Abstract
Carboxypeptidase A6 (CPA6) is an extracellular matrix-bound metallocarboxypeptidase (CP) that has been implicated in Duane syndrome, a neurodevelopmental disorder in which the lateral rectus extraocular muscle is not properly innervated. Consistent with a role in Duane syndrome, CPA6 is expressed in a number of chondrocytic and nervous tissues during embryogenesis. To better characterize the enzymatic function and specificity of CPA6 and to compare this with other CPs, CPA6 was expressed in HEK293 cells and purified. Kinetic parameters were determined using a panel of synthetic carboxypeptidase substrates, indicating a preference of CPA6 for large hydrophobic C-terminal amino acids and only very weak activity toward small amino acids and histidine. A quantitative peptidomics approach using a mixture of peptides representative of the neuropeptidome allowed the characterization of CPA6 preferences at the P1 substrate position and suggested that small and acidic P1 residues significantly inhibit CPA6 cleavage. Finally, a comparison of available kinetic data for CPA enzymes shows a gradient of specificity across the subfamily, from the very restricted specificity of CPA2 to the very broad activity of CPA4. Structural data and modeling for all CPA/B subfamily members suggests the structural basis for the unique specificities observed for each member of the CPA/B subfamily of metallocarboxypeptidases.
Keywords: Carboxypeptidase, Enzyme Kinetics, Metalloprotease, Peptidases, Proteomics
Introduction
The M14 family of metallocarboxypeptidases (CPs)2 is a large family of enzymes that functions in the cleavage of amino acids from the C termini of a variety of peptide and protein substrates (1). This family can be divided into three subfamilies based on sequence and structural similarities. The CPA/B subfamily was the first to be described, each member containing a well conserved CP domain as well as an N-terminal prodomain that is proteolytically removed for full activity (2, 3). Members of the CPN/E subfamily do not have a prodomain, but rather have a C-terminal transthyretin-like subdomain thought to be involved in protein folding (4). CPE is one well characterized member of this subfamily that processes neuropeptides and hormones in the secretory pathway (5). The cytosolic carboxypeptidase subfamily is the most recently identified, each member containing conserved CP and N-terminal domains and, in many cases, large N- and C-terminal extensions (6, 7). Recent work suggests a role for these enzymes in cytosolic peptide degradation and autophagy (8).
The CPA/B subfamily is composed of nine members, six having specificity toward C-terminal aliphatic and aromatic amino acids (CPA1–6), two having specificity toward C-terminal basic amino acids (CPB1, CPB2), and one having predicted specificity toward C-terminal acidic amino acids (CPO) (1, 3, 9). CPA1 is the best characterized member of this subfamily, as it is a highly expressed pancreatic enzyme involved in food digestion in the gut and was one of the first proteins to have its crystal structure solved (10, 11). In recent years more has been learned about the enzymatic specificity, gene expression, and physiological roles of other members of this subfamily. X-ray crystal structures have been solved for five of the nine members of this subfamily (3, 12–17). CPA2 and CPB1 are pancreatic metallocarboxypeptidases that function in the digestion of food (18). The role of CPA3 in the protective responses of mast cells has been characterized (19). CPB2, which is also known as thrombin-activatable fibrinolysis inhibitor, has been extensively studied for its role in reducing fibrinolysis (20, 21). Recently, the enzymatic characteristics of CPA4, an enzyme implicated in prostate cancer (22), have been determined and compared with those of CPA1 and CPA2 (23).
CPA6 was first identified in a bioinformatics search for additional members of the M14 metallo-CP family in the human genome (9). Soon after its identification, the CPA6 gene was shown to be present within a previously identified Duane syndrome genomic locus and disrupted in a Duane syndrome patient (24). This implicated CPA6 in the etiology of Duane syndrome, which is a neurodevelopmental disorder in which the sixth cranial nerve does not innervate its target extraocular muscle, the lateral rectus, resulting in a defect in eye abduction (25). Analysis of the CPA6 mRNA expression pattern in the mouse indicated that CPA6 is found in a number of tissues in the embryonic E14.5 mouse, including the developing vertebrae, dorsal root ganglia, skin, cerebellum, and a condensation posterior to the eye (26). We have recently shown in zebrafish that this tissue condensation near the eye is developing chondrogenic tissue near the lateral rectus muscle that may have a role in the innervation of this muscle.3 CPA6 may also play a role in developmental processes in the adult. CPA6 expression is found in the adult mouse olfactory bulb (26), where neurons develop continuously throughout adulthood, as well as in human and mouse bone marrow and the chicken Bursa of Fabricius (Unigene data base), both locations of B-cell maturation.
CPA6 is a secreted enzyme that binds tightly to the extracellular matrix (ECM) (27), suggesting that endogenous substrates of CPA6 might also be found at the ECM. CPA6 was predicted to have specificity for hydrophobic C-terminal amino acids (9). This was confirmed in a preliminary characterization of CPA6, which investigated the enzymatic activity of CPA6 in the ECM and did not use purified enzyme (27). Furthermore, these previous studies were not quantitative and did not provide kinetic values for substrate cleavage, which are important for comparing among related enzymes. Here we report the purification of human CPA6 and the characterization of its enzymatic activity and specificity. We compare these results with similar experiments using CPA1, CPA2, and CPA4 and perform modeling of the substrate binding pocket of CPA6 to compare with x-ray crystal structure information available for related enzymes. Taken together, these studies provide a comprehensive analysis of the substrate specificity of CPA6 as well as a broader perspective on metallo-CPs in general.
EXPERIMENTAL PROCEDURES
Purification of CPA6
Human CPA6, containing C-terminal HA and His6 tags (hCPA6-HAH6), was stably expressed in HEK293 cells (American Type Culture Collections (ATCC), Manassas, VA) grown in minimum essential medium (Mediatech, Inc., Manassas, VA) supplemented with 10% horse serum, glutamine, and penicillin/streptomycin. Over a period of 10 days these cells were adapted to SFM4 HEK293 serum-free medium (Thermo Scientific Hyclone, Logan, UT) supplemented with 0.5% horse serum (serum free medium resulted in significantly lower expression levels) and then transferred to spinner flasks for suspension culture (maximum 0.5 liters in a 2-liter flask for proper aeration). Medium was collected and replaced each week.
