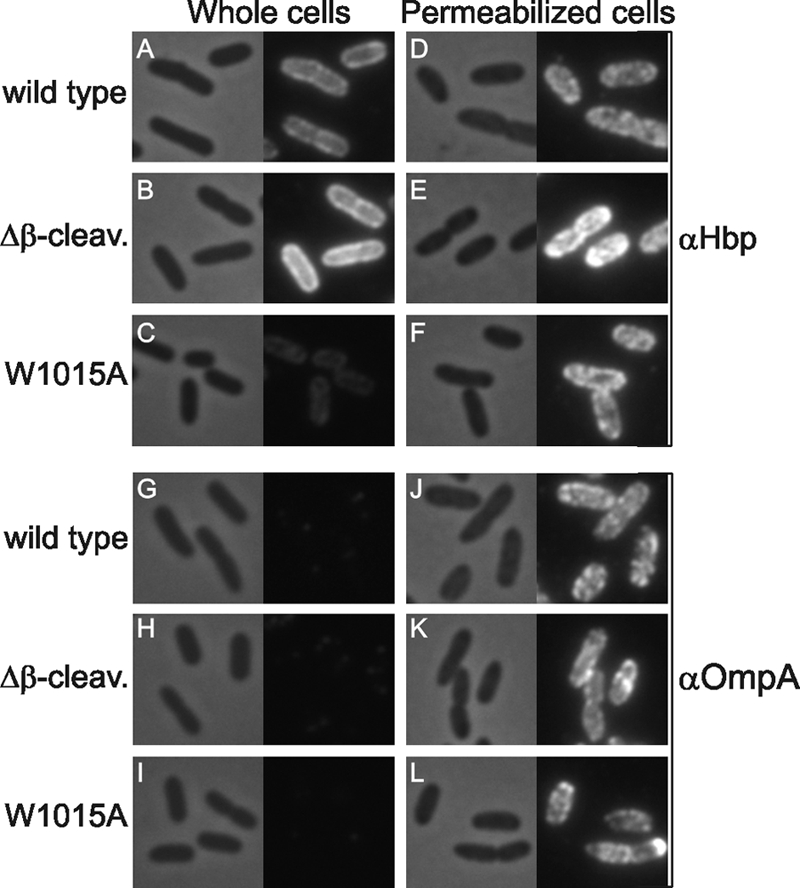
HbpW1015A cannot be detected at the cell surface by immunofluorescence. MC1061degP::S210A cells expressing Hbp or the indicated mutant Hbp derivatives or carrying an empty vector (pEH3) were collected 1 h after induction with 200 μm IPTG and subjected to indirect immunofluorescence using a polyclonal antiserum against either the Hbp passenger or OmpA and subsequently a Cy3-labeled conjugate (right panels). In addition, the corresponding fields are shown by phase-contrast microscopy (left panels). The OM of half of the cells was permeabilized prior to labeling, as indicated and described under “Experimental Procedures.” Δβ-cleav., Δβ-cleavage.
HbpW1015A Is Only Partly Associated with BamA and SurA
By using a cross-linking and immunopurification approach, we have previously shown that Hbp-L110C/G348C is a translocation intermediate that is associated with the periplasmic chaperone SurA and the BamA and BamB subunits of the Bam complex (30). Although the role of SurA and Bam in AT biogenesis is not clear, these factors are known to catalyze the insertion and assembly of OM proteins in general (32). To further characterize the stage at which HbpW1015A secretion is blocked, we investigated its molecular contacts by chemical cross-linking followed by a pulldown approach as described for the Hbp-L110C/G348C mutant (30), Briefly, MC1061degP::S210A cells expressing W1015A were cross-linked using the Lys-specific thiol-cleavable reagent DSP. After the cross-linking reaction, membrane fractions were isolated, solubilized, and subjected to immunopurification using immobilized Hbp passenger antibodies. Hbp and associated proteins were eluted in sample buffer containing DTT to dissociate the cross-linked partner proteins. As controls, the same host cells expressing wild-type Hbp, Hbp-L110C/G348C, or no Hbp at all were used. As observed before (30), four proteins were specifically cross-linked to and co-eluted with the Hbp-L110C/G348C intermediate and not with wild-type Hbp. These proteins have been identified by peptide mass fingerprinting as BamA, SurA, and BamB and confirmed by immunoblotting (Fig. 7B and data not shown). A portion of the accumulated pro-form is cleaved during or after the pulldown procedure (compare Fig. 7A, lane 5, with Fig. 2B, lane 11), as has been observed before (30). In contrast to wild-type Hbp, pulldown treatment of the cross-linked W1015A mutant shows that although similar amounts of Hbp were precipitated, significantly reduced amounts of BamA, SurA, and BamB co-eluted with the pro-form (Fig. 7A, lane 5). Taken together with the localization data, the results might suggest that the W1015A mutant is not yet fully engaged with the Bam machinery. Hence, W1015A might represent an earlier intermediate in the biogenesis of Hbp as compared with the translocation intermediate Hbp-L110C/G348C that is partly exposed at the cell surface and in closer contact with the Bam and SurA factors (8).
