Abstract
We have previously reported that thrombin-activatable fibrinolysis inhibitor (TAFI) exhibits intrinsic proteolytic activity toward large peptides. The structural basis for this observation was clarified by the crystal structures of human and bovine TAFI. These structures evinced a significant rotation of the pro-domain away from the catalytic moiety when compared with other pro-carboxypeptidases, thus enabling access of large peptide substrates to the active site cleft. Here, we further investigated the flexible nature of the pro-domain and demonstrated that TAFI forms productive complexes with protein carboxypeptidase inhibitors from potato, leech, and tick (PCI, LCI, and TCI, respectively). We determined the crystal structure of the bovine TAFI-TCI complex, revealing that the pro-domain was completely displaced from the position observed in the TAFI structure. It protruded into the bulk solvent and was disordered, whereas TCI occupied the position previously held by the pro-domain. The authentic nature of the presently studied TAFI-inhibitor complexes was supported by the trimming of the C-terminal residues from the three inhibitors upon complex formation. This finding suggests that the inhibitors interact with the active site of TAFI in a substrate-like manner. Taken together, these data show for the first time that TAFI is able to form a bona fide complex with protein carboxypeptidase inhibitors. This underlines the unusually flexible nature of the pro-domain and implies a possible mechanism for regulation of TAFI intrinsic proteolytic activity in vivo.
Keywords: Blood Coagulation Factors, Enzyme Inactivation, Enzyme Inhibitors, Fibrinolysis, Metalloprotease, Protease, Protein Structure, X-ray Crystallography
Introduction
Pancreatic carboxypeptidase A1 is the archetype of the M14 metallocarboxypeptidase (MCP)3 family (1), also referred to as the funnelin tribe of metallopeptidases (2). The members of this family are subdivided into A/B or N/E MCPs on the basis of primary structure and structural homologies (3). They are also classified according to their substrate preference for C-terminal hydrophobic (A-type) or basic (B-type) amino acid residues. Thrombin-activatable fibrinolysis inhibitor (TAFI) (EC 3.4.17.20; UniProt Q96IY4) is a paralogue of pancreatic pro-carboxypeptidase B1 (pro-CPB1), pro-carboxypeptidases A1 (pro-CPA1), and A2 (pro-CPA2) and is also known as plasma pro-carboxypeptidase B2, R, or U (4–9). Similarly to other B-type MCPs, TAFI possesses an N-terminal pro-domain that functions as a gatekeeper preventing substrate access to a preformed active site (for a review, see Refs. 2, 3). In human and porcine pancreatic pro-CPB1, access to the active site is completely shielded, resulting in no detectable intrinsic enzymatic activity of the zymogen (10). This is in contrast to human pro-CPA1 and pro-CPA2, which allow cleavage of small peptide substrates (11). Equally, pro-carboxypeptidase B from Pacific dogfish and African lungfish exhibit intrinsic activity toward small substrate molecules (12, 13). Likewise, TAFI interferes with carboxypetidase N assays (14), and is capable of cleaving larger peptide substrates due to its low but continuous carboxypeptidase activity (15). These variations in the level of intrinsic activity between the different pro-carboxypeptidases may be explained by structural differences of their activation peptides and the adjustment between pro-domain and mature enzyme moiety (10, 16). In particular, the presence of a strong salt-bridge between Asp-41 of the pro-domain (human CPB numbering) and a S1′ substrate-binding site residue (Arg-145) in the catalytic moiety of pro-CPB1 has been attributed a central role in explaining the lack of intrinsic activity (17).
The 92-residue pro-domain of TAFI contains four N-linked glycan structures (18). This is the only MCP pro-domain that is glycosylated, thus suggesting biological significance. In this context, it has been speculated that the carbohydrates aid in stabilization and solubility of the enzyme and may affect the intrinsic proteolytic activity (15, 19). In addition, the majority of the identified transglutaminase amine acceptor sites are also located in the pro-domain, thus enabling TAFI to be cross-linked to other proteins during coagulation/fibrinolysis (18, 20, 21).
The pro-domain of TAFI can be removed by trypsin-like proteases through proteolysis of the Arg-92—Ala-93 peptide bond, forming the mature form, TAFIa (5, 22–26). Following this cleavage event, the activity is increased significantly but at the expense of a dramatic decrease in thermodynamic stability, which renders TAFIa inactive within minutes both in vitro and in the bloodstream (5, 25, 27–30). During this burst of activity, TAFIa modulates hemostasis by removing surface-exposed lysine residues from the newly formed clot during the fibrinolysis step (5, 22–25), thus functioning as an inhibitor of fibrinolysis (7, 27, 31–34). TAFIa has also been implicated in other processes including wound healing (35), blood pressure regulation (36–38), inflammation (39, 40), sepsis (41), endometriosis (42), and pulmonary fibrosis (43).