Four liters of conditioned medium was supplemented with 500 mm NaCl and 0.1% Nonidet P-40 (Nonidet P-40) and incubated batch-wise with 5 ml in Talon metal-affinity resin (Clontech, Mountain View, CA) for 2 h at 4 °C. Resin was removed by gravity filtration through filter paper, washed extensively with wash buffer (50 mm sodium phosphate, pH 7.0, 500 mm NaCl, 0.1% Nonidet P-40), transferred to a column, and eluted with 50 ml of elution buffer (50 mm sodium acetate, pH 5.0, 1.0 m NaCl, 0.1% Nonidet P-40). Eluate was immediately diluted 8-fold into 1.0 m Tris-HCl, pH 7.8, to neutralize. The Talon column eluate was diluted 5-fold in 0.1% Nonidet P-40 (to reduce NaCl ≤200 mm) and loaded onto a 1-ml HiTrap Heparin HP column (GE Healthcare). The column was washed with 50 ml of wash buffer (50 mm Tris-HCl, pH 7.5, 400 mm NaCl, 0.1% Nonidet P-40), followed by a brief detergent-free wash and elution with 50 mm Tris, pH 7.5, and 600–1000 mm NaCl. CPA6 eluted at NaCl concentrations of 800 mm or greater.
Carboxypeptidase Assays
Most of the 3-(2-furyl)acryloyl peptide (FA) carboxypeptidase substrates used in the present study were synthesized as described (23); some were purchased from Bachem (Torrance, CA). Substrates were dissolved in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl. Human CPA6 was purified from stably expressing HEK293 cells as described above. Bovine CPA1 was purchased from Sigma. Cleavage of substrate was performed at 25 °C using a total volume of 100 μl in a polystyrene 96-well plate and was measured as a decrease in absorbance at 342 nm. Assays were performed two to four times in triplicate and kinetic parameters were determined by fitting to a Michaelis-Menten curve using nonlinear regression analysis. When determining the pH optimum, substrate was dissolved in 50 mm Tris acetate buffer containing 150 mm NaCl at the indicated pH values.
Peptidomics
Quantitative peptidomics experiments were performed as described (23) with minor modifications. In brief, peptides purified from mouse brain were incubated for 90 min with 100, 10, or 1 nm purified CPA6, or incubated in the absence of enzyme. Following the incubation, the reaction was quenched, peptides were labeled with stable isotopic tags, pooled, and subjected to liquid chromatography and mass spectrometry, as described (23). In the present study, the peptidomics analysis was performed twice, each time testing 3 concentrations of enzyme and one control incubation, but with different isotopic tags used for each enzyme concentration; this was done to control for potential variations in the labeling efficiency with the isotopic reagents.
Modeling
Models were made using SWISS-MODEL (expasy) (28) and incorporate active site side chain rotamers most consistent with known CPA structures. Ramachandran plots were produced for all models to verify proper amino acid stereochemistry, and local and overall model quality was verified using Prosa-web. All images were drawn using PyMol.
RESULTS
Initially, several expression systems were tested for the production of enzymatically active CPA6. A number of CPs related to CPA6 have been expressed and secreted in high levels from insect cells using a baculovirus expression system (CPA5 (9), CPE (29), and CPM (30)), and from Pichia pastoris yeast cells (CPA4 (31) and CPB1 (32)). However, neither of these systems was successful for CPA6. Expression of a His6-tagged CPA6 in P. pastoris was not detected in either cell extracts or in the medium. Although CPA6 was strongly expressed in Sf9 insect cells using the baculovirus expression system, protein was not secreted in appreciable quantities. The small amount of intracellular CPA6 that was soluble in Sf9 cells could not be purified on the metal chelate resin, suggesting it did not have an exposed His6 tag due to improper folding. Previously, CPA6 was found to be secreted from HEK293T cells in an active form, in which the propeptide was cleaved (27). In these previous studies, CPA6 was largely retained in the extracellular matrix of the HEK293T cells. Therefore, we stably expressed CPA6 in mammalian HEK293 cells and tested if the protein was secreted into the medium when cells were grown in suspension. It was found that CPA6 was secreted into low serum-containing medium as an active enzyme (supplemental Fig. S1).
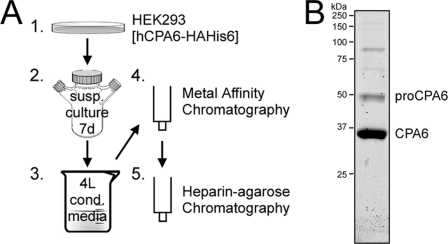
CPA6 was purified through a two-step affinity chromatography protocol (Fig. 1A and supplemental Fig. S1). The first purification step was metal affinity chromatography, making use of a His6 tag at the C terminus of CPA6. To avoid the enzymatic removal of the His6 tag by CPA6, as discovered in a previous study (27), we supplemented the medium with 2 mm benzylsuccinic acid, a CPA-specific enzyme inhibitor. HEK293-conditioned medium was stirred batchwise with Talon metal affinity resin. The resin was then collected, washed, and eluted in low pH buffer. Because CPA6 binds with high affinity to heparin (27) the Talon resin eluate was passed through a heparin-agarose affinity chromatography column and CPA6 was eluted in high salt. The resulting product was analyzed by SDS-PAGE and Coomassie Blue staining and determined to be greater than 95% pure, showing the presence of major bands at 35 and 50 kDa, corresponding to the active enzyme and proenzyme, respectively (Fig. 1B). Several weak high molecular weight bands were also seen, but could not be eliminated even by size exclusion chromatography. These may be oligomeric forms of CPA6.
FIGURE 1.
Purification of human CPA6. A, the scheme used to express and purify CPA6 involved stable expression in HEK293 cells (1), large-scale suspension culture in spinner flasks (2), collection of conditioned medium (3) and affinity chromatography using metal-affinity (4) and heparin (5) columns. B, purified CPA6 was resolved by SDS-PAGE and analyzed by Coomassie Blue staining.
To confirm the correct folding and tertiary structure of the enzyme, purified CPA6 was passed through an affinity column composed of potato carboxypeptidase inhibitor (PCI) coupled to Sepharose 4B. PCI is a specific and potent inhibitor of CPA/B enzymes through extensive surface and active site interactions (33), and has been found to inhibit CPA6 with a Ki in the low nanomolar range (27). All CPA6 loaded onto the PCI-Sepharose column bound strongly, as judged by Western blot and enzymatic activity of the flow-through and washes, and eluted in pH 12 buffer (supplemental Fig. S2). Although the high pH eluate was immediately neutralized, it had no detectable enzymatic activity, likely due to this brief exposure to strongly alkaline conditions. Therefore, this affinity column was not useful in the purification scheme. However, the PCI binding strongly suggests correct folding of the enzyme.
The pH optimum and kinetic properties of CPA6 were evaluated using a panel of CP substrates recently synthesized in our laboratory (23). These substrates consisted of the 3-(2-furyl)acryloyl (FA) chromogenic group conjugated to a C-terminal dipeptide (34). As a penultimate phenylalanine is thought to be preferred by CPA6 (27), each FA substrate contained a penultimate phenylalanine followed by a hydrophobic C-terminal amino acid (Phe, Tyr, Trp, Met, Leu, Ile, Val, Ala, or His).