FIGURE 7.
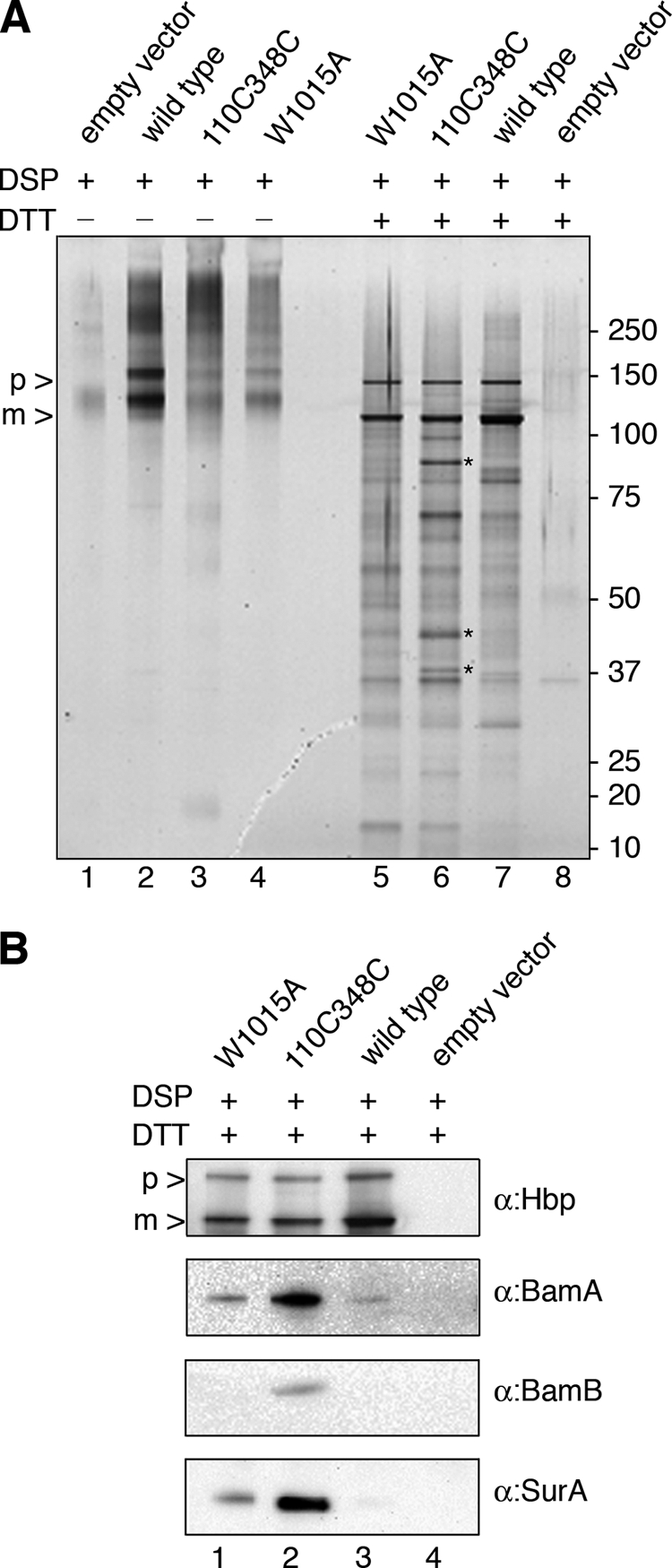
Contacts of the HbpW1015A. A, silver-stained polyacrylamide gel of samples of cross-linked cells obtained by immunopurification using anti-Hbp antibody. Cells expressing the indicated Hbp derivatives were incubated with DSP. Crude membrane fractions were isolated, solubilized, and immunopurified with anti-Hbp. Eluates were treated with DTT to cleave DSP prior to SDS-PAGE and silver staining. The asterisks indicate from top to bottom the specific cross-linked products BamA, SurA, and BamB pulled down with Hbp-L110C/G348C. p, pro-Hbp; m, mature Hbp. B, immunoblot analysis of samples obtained as described under A using antibodies against the Hbp passenger, BamA, BamB, and SurA. The antibodies against Hbp recognize both pro-Hbp and mature Hbp, which are indicated on the left side of the panels as p and m, respectively.