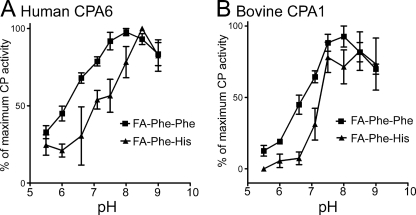
The pH optimum for purified human CPA6 was determined using the chromogenic substrate FA-Phe-Phe and compared with the pH optimum of bovine CPA1. Both enzymes exhibited an optimum at pH 7.5–8.0 (Fig. 2), consistent with a role outside of the secretory system. Outside of this optimum CPA6 exhibited a broader range of activity than that seen for CPA1. The pH optimum of both enzymes for the substrate FA-Phe-His was also determined. Differences were suspected for this substrate as the side chain of histidine has a pKa of 6.04 and therefore would be protonated at lower pH values. The preference of both CPA1 and CPA6 for uncharged substrates was supported here by the observation of a narrower pH optimum shifted slightly toward a higher pH (Fig. 2).
FIGURE 2.
pH optimum of purified human CPA6. Human CPA6 (A) or bovine CPA1 (B) were incubated with 0.2 mm FA-Phe-Phe and FA-Phe-His at the indicated pH at 25 °C. Enzyme activity was measured as the change in absorbance at 340 nm, initial reaction rates were determined and shown as the percent of maximum rate. Error bars indicate standard error of the mean.
A comparison of the Km and kcat of CPA6 in the binding and cleavage of a number of different C-terminal amino acids was performed using the above described panel of CP substrates, along with the commercially available FA-Arg-Leu. The substrates fell into three groups in regards to their ability to be cleaved by CPA6 (Table 1). Substrates with a penultimate Phe and C-terminal Phe, Tyr, Leu, Trp, and Met exhibited Km values in the range of 100–800 μm and kcat values of 4–10/s. Two substrates, FA-Arg-Leu and FA-Phe-Ile, exhibited very low enzyme affinities with Km in the range of 2000–3000 μm, but yet with turnover numbers similar to the previous group. Finally, the last three substrates, with penultimate Phe and C-terminal His, Val, and Ala exhibited very low kcat (<0.5/s) and variable affinities.
TABLE 1.
Kinetic constants for hydrolysis of synthetic substrates by CPA6
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 ± S.E. | μm ± S.E. | mm−1s−1 ± S.E. | |
| FA-Phe-Phe | 9.94 ± 1.58 | 266 ± 23 | 37.0 ± 3.4 |
| FA-Phe-Tyr | 2.84 ± 0.35 | 100 ± 10 | 29.2 ± 3.9 |
| FA-Phe-Leu | 4.67 ± 0.35 | 386 ± 127 | 19.2 ± 5.6 |
| FA-Phe-Trp | 4.13 ± 0.41 | 339 ± 60 | 15.4 ± 3.8 |
| FA-Phe-Met | 4.03 ± 0.88 | 786 ± 86 | 6.64 ± 0.92 |
| FA-Arg-Leu | 9.29 ± 1.10 | 1990 ± 240 | 4.76 ± 0.41 |
| FA-Phe-Ile | 1.91 ± 0.40 | 3070 ± 730 | 0.648 ± 0.048 |
| FA-Phe-His | 0.287 ± 0.009 | 723 ± 83 | 0.505 ± 0.084 |
| FA-Phe-Val | 0.162 ± 0.034 | 367 ± 26 | 0.434 ± 0.061 |
| FA-Phe-Ala | 0.403 ± 0.067 | 2330 ± 380 | 0.174 ± 0.015 |
The above results showed the ability of CPA6 to cleave different C-terminal (P1′) amino acids, focusing on aliphatic/aromatic residues likely to be substrates. These results gave little information on the effects of penultimate (P1) amino acids or those even farther from the C terminus. Therefore we applied a quantitative peptidomics technique to address this question. Different amounts of purified CPA6 enzyme were incubated with a peptide mixture extracted from mouse brain, representative of the peptidome of the mouse brain that might be encountered by secreted CPA6. After incubation with enzyme, the peptides were differentially labeled with isotopic tags, combined, and analyzed by liquid chromatography/mass spectrometry (LC-MS) (23, 35, 36).
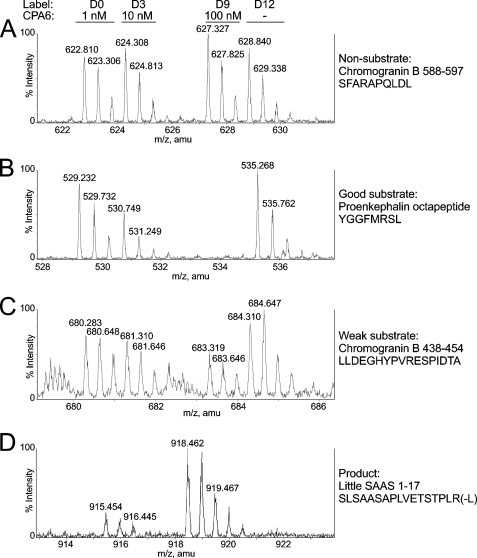
Over 100 peptides were detected through these LC-MS analyses, with close to 50 being identified through a combination of tandem mass spectrometry (MS/MS) and close matches with previously identified peptides; the criteria used for the matches included an observed monoisotopic mass within 0.004% of the theoretical mass, an expected charge equal to the number of basic residues plus the N terminus, and a correct number of isotopic tags incorporated. A number of peptides were identified in multiple LC-MS runs. In many cases the peptides in the peak set exhibited roughly equal peak heights, indicating that these peptides were not substrates or products of CPA6 under the reaction conditions used (Fig. 3A and supplemental Table S1). Some peptides exhibited a decrease in peak intensity upon incubation with medium amounts of enzyme and complete or near complete decrease with high amounts of enzyme; these are good substrates for CPA6 (Fig. 3B and Table 2). Some peptides showed little decrease in intensity with medium amounts of enzyme and no more than 70% decrease in intensity upon incubation with high amounts of enzyme; these are weak substrates of CPA6 (Fig. 3C and Table 3). Sometimes peak intensities increased with increasing amounts of CPA6 enzyme. These peptides are products formed upon cleavage of a substrate by CPA6 (Fig. 3D and Table 4).
FIGURE 3.
Identification of CPA6 peptide substrates by quantitative peptidomics. Peptides were extracted from mouse brain, digested with different amounts of purified CPA6, labeled with isotopic tags (D0 = 1 nm CPA6; D3 = 10 nm CPA6; D9 = 100 nm CPA6; D12 = no enzyme) and analyzed by LC-MS/MS for quantitative peptidomics analysis. Examples of representative data are shown for (A) non-substrates, (B) good substrates, (C) weak substrates, and (D) products.
TABLE 2.
Good substrates of CPA6
| Protein name | Peptide name | Sequence | Za | T | Obs M | Theor M | ppm | Ratio, CPA6/no enzyme |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Low CPA6 | Med CPA6 | High CPA6 | ||||||||
| Chromogranin B | 600–613 | QYDGVAELDQLLHY | 2 | 1 | 1662.80 | 1662.79 | 4 | 1.03 | 0.69 | 0.00 |
| Chromogranin B | 64–86 | SGKEVKGEEKGENQNSKFEVRLL | 6 | 5 | 2604.38 | 2604.35 | 11 | 0.76 | 0.33 | 0.00 |
| Proenkephalin | Leu-Enkephalin | YGGFL | 1 | 1 | 555.27 | 555.27 | −7 | 1.10 | 0.75 | 0.11 |
| Proenkephalin | Met-Enkephalin | YGGFM | 1 | 1 | 573.23 | 573.23 | 0 | 0.81 | 0.52 | 0.06 |
| Proenkephalin | Heptapeptide | YGGFMRF | 2 | 1 | 876.37 | 876.39 | −25 | 0.73 | 0.00 | 0.00 |
| Proenkephalin | Octapeptide | YGGFMRSL | 2 | 1 | 929.43 | 929.45 | −23 | 0.95 | 0.48 | 0.00 |
| Pro-SAAS | Little SAAS | SLSAASAPLVETSTPLRL | 2 | 1 | 1811.99 | 1812.01 | −12 | 0.95 | 0.41 | 0.08 |
| Secretogranin II | 300–316 | ESKDQLSEDASKVITYL | 3 | 3 | 1924.94 | 1924.96 | −11 | 0.79 | 0.42 | 0.00 |
a Z, charge; T, number of TMAB tags; Obs M, observed mass; Theor M, theoretical mass; ppm, difference in parts per million between observed mass and theoretical mass; ratio indicates the peak intensity observed for peptide incubated with CPA6 divided by the peak intensity for the same peptide incubated without enzyme.
TABLE 3.
Weak substrates of CPA6
See Table II for abbreviation definitions.
| Protein name | Peptide name | Sequence | Z | T | Obs M | Theor M | ppm | Ratio, CPA6/no enzyme |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Low CPA6 | Med CPA6 | High CPA6 | ||||||||
| Cathepsin D | 138–155 | YTVFDRDNNRVGFANAVV | 3 | 1 | 2056.02 | 2056.01 | 3 | 1.31 | 1.08 | 0.32 |
| Chromogranin B | 438–446 | LLDEGHYPV | 2 | 1 | 1041.51 | 1041.52 | −8 | 1.03 | 0.97 | 0.57 |
| Chromogranin B | 438–454 | LLDEGHYPVRESPIDTA | 3 | 1 | 1910.93 | 1910.93 | 0 | 0.82 | 0.58 | 0.36 |
| Myelin basic protein | N-terminal fragment | Ac-ASQKRPSQRSKYLA | 4 | 2 | 1660.96 | 1660.91 | 31 | 0.67 | 0.41 | 0.37 |
| Myelin basic protein | N-terminal fragment | Ac-ASQKRPSQRSKYLATA | 4 | 2 | 1833.02 | 1832.99 | 16 | 0.77 | 0.56 | 0.31 |
| Peptidylprolyl isomerase A | 26–39 | ADKVPKTAENFRAL | 4 | 3 | 1558.85 | 1558.85 | 5 | 0.91 | 0.62 | 0.34 |
| Prodynorphin | Dynorphin A10–17 | PKLKWDNQ | 3 | 3 | 1027.56 | 1027.54 | 12 | 0.86 | 0.63 | 0.73 |
| Proenkephalin | 218–228 | VGRPEWWMDYQ | 2 | 1 | 1465.65 | 1465.64 | 0 | 1.18 | 0.88 | 0.48 |
| Pro-SAAS | PEN | SVDQDLGPEVPPENVLGALLRV | 3 | 1 | 2316.23 | 2316.23 | 1 | 0.89 | 0.89 | 0.72 |
| Thioredoxin 1 | N-terminal fragment | VKLIESKEAFQEAL | 3 | 3 | 1603.84 | 1603.88 | −26 | 1.12 | 0.71 | 0.45 |
TABLE 4.
Products of CPA6
See Table II for abbreviation definitions.
| Protein name | Peptide name | Sequence | Cleaved | Z | T | Obs M | Theor M | ppm | Ratio, CPA6/no enzyme |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low CPA6 | Med CPA6 | High CPA6 | |||||||||
| Macrophage migration inhibitory factor | 28–47 | AQATGKPAQYIAVHVVPDQL | M | 3 | 2 | 2105.05 | 2105.13 | −37 | >3 | >3.6 | >4.4 |
| Procholecystokinin | 46–60 | AVLRTDGEPRARLGA | L | 4 | 1 | 1580.92 | 1580.87 | 29 | 1.24 | 1.18 | 2.41 |
| Procholecystokinin | 46–61 | AVLRTDGEPRARLGAL | L | 4 | 1 | 1694.01 | 1693.97 | 24 | 1.40 | 3.00 | 3.80 |
| Pro-SAAS | Little SAAS 1–17 | SLSAASAPLVETSTPLR | L | 2 | 1 | 1698.91 | 1698.91 | −2 | NDa | >5 | >20 |
a ND, not detectable.
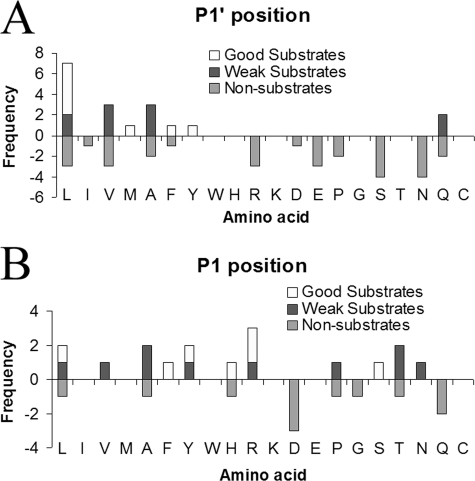
Analysis of both the C-terminal and penultimate residues of CPA6 substrate and non-substrate peptides indicated preferences at both positions, with preferred substrates having large hydrophobic C termini and large hydrophobic or basic penultimate residues. Specifically, good substrates of CPA6 contained C-terminal Tyr, Leu, Met, and Phe (Fig. 4A) and penultimate amino acids in these substrates were, with one exception, large hydrophobic or basic residues (Table 2 and Fig. 4B). Analysis of the downstream residues of the products indicated that formation of these peptides required cleavage of C-terminal Leu and Met (Table 4). Weak substrates of CPA6 contained C-terminal Val, Ala, Gln, and Leu (Fig. 4A). In the two cases in which a C-terminal Leu was a poor substrate, the penultimate residue was Ala (Table 3). No peptides with C-terminal basic or acidic residues were identified as substrates. However, many such peptides were detected in the study, and all were found to be non-substrates. These non-substrates included peptides with C-terminal Arg, Glu, Asp, Asn, Ser, and Pro (supplemental Table S1), consistent with predictions that CPA6 would not cleave charged or polar residues. However, some of the non-substrates contained C-terminal amino acids that had been found to be substrates in other peptides or chromogenic substrates; these residues included Leu, Ile, Val, Ala, Phe, and Gln (Fig. 4A). In cases in which the C-terminal residue was Leu or Phe, a residue known to be efficiently cleaved by CPA6, the penultimate residue was either Asp or Pro (supplemental Table S1). In cases in which the C-terminal residue was not a large hydrophobic residue, but yet known to be cleaved (Val, Gln, Ile, and Ala), the penultimate residue appeared to determine cleavage, with Gln, Thr, His, Gly, Ala, as well as Asp and Pro being unfavorable for cleavage (Fig. 4B). On occasion non-substrate peptides were found with P1 and P1′ residues that were conductive to cleavage in other substrate peptides. These may have amino acids in P2 and P3 positions that impart specificity.