DISCUSSION
The C-terminal region of the AT passenger domain just upstream of the β-domain has been implicated in the folding and secretion across the OM, but the precise function of this so-called AC domain has remained unclear (22). Here, we have identified in the SPATE Hbp a conserved Trp at position 1015 that is critical for translocation of the passenger across the OM. Non-conservative mutations at this position blocked secretion and provoked degradation of Hbp in the periplasm by DegP. Furthermore, chemical cross-linking studies indicated that mutating Trp1015 did not yet fully engage the Bam machinery required for OM translocation. We showed that non-conservative substitutions of this residue stall secretion at an early stage, before initiation of OM translocation.
Around 1600 ATs have been identified in bacterial genome sequences to date. Although there is a large variation in sequence and function, available structural data and structure prediction analyses suggest that the majority of secreted passenger domains have an elongated β-helix core (15, 20–22). This core consists of stacked triangular rungs, each of which contains three β-strands connected by loops. These form a very stable scaffold to which functional domains are connected. For Hbp, this is the globular serine protease domain at the N terminus and a small domain of unknown function that protrudes from the β-stem (13). How and where the passenger folds during biogenesis and OM translocation remains unknown. However, most of the available evidence suggests that translocation of the passenger is a vectorial process in which the β-domain functions as a protein-conducting channel (2, 3). Recent in vitro unfolding/refolding studies revealed that the pertactin β-helix refolds to native protein via an intermediate state with a folded C terminus and unfolded N terminus (20, 27). In Hbp, the Trp1015 residue is located in the cap region that folds to cover the C-terminal end of the β-helical stem of the passenger (Fig. 1B). The side chain points inward in the β-helical lumen and stacks with other aromatic residues farther up in the stem. Sequence alignments of SPATEs and related serine protease ATs showed that a Trp residue is most often found at this position, although, for example, the Haemophilus influenzae ATs IgA protease (14) and Hap (46) have a Tyr and Phe at this position, respectively. β-Helix structures are usually capped both at the C terminus and the N terminus to prevent inappropriate interactions with the hydrophobic core and the unpaired strands (16). The capping structures are varied but usually contain long aliphatic residues such as Tyr or Trp to fill the helix interior. In the Hbp structure, Trp1015 appears to fulfill this function, pointing inward from an antiparallel β-hairpin that follows a disruption of the β-helix structure at Ala979/Pro980. It should be noted that the structures of the cap regions of crystallized ATs are relatively different, which might explain the different functions in AT biogenesis described (11).
The central position of this aromatic residue in the cap domain suggests that it may be involved in the nucleation of the processive folding at the extracellular side of the OM to drive translocation of the passenger across (16, 23, 25, 47). Also, other aromatic residues located closely downstream from Trp1015 in the center of the β-helix appear important for efficient secretion, although to a lesser extent. This would be expected for a sequential role of the aromatic residues in the core during the C- to N-terminal folding of the β-helix region at the cell surface after a hairpin in the β-domain is formed. In accordance with this supposition, for an intermediate of pertactin that was stalled during translocation halfway across the passenger domain, it was shown that the C terminus had crossed the OM (27).