FIGURE 4.
Peptidomics analysis of CPA6 preferences at substrate P1′ and P1 positions. A, analysis of the P1′ residue. All good substrates have C-terminal Leu, Met, Phe, or Tyr, whereas weak substrates have C-terminal Leu, Val, Ala, or Gln. Many different C-terminal amino acids can be found in the non-substrate category. B, analysis of the P1 residue of peptides with permissive P1′ residues. Altogether, 28 peptides were detected with C-terminal hydrophobic residues (Leu, Ile, Val, Met, Ala, Phe, and Tyr) or Gln; all of these residues were found to be cleaved either for some peptides in the peptidomics analysis, or with small synthetic substrates. Of these 28, only 7 were good substrates, 10 were weak substrates, and 11 were not cleaved. Analysis of the sequences of these peptides indicates that most good substrates contain hydrophobic or basic amino acids in the P1 position, whereas the majority of non-substrates contain Asp or Gln in this position.
With this data on the specificity of CPA6, and recently published data regarding the specificity of CPA1, -2, and -4 obtained from studies performed using the same batch of synthetic substrates (23), we now have comparable data on the enzymatic activity and specificity of four of the six mammalian CPA enzymes (supplemental Table S2). Based on this comparison, it appears that the activity of CPA6 toward its better substrates is about 10–100-fold lower than the activities reported for CPA1, -2, and -4. Because a metal affinity resin was used in the purification of CPA6, the possibility remained that a portion of CPA6 was properly folded but enzymatically inactive due to chelation of the critical active site zinc. This possibility was investigated by incubating purified CPA6 with nanomolar concentrations of zinc, which would be expected to restore activity if zinc chelation was a problem. No large changes in CPA6 activity were detected with nanomolar levels of zinc (supplemental Fig. S3). However, as for many CPs (37), moderate concentrations of zinc (10 μm to 1 mm) resulted in inhibition of CPA6 activity (supplemental Fig. S3). Another possible explanation for the weak CPA6 activity was that CPA6 might require interaction with components of the ECM for optimal activity. Because ECM-bound CPA6 was previously solubilized by heparin (27), likely by competing with heparin sulfate proteoglycans for interaction with CPA6, heparin was added to enzymatic reactions of CPA6 and the FA-Phe-Phe substrate. No change in activity was detected (results not shown). Finally, a direct comparison of enzymatic activity was made between equivalent molar amounts of purified CPA6 and CPA6 found within its native environment of the ECM following secretion from HEK293T cells. Dilutions of purified HA-tagged CPA6 were analyzed by Western blot alongside ECM extracts from cells transfected with the same HA-tagged CPA6 to determine the approximate amount of CPA6 present within the ECM. Both ECM-bound and purified CPA6 were then incubated with 0.4 mm FA-Phe-Phe substrate and activities were compared (supplemental Fig. S4). These results suggested that CPA6 exhibited comparable enzymatic activity under both conditions. Thus, components of the ECM did not appear to have a large effect on CPA6 enzyme activity. Furthermore, this result indicated that the large difference in activity between purified CPA6 and other CPA enzymes was not due to inactivation of the CPA6 during purification.
When the absolute reaction rate was disregarded and the reaction rates of each enzyme were compared across the panel of substrates relative to FA-Phe-Phe, these four peptidases could be arranged in a hierarchy of substrate cleavage, CPA2 < CPA6 < CPA1 < CPA4, from very narrow to quite broad substrate specificity (corresponding, to a large extent, to Km differences for each substrate). Although CPA2 had very restricted specificity for aromatic amino acids such as Phe and Trp, CPA6 was able to cleave aromatic amino acids as well as Leu and Met, with negligible activity toward other substrates. CPA1 appeared to cleave practically all amino acids tested, albeit with reduced activity toward Trp, Ala, and Met. Finally, CPA4 had specificity similar in nature to CPA1, but with the additional ability to cleave Met.
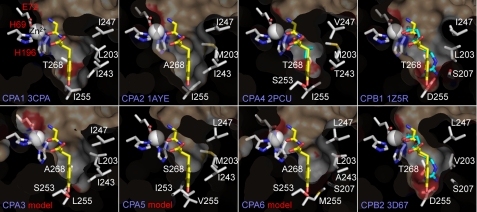
An explanation for differences in substrate specificity exhibited by CPA1, -2, -4, and -6 was likely to be found in the binding pockets of these enzymes. X-ray crystal structures available for CPA1, -2, and -4, as well as for CPB1 and -2, were analyzed and used to model the active sites of CPA3, -5, and -6 (Fig. 5 and Table S3). The strong affinity of CPA2 for large aromatic amino acids could be seen in a much larger active site pocket when compared with other CPs, given more space by Ala268, rather than Thr268 as seen in CPA1 and CPA4 (Fig. 5). The exclusion of smaller branched amino acids such as Leu and Ile by CPA2 might be mediated by the presence of the longer Met203 at the neck of the pocket, rather than Leu203 of CPA1 (Fig. 5). This Met203 was also present in CPA4 and CPA5 (Fig. 5 and Table S3) and might account for the reduced affinity of CPA4 for Val and Ile when compared with CPA1. Although CPA1 and CPA2 had a Gly at the bottom of the active site pocket in position 253 (not shown), several enzymes had either a Ser (CPA3, CPA4, and CPA6) or an Ile (CPA5; Fig. 5), either of which should interfere with the binding of very large amino acids such as Trp. Finally, it might be noted that the bottom of the active site pocket in both CPA4 and CPA6 may exhibit significant electronegativity due to the presence of a number of polar residues: Thr243, Thr268, and Ser253 of CPA4, and Ser207 and Ser253 of CPA6. In fact, Ser207 was found in both CPB1 and CPB2 and appears to contribute to the electronegative pocket suitable for binding basic residues in these enzymes. This suggests a possible reason for the cleavage of polar amino acids such as Gln by CPA4 and -6 and for the relatively high affinity of CPA6 for FA-Phe-Tyr when compared with other substrates. The significance of a unique Met at position 255 at the base of the specificity pocket in CPA6 is unknown at this time. Modeling does not suggest any function, as it appears that the amino acid at position 253 may play a more important role in the nature of the specificity pocket in A-like enzymes. In addition, this residue in CPA6 orthologs in a number of fishes (fugu, stickleback, and medaka) is replaced by Ile, as found in CPA1, -2, and -4.