Studies on diverse autotransporters, such as BrkA (23), SSP (25), the adhesin involved in diffuse adherence (AIDA) (48), and EspP (24), have indicated that the relatively conserved C-terminal ∼100 amino acids of the AC domain are important for folding and/or secretion. Work in the group of Fernandez (29) has shown that deletion of the BrkA AC domain, consisting of residues 601–692 at the extreme C terminus, does not inhibit translocation in itself but decreases the stability of the secreted passenger because it becomes sensitive to proteases in the OM. Strikingly, surface expression of the BrkA C terminus in trans restored the stability of the surface-exposed truncated BrkA mutant, suggesting that the AC domain could act after translocation as a scaffold from which folding is initiated. Similar results were obtained for the serine protease SSP (25) and the adhesin AIDA (48). In Hbp and other related ATs that appear conserved in this region, the conserved Trp at position 1015 might be crucial to make the first turn in the protein backbone to enable the processive stacking of the β-strands to create the native β-helix. At least it would prevent backsliding of the passenger across the presumably narrow conduit in the OM in a Brownian ratchet mechanism. In this respect, it is of interest that in vitro, the C terminus of the β-helix of both pertactin and the plasmid encoded toxin (Pet) folds faster and into a more rigid conformation than the N-terminal region (20, 47). The importance of residues that are buried in the hydrophobic core in processive folding seems to be a general feature of β-helical segments. In the P22 tailspike protein, bulky hydrophobic side chains appear to be essential for folding (17).
In the SPATE EspP, mutations in the C terminus had a drastic effect on translocation across the OM per se (24). More specifically, a region was identified corresponding to the cap and the last rungs of the EspP passenger, which is rich in conserved hydrophobic residues (24). Most residues appeared critical for efficient secretion and hence were called the hydrophobic secretion facilitator (HSF) domain. Our results are in agreement with a role for the extreme C terminus of the passenger domain in secretion itself rather than in folding after secretion because we found that the W1015A mutant is not detectably exposed at the cell surface and is degraded in the periplasm. It points toward a failure to fold the passenger into a translocation-competent state, possibly because the β-hairpin that nucleates passenger folding is strongly destabilized.
Cross-linking experiments have shown that translocation of AT passengers occurs in the direct vicinity of the Bam complex during OM translocation (30, 31). The Bam complex may keep the β-domain in an expanded conformation to facilitate transfer of small folded elements in the passenger domain. However, a concerted action of the Bam complex in the insertion of the β-domain and transfer of the passenger and even a direct pore function for the Bam complex cannot yet be excluded. Although the precise mechanism of passenger translocation is still controversial, several studies have indicated that its flexibility is limited and not compatible with the transfer of a folded β-stem or sizeable folded domains fused to it (6, 8–10). Hence, considerable folding is thought to occur upon translocation across the OM. Consistently, translocation intermediates that are partly present in the periplasm are readily degraded by DegP, indicating that they have not yet acquired their stable native conformation (6, 8–10). Under normal circumstances, degradation in the periplasm may be prevented by the rapid transit and the binding of chaperones such as SurA and Skp (30, 31, 49). However, when these chaperones are titrated by (partly) trapped intermediates in the periplasm, it may trigger the extracellular stress response and lead to up-regulation of the protease DegP.
At which stage does W1015A block secretion? Previously, we have shown that an Hbp passenger domain with a large disulfide bonded loop (Hbp-L110C/G348C) and an Hbp chimera with a folded calmodulin moiety form translocation intermediates that appear to be stalled halfway through the OM as they are sensitive to DegP but that they are also in part exposed at the cell surface (8). In contrast, W1015A appears to represent an intermediate that is stalled at an earlier step. It is also membrane-associated and sensitive to DegP but not yet exposed to the cell surface. Also, although the Hbp-L110C/G348C is extensively cross-linked to BamA, BamB, and SurA (30), this is not the case for W1015A. Although interpretation of the lack of cross-linking requires caution, it could mean that W1015A has a less intimate or less ordered contact with the Bam complex. This could reflect an enhanced flexibility of the W1015A passenger, which is not fixed in a translocation-competent state by a folded AC domain or even a contact between Bam and Trp1015 itself, although there is no evidence to support such a model. Alternatively, in a concerted translocation model, Trp1015 might be important for acquiring a passenger-β-domain conformation that is competent for a coupled Bam-mediated insertion and translocation event.
Supplementary Material
Acknowledgments
We are grateful to H. B. van den Berg van Saparoea and J. Verheul for technical assistance. We acknowledge W. Bitter and Z. Yu for critical reading of the manuscript. J. Samuelson, J. Luo, T. Silhavy, R. Kolter, V. Koronakis, J.-W. deGier, and J. Tommassen are acknowledged for providing strains and antisera.
This work was supported by an “Aard en Levens Wetenschappen Open Program” grant from the Netherlands Organization for Scientific Research (NWO) (to A. S. and W. S. P. J.). and a Mosaic Grant from NWO (to Z. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- AT
- autotransporter
- AC
- autochaperone
- IM
- inner membrane
- OM
- outer membrane
- Hbp
- hemoglobin protease
- DSP
- dithiobis(succinimidyl) propionate
- SPATE
- serine protease autotransporters of Enterobacteriaceae
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- HSP
- high speed pellet
- Lep
- Leader peptidase.
REFERENCES
- 1.Henderson I. R., Nataro J. P. (2001) Infect. Immun. 69, 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein H. D. (2007) Trends Microbiol. 15, 441–447 [DOI] [PubMed] [Google Scholar]
- 3.Hodak H., Jacob-Dubuisson F. (2007) Res. Microbiol. 158, 631–637 [DOI] [PubMed] [Google Scholar]
- 4.Henderson I. R., Navarro-Garcia F., Nataro J. P. (1998) Trends Microbiol. 6, 370–378 [DOI] [PubMed] [Google Scholar]
- 5.Dautin N., Bernstein H. D. (2007) Annu. Rev. Microbiol. 61, 89–112 [DOI] [PubMed] [Google Scholar]
- 6.Barnard T. J., Dautin N., Lukacik P., Bernstein H. D., Buchanan S. K. (2007) Nat. Struct. Mol. Biol. 14, 1214–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oomen C. J., van Ulsen P., van Gelder P., Feijen M., Tommassen J., Gros P. (2004) EMBO J. 23, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jong W. S., ten Hagen-Jongman C. M., den Blaauwen T., Slotboom D. J., Tame J. R., Wickström D., de Gier J. W., Otto B. R., Luirink J. (2007) Mol. Microbiol. 63, 1524–1536 [DOI] [PubMed] [Google Scholar]
- 9.Skillman K. M., Barnard T. J., Peterson J. H., Ghirlando R., Bernstein H. D. (2005) Mol. Microbiol. 58, 945–958 [DOI] [PubMed] [Google Scholar]
- 10.Brandon L. D., Goldberg M. B. (2001) J. Bacteriol. 183, 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley P., Charles I. G., Fairweather N. F., Isaacs N. W. (1996) Nature 381, 90–92 [DOI] [PubMed] [Google Scholar]
- 12.Gangwer K. A., Mushrush D. J., Stauff D. L., Spiller B., McClain M. S., Cover T. L., Lacy D. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16293–16298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto B. R., Sijbrandi R., Luirink J., Oudega B., Heddle J. G., Mizutani K., Park S. Y., Tame J. R. (2005) J. Biol. Chem. 280, 17339–17345 [DOI] [PubMed] [Google Scholar]
- 14.Johnson T. A., Qiu J., Plaut A. G., Holyoak T. (2009) J. Mol. Biol. 389, 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins J., Pickersgill R. (2001) Prog. Biophys. Mol. Biol. 77, 111–175 [DOI] [PubMed] [Google Scholar]
- 16.Kajava A. V., Steven A. C. (2006) Adv. Protein Chem. 73, 55–96 [DOI] [PubMed] [Google Scholar]
- 17.Simkovsky R., King J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3575–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamen D. E., Griko Y., Woody R. W. (2000) Biochemistry 39, 15932–15943 [DOI] [PubMed] [Google Scholar]
- 19.Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 20.Junker M., Schuster C. C., McDonnell A. V., Sorg K. A., Finn M. C., Berger B., Clark P. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajava A. V., Steven A. C. (2006) J. Struct. Biol. 155, 306–315 [DOI] [PubMed] [Google Scholar]