FIGURE 5.
Comparative modeling of CPA6 and other CPA/B enzymes suggests residues critical for substrate binding. CPA/B residues involved in forming the specificity pockets (having side chains within 6 Å of bound substrate in 3CPA structure) of all members of the subfamily are shown. As is the convention in the field, all residues are numbered according to the corresponding active site residues in active bovine CPA1 (following propeptide cleavage). The GY dipeptide ligand (yellow) from the 3CPA crystal structure is overlaid on each structure for comparison, partially hiding residue 268. The CPA4 2PCU and CPB2 3D67 structures are shown with co-crystallized ligands aspartate and GEMSA, respectively, shown in cyan. GEMSA is also modeled into the CPB1 active site for comparison sake.
DISCUSSION
The metallocarboxypeptidase family is a large family of proteases, many with overlapping substrate specificity profiles. For example, six different CPs (CPD, CPE, CPZ, CPN, CPB1, and CPB2) are all able to cleave C-terminal basic residues Arg and Lys (38). Their unique functions are thought to depend largely on unique spatiotemporal and subcellular distributions. In a similar manner, mammalian genomes contain six CPA genes all producing enzymes able to cleave C-terminal hydrophobic amino acids (9). Many of these genes have unique expression profiles and all of their products are regulated through propeptide cleavage.
Although the CPAs are often thought to have similar substrate specificity toward hydrophobic amino acids, it has been known for some time that CPA2 is unique in that it has a strong preference for large amino acids such as tryptophan and phenylalanine (39). Recently, the substrate specificity of CPA1, CPA2, and CPA4 toward a large number of hydrophobic amino acids was compared, indicating a number of differences that may be critical in understanding the natural substrates of these enzymes (23). This study confirmed the preference of CPA2 for large hydrophobic amino acids, and showed a rather broad cleavage spectrum for CPA4. CPA1 also exhibited broad substrate specificity, but when compared with CPA4 showed significantly lower activity toward Met and Ile when compared with its activity toward Phe.
In the present study we have purified CPA6 and determined its substrate specificity. We show that CPA6, rather than cleaving any hydrophobic amino acid, exhibits substrate cleavage preferences for large hydrophobic amino acids. Reaction rates (kcat/Km) for the cleavage of nine different C-terminal hydrophobic amino acids by CPA6 spanned 3 orders of magnitude, with C-terminal Ile, Val, Ala, and His exhibiting reaction rates ∼100-fold lower than the bulkier amino acids Phe, Leu, Tyr, Met, and Trp. Our results also suggest that CPA6 does not cleave substrates with Gly, Pro, Asp, or Glu in the penultimate position. This is similar to many other CPs which, regardless of C-terminal specificity, have been shown not to cleave substrates containing a penultimate Pro (CPM (40), thrombin-activatable fibrinolysis inhibitor (41), CPZ (42), and CPA4 (23)). CPA4 has also been been shown not to cleave substrates with acidic residues in the penultimate P1 position (23).
It was surprising that C-terminal histidine was such a poor substrate for CPA6 in the present study. Previously we had found that CPA6 bound to the ECM cleaved histidine as part of a C-terminal His6 tag (27). Our current results suggest two possibilities: a His6 tag as a part of a larger protein is a better substrate for CPA6 than small chromogenic substrates, and/or the localization of a substrate to the ECM, possibly at locally high concentrations, is a stronger determinant for cleavage by CPA6 than simply C-terminal amino acid affinity. This second possibility is supported by another observation; although a CP inhibitor was typically included in the culture medium of the CPA6-expressing HEK293 cells to prevent His6 tag removal, the absence of this inhibitor in suspension culture did not result in large-scale removal of His6 tags.3 This was in contrast to results obtained from CPA6 bound to ECM, in which most histidines were cleaved in the absence of the CP inhibitor (27).
Because CPA6 is present in the ECM and expressed during development, it is possible that it processes proteins or peptides involved in morphogenesis. Several extracellular morphogens have conserved hydrophobic C-terminal amino acids that might be cleaved by an enzyme such as CPA6; examples include Wnt1, Wnt6, several bone morphogenetic proteins, semaphorin 3a, and fibroblast growth factor-4 and -6. A few of these or their relatives have been shown to be functionally modified through C-terminal proteolytic processing. For example, the C-terminal Arg residue of Wnt4 is cleaved by CPZ (43), an ECM metallocarboxypeptidase with specificity toward basic amino acids (42, 44, 45). This processing step enhances the activity of Wnt4 and its effect on growth plate chondrocyte terminal differentiation (43). Wnt4 is a member of the large highly conserved family of ECM-bound Wnt ligands. Two members of this family, Wnt1 and Wnt6, have C-terminal sequences similar to Wnt4, but with a hydrophobic Leu at their C termini rather than the Arg found in Wnt4 and many other members. Removal of this C-terminal Leu by an extracellular enzyme such as CPA6 might activate Wnt1 and Wnt6 in a manner similar to Wnt4. Wnt6 in particular has an expression pattern in the developing limbs and somites consistent with that observed for CPA6 (46–48).
A number of potential neuropeptide substrates were identified in the present study. These included peptides derived from proenkephalin (Leu-enkephalin, Met-enkephalin, heptapeptide, and octapeptide), pro-SAAS (Little SAAS and PEN), and chromogranin B (600–613, 64–86, 438–446, and 438–454). The mRNAs encoding all of these proteins are expressed in the mitral and granular cell layers of the mouse olfactory bulb (Allen brain atlas), which is the major location of CPA6 mRNA in adult mouse (26). Thus, these identified peptide substrates of CPA6 are possible in vivo targets of CPA6. However, little is known about the biological functions of many of these peptides. Removal of C-terminal Leu and Met from enkephalin virtually eliminates activity (49); however, a role for enkephalins in the olfactory bulb is unknown at this time. Pro-SAAS-derived peptides are not yet well characterized, although they may be involved in body weight regulation (50). The physiological effect of C-terminal Leu removal from Little SAAS is not known. A number of peptides have been identified from chromogranin B, some proposed to have antimicrobial functions, and others of unknown function (51). Chromogranin B 600–613, shown in this study to be cleaved by CPA6, has been previously found to undergo carboxypeptidase-like cleavages resulting in C-terminal-truncated forms of this peptide (52). However, to our knowledge the functions of these peptides remain unknown.