- 22.Yen Y. T., Kostakioti M., Henderson I. R., Stathopoulos C. (2008) Trends Microbiol. 16, 370–379 [DOI] [PubMed] [Google Scholar]
- 23.Oliver D. C., Huang G., Nodel E., Pleasance S., Fernandez R. C. (2003) Mol. Microbiol. 47, 1367–1383 [DOI] [PubMed] [Google Scholar]
- 24.Velarde J. J., Nataro J. P. (2004) J. Biol. Chem. 279, 31495–31504 [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi Y., Nishiyama M., Horinouchi S., Beppu T. (1994) J. Biol. Chem. 269, 32800–32806 [PubMed] [Google Scholar]
- 26.May K. L., Morona R. (2008) J. Bacteriol. 190, 4666–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junker M., Besingi R. N., Clark P. L. (2009) Mol. Microbiol. 71, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 28.Jacob-Dubuisson F., Fernandez R., Coutte L. (2004) Biochim. Biophys. Acta 1694, 235–257 [DOI] [PubMed] [Google Scholar]
- 29.Oliver D. C., Huang G., Fernandez R. C. (2003) J. Bacteriol. 185, 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauri A., Soprova Z., Wickström D., de Gier J. W., Van der Schors R. C., Smit A. B., Jong W. S., Luirink J. (2009) Microbiology 155, 3982–3991 [DOI] [PubMed] [Google Scholar]
- 31.Ieva R., Bernstein H. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. (2009) Nat. Rev. Microbiol. 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 33.Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006) PLoS Biol. 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casadaban M. J., Cohen S. N. (1980) J. Mol. Biol. 138, 179–207 [DOI] [PubMed] [Google Scholar]
- 35.Spiess C., Beil A., Ehrmann M. (1999) Cell 97, 339–347 [DOI] [PubMed] [Google Scholar]
- 36.Miller J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, Section 25.3, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: § [Google Scholar]
- 37.Hashemzadeh-Bonehi L., Mehraein-Ghomi F., Mitsopoulos C., Jacob J. P., Hennessey E. S., Broome-Smith J. K. (1998) Mol. Microbiol. 30, 676–678 [DOI] [PubMed] [Google Scholar]
- 38.Otto B. R., van Dooren S. J., Dozois C. M., Luirink J., Oudega B. (2002) Infect. Immun. 70, 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., Luirink J. (2000) EMBO J. 19, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robichon C., Vidal-Ingigliardi D., Pugsley A. P. (2005) J. Biol. Chem. 280, 974–983 [DOI] [PubMed] [Google Scholar]
- 41.Den Blaauwen T., Aarsman M. E., Vischer N. O., Nanninga N. (2003) Mol. Microbiol. 47, 539–547 [DOI] [PubMed] [Google Scholar]
- 42.Tsai H. H., Gunasekaran K., Nussinov R. (2006) Structure 14, 1059–1072 [DOI] [PubMed] [Google Scholar]
- 43.Otto B. R., van Dooren S. J., Nuijens J. H., Luirink J., Oudega B. (1998) J. Exp. Med. 188, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dooren S. J., Tame J. R., Luirink J., Oudega B., Otto B. R. (2001) FEMS Microbiol. Lett. 205, 147–150 [DOI] [PubMed] [Google Scholar]
- 45.Tajima N., Kawai F., Park S. Y., Tame J. R. (2010) J. Mol. Biol. 402, 645–656 [DOI] [PubMed] [Google Scholar]
- 46.Fink D. L., St. Geme J. W., 3rd (2003) J. Bacteriol. 185, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renn J. P., Clark P. L. (2008) Biopolymers 89, 420–427 [DOI] [PubMed] [Google Scholar]
- 48.Konieczny M. P. J., Benz I., Hollinderbäumer B., Beinke C., Niederweis M., Schmidt M. A. (2001) Antonie Van Leeuwenhoek 80, 19–34 [DOI] [PubMed] [Google Scholar]
- 49.Purdy G. E., Fisher C. R., Payne S. M. (2007) J. Bacteriol. 189, 5566–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.