Ultimately it is the determination of the unique substrate specificities of each CP enzyme and the identification of substrates that will allow us to understand the functions of these enzymes. One step toward this understanding has been the clarification of the structural basis for the activity and specificity of CPs through x-ray crystallography. Although many members of the CPA/B subfamily of CPs have had their structures solved, CPA3, CPA5, and CPA6 have not. Previous studies have reported structural models for these enzymes (9, 53), but focused largely on overall tertiary structure rather than analysis of the enzyme active sites. Here we have described a detailed analysis of the modeled substrate binding pockets of CPA3, CPA5, and CPA6 alongside crystallized members of this subfamily. Although we have kinetic data for CPA6 to support the modeling, no such data yet exists for CPA3 and CPA5. However, these structural models might be used to predict the substrate specificity of CPA3 and CPA5. The specificity pocket of CPA3 (Fig. 5) suggests similarity to the substrate binding profile of CPA1 and CPA6, with Ala268 making the pocket deeper, yet Ser253 making it shorter, resulting in exclusion of amino acids at the extremes in size. Experiments have shown CPA3 to be able to cleave the C-terminal residues of neurotensin (Leu), endothelin-1 (Trp), sarafotoxin 6b (Ile-Trp), kinetensin (Leu), Leu-enkephalin (Leu), and xenopsin (Leu) (54). However, another study indicated that purified human CPA3 displayed no activity toward carboxyl-terminal Trp or Ala, but more activity than bovine CPA1 against carboxyl-terminal Leu residues and about equal activity toward Phe and Tyr residues (55), suggesting that, indeed, intermediate sized residues may be optimal. CPA5, in contrast to CPA3, must certainly prefer smaller substrates, as it contains a Ser268 restricting depth, Ile253 restricting length, and Met203 restricting width (Fig. 5). At this point there is little data regarding the specificity of CPA5 although it has been shown to cleave FA-Arg-Leu (9).
In conclusion, the data presented confirm a role for CPA6 in the cleavage of large hydrophobic amino acids. These data also suggest that less optimal substrates, including His and small hydrophobic amino acids, are also cleaved by CPA6, possibly more so if substrates containing these C-terminal amino acids are proteins bound to the ECM along with CPA6. We show that the substrate P1 residue has a large effect on cleavage, with small and acidic residues significantly inhibiting this process. Finally, modeling illustrates the composition of the specificity pocket of CPA6, as well as CPA3 and CPA5, and has enabled some predictions to be made in regard to optimal substrates for these enzymes.
Supplementary Material
Acknowledgments
We thank Dr. Jonathan Backer for the use of his spinner flask system and technical advice and Prof. F. X. Aviles for the generous gift of the PCI-Sepharose resin.
This work was supported, in whole or in part, by National Institutes of Health Grant DA-004494 (to L. D. F.) and postdoctoral fellowships (to P. J. L.) from the Natural Sciences and Engineering Research Council of Canada and the National Eye Institute of the National Institutes of Health Grant EY-194332 from the NEI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S4.
P. J. Lyons and L. D. Fricker, unpublished data.
- CP
- carboxypeptidase
- ECM
- extracellular matrix
- FA
- 3-(2-furyl)acryloyl
- PCI
- potato carboxypeptidase inhibitor
- LC
- liquid chromatography.
REFERENCES
- 1.Arolas J. L., Vendrell J., Aviles F. X., Fricker L. D. (2007) Curr. Pharm. Des. 13, 349–366 [DOI] [PubMed] [Google Scholar]
- 2.Ventura S., Gomis-Rüth F. X., Puigserver A., Avilés F. X., Vendrell J. (1997) Biol. Chem. 378, 161–165 [PubMed] [Google Scholar]
- 3.Gomis-Rüth F. X. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 319–345 [DOI] [PubMed] [Google Scholar]
- 4.Gomis-Rüth F. X., Companys V., Qian Y., Fricker L. D., Vendrell J., Avilés F. X., Coll M. (1999) EMBO J. 18, 5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fricker L. D., Leiter E. H. (1999) Trends Biochem. Sci. 24, 390–393 [DOI] [PubMed] [Google Scholar]
- 6.Kalinina E., Biswas R., Berezniuk I., Hermoso A., Aviles F. X., Fricker L. D. (2007) FASEB J. 21, 836–850 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez de la Vega M., Sevilla R. G., Hermoso A., Lorenzo J., Tanco S., Diez A., Fricker L. D., Bautista J. M., Avilés F. X. (2007) FASEB J. 21, 851–865 [DOI] [PubMed] [Google Scholar]
- 8.Berezniuk I., Sironi J., Callaway M. B., Castro L. M., Hirata I. Y., Ferro E. S., Fricker L. D. (2010) FASEB J. 24, 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei S., Segura S., Vendrell J., Aviles F. X., Lanoue E., Day R., Feng Y., Fricker L. D. (2002) J. Biol. Chem. 277, 14954–14964 [DOI] [PubMed] [Google Scholar]
- 10.Rupley J. A., Neurath H. (1960) J. Biol. Chem. 235, 609–615 [PubMed] [Google Scholar]
- 11.Hartsuck J. A., Ludwig M. L., Muirhead H., Steitz T. A., Lipscomb W. N. (1965) Proc. Natl. Acad. Sci. U.S.A. 53, 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christianson D. W., Lipscomb W. N. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 7568–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sáez I., Reverter D., Vendrell J., Avilés F. X., Coll M. (1997) EMBO J. 16, 6906–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayés A., Fernández D., Solà M., Marrero A., García-Piqué S., Avilés F. X., Vendrell J., Gomis-Rüth F. X. (2007) Biochemistry 46, 6921–6930 [DOI] [PubMed] [Google Scholar]
- 15.Adler M., Bryant J., Buckman B., Islam I., Larsen B., Finster S., Kent L., May K., Mohan R., Yuan S., Whitlow M. (2005) Biochemistry 44, 9339–9347 [DOI] [PubMed] [Google Scholar]
- 16.Marx P. F., Brondijk T. H., Plug T., Romijn R. A., Hemrika W., Meijers J. C., Huizinga E. G. (2008) Blood 112, 2803–2809 [DOI] [PubMed] [Google Scholar]
- 17.Sanglas L., Valnickova Z., Arolas J. L., Pallarés I., Guevara T., Solà M., Kristensen T., Enghild J. J., Aviles F. X., Gomis-Rüth F. X. (2008) Mol. Cell 31, 598–606 [DOI] [PubMed] [Google Scholar]
- 18.Clauser E., Gardell S. J., Craik C. S., MacDonald R. J., Rutter W. J. (1988) J. Biol. Chem. 263, 17837–17845 [PubMed] [Google Scholar]
- 19.Schneider L. A., Schlenner S. M., Feyerabend T. B., Wunderlin M., Rodewald H. R. (2007) J. Exp. Med. 204, 2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouma B. N., Mosnier L. O. (2006) Ann. Med. 38, 378–388 [DOI] [PubMed] [Google Scholar]
- 21.Willemse J. L., Heylen E., Nesheim M. E., Hendriks D. F. (2009) J. Thromb. Haemost. 7, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., Reed C. P., Zhang J. S., Shridhar V., Wang L., Smith D. I. (1999) Cancer Res. 59, 2981–2988 [PubMed] [Google Scholar]
- 23.Tanco S., Zhang X., Morano C., Avilés F. X., Lorenzo J., Fricker L. D. (2010) J. Biol. Chem. 285, 18385–18396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pizzuti A., Calabrese G., Bozzali M., Telvi L., Morizio E., Guida V., Gatta V., Stuppia L., Ion A., Palka G., Dallapiccola B. (2002) Invest. Ophthalmol. Vis. Sci. 43, 3609–3612 [PubMed] [Google Scholar]
- 25.Gutowski N. J. (2000) Eur. J. Neurol. 7, 145–149 [DOI] [PubMed] [Google Scholar]
- 26.Fontenele-Neto J. D., Kalinina E., Feng Y., Fricker L. D. (2005) Brain Res. Mol. Brain Res. 137, 132–142 [DOI] [PubMed] [Google Scholar]
- 27.Lyons P. J., Callaway M. B., Fricker L. D. (2008) J. Biol. Chem. 283, 7054–7063 [DOI] [PubMed] [Google Scholar]
- 28.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 29.Qian Y., Varlamov O., Fricker L. D. (1999) J. Biol. Chem. 274, 11582–11586 [DOI] [PubMed] [Google Scholar]
- 30.Tan F., Balsitis S., Black J. K., Blöchl A., Mao J. F., Becker R. P., Schacht D., Skidgel R. A. (2003) Biochem. J. 370, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallarès I., Bonet R., García-Castellanos R., Ventura S., Avilés F. X., Vendrell J., Gomis-Rüth F. X. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3978–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura S., Villegas V., Sterner J., Larson J., Vendrell J., Hershberger C. L., Avilés F. X. (1999) J. Biol. Chem. 274, 19925–19933 [DOI] [PubMed] [Google Scholar]
- 33.Vendrell J., Querol E., Avilés F. X. (2000) Biochim. Biophys. Acta 1477, 284–298 [DOI] [PubMed] [Google Scholar]
- 34.Peterson L. M., Holmquist B., Bethune J. L. (1982) Anal. Biochem. 125, 420–426 [DOI] [PubMed] [Google Scholar]
- 35.Fricker L. D., Lim J., Pan H., Che F. Y. (2006) Mass. Spectrom. Rev. 25, 327–344 [DOI] [PubMed] [Google Scholar]
- 36.Morano C., Zhang X., Fricker L. D. (2008) Anal. Chem. 80, 9298–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez-Ortiz M., Gomis-Rüth F. X., Huber R., Avilés F. X. (1997) FEBS Lett. 400, 336–340 [DOI] [PubMed] [Google Scholar]
- 38.Reznik S. E., Fricker L. D. (2001) Cell Mol. Life Sci. 58, 1790–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardell S. J., Craik C. S., Clauser E., Goldsmith E. J., Stewart C. B., Graf M., Rutter W. J. (1988) J. Biol. Chem. 263, 17828–17836 [PubMed] [Google Scholar]
- 40.Deiteren K., Surpateanu G., Gilany K., Willemse J. L., Hendriks D. F., Augustyns K., Laroche Y., Scharpé S., Lambeir A. M. (2007) Biochim. Biophys. Acta 1774, 267–277 [DOI] [PubMed] [Google Scholar]
- 41.Willemse J., Leurs J., Verkerk R., Hendriks D. (2005) Anal. Biochem. 340, 106–112 [DOI] [PubMed] [Google Scholar]
- 42.Novikova E. G., Fricker L. D. (1999) Biochem. Biophys. Res. Commun. 256, 564–568 [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Shao Y. Y., Ballock R. T. (2009) J. Bone Miner Res. 24, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novikova E. G., Reznik S. E., Varlamov O., Fricker L. D. (2000) J. Biol. Chem. 275, 4865–4870 [DOI] [PubMed] [Google Scholar]
- 45.Song L., Fricker L. D. (1997) J. Biol. Chem. 272, 10543–10550 [DOI] [PubMed] [Google Scholar]
- 46.Geetha-Loganathan P., Nimmagadda S., Christ B., Huang R., Scaal M. (2010) BMC Dev. Biol. 10, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witte F., Dokas J., Neuendorf F., Mundlos S., Stricker S. (2009) Gene Expr. Patterns 9, 215–223 [DOI] [PubMed] [Google Scholar]
- 48.Schmidt C., Stoeckelhuber M., McKinnell I., Putz R., Christ B., Patel K. (2004) Dev. Biol. 271, 198–209 [DOI] [PubMed] [Google Scholar]
- 49.Morgan B. A., Smith C. F., Waterfield A. A., Hughes J., Kosterlitz H. W. (1976) J. Pharm. Pharmacol. 28, 660–661 [DOI] [PubMed] [Google Scholar]
- 50.Morgan D. J., Wei S., Gomes I., Czyzyk T., Mzhavia N., Pan H., Devi L. A., Fricker L. D., Pintar J. E. (2010) J. Neurochem. 113, 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helle K. B. (2010) Results Probl. Cell Differ. 50, 21–44 [DOI] [PubMed] [Google Scholar]
- 52.Dillen L., Boel S., De Potter W. P., Claeys M. (1992) Biochim. Biophys. Acta 1120, 105–112 [DOI] [PubMed] [Google Scholar]
- 53.Springman E. B., Dikov M. M., Serafin W. E. (1995) J. Biol. Chem. 270, 1300–1307 [DOI] [PubMed] [Google Scholar]
- 54.Pejler G., Knight S. D., Henningsson F., Wernersson S. (2009) Trends Immunol. 30, 401–408 [DOI] [PubMed] [Google Scholar]
- 55.Natsuaki M., Stewart C. B., Vanderslice P., Schwartz L. B., Natsuaki M., Wintroub B. U., Rutter W. J., Goldstein S. M. (1992) J. Invest. Dermatol. 99, 138–145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.