The crystal structures of human and bovine TAFI revealed the molecular basis for the thermodynamic instability of the mature protease as compared with its related, but more robust pancreatic counterparts (44, 45). Furthermore, the incentive for the intrinsic proteolytic activity of the protein was revealed. The TAFI pro-domain was partially rotated away from the active site, exposing the catalytic residues to the solvent. Additionally, the corresponding salt-bridge in pro-CPB, located between the pro-domain and the substrate binding site in the catalytic moiety, was missing in TAFI, thus unlocking the active site (44, 45).
Protein inhibitors of MCPs described to date include: potato carboxypeptidase inhibitor (PCI), leech carboxypeptidase inhibitor (LCI), tick carboxypeptidase inhibitor (TCI), Ascaris carboxypeptidase inhibitor (ACI), and latexin (46–50). The use of PCI, LCI, and TCI has been shown to delay fibrinolysis during in vitro clot lysis assays through inhibition of TAFIa (3). Latexin, the only reported endogenous inhibitor, inhibits TAFIa in vitro but it is most likely not physiologically relevant as the two proteins are found in different tissues (50). The recently published structures of bovine and human TAFIa in complex with TCI show structural similarity with the pancreatic carboxypeptidase complexes of this inhibitor (51–53). Isolated from the tick, Rhipicephalus bursa, TCI is a physiologically relevant inhibitor of TAFI because ticks are hematophagous animals known to harbor compounds that interfere with the hemostatic system of their hosts.
The recent advances in the studies of the TAFI structure prompted us to explore the flexibility of the pro-domain by using protein inhibitors. Here we show that PCI, LCI, and TCI all bind to TAFI and significantly decrease or abolish its intrinsic proteolytic activity, suggesting that the activation peptide is displaced. This was substantiated by the crystal structure of bovine TAFI in complex with TCI, which revealed that the pro-domain was indeed relocated from its original position, no longer shielding the access to the active site. The functional implications of this highly mobile pro-domain are unclear. However, it cannot be ruled out that the position of the activation peptide may be influenced by endogenous factors or conditions, thus unleashing the full proteolytic potential of TAFI. Such a mechanism has not been described hitherto, but must be considered in light of the unusual flexible nature of the pro-domain.
EXPERIMENTAL PROCEDURES
Reagents and Proteins
Hippuryl-Arg, 1,10-phenantroline, cyanuric chloride, and 1,4-dioxane were purchased from Sigma. Human TAFI was purified from normal plasminogen-depleted plasma (Skejby blood bank, Aarhus University Hospital, Denmark) as described previously (5, 18, 26). Bovine TAFI was purified from one single cow as described previously (52). Recombinant PCI, LCI, and TCI were produced in Escherichia coli and purified and quantified as previously reported (46, 48). Human plasminogen was purified by affinity chromatography by using ECH-lysine-Sepharose (GE Healthcare) as described previously (54). Human TAFI polyclonal rabbit antiserum was raised commercially (Pel-Freez).
Amino Acid Analysis
The concentration of TAFI was determined by quantitative amino acid analysis (55). The analysis was performed in triplicates by using ∼2 μg of purified bovine or human TAFI. The samples were dried in 500-μl polypropylene vials, the lids were punctured, and the vials placed in a 25-ml glass vial equipped with a MinInert valve (Pierce Biotechnology). 200 μl of 6 n HCl containing 0.1% phenol was placed in the bottom of the glass and blown with argon before a vacuum was applied. The samples were incubated at 110 °C for 18 h. The samples were subsequently re-dissolved in 50 μl of 0.2 m sodium citrate loading buffer at pH 2.2 (Biochrom, Cambridge, UK), transferred into microvials and loaded onto a BioChrom 30 amino acid analyzer (Biochrom). Data analysis was performed using in-house developed software.
Inhibition of Intrinsic TAFI Activity by Protein Inhibitors
The ability of PCI, LCI, and TCI to inhibit the intrinsic TAFI activity was analyzed as described previously (19, 21). Briefly, 15 μl of TAFI (0.37 μm, final concentration) in 50 mm Tris-HCl, pH 7.5 was titrated using PCI, LCI, or TCI (ranging from 0 to 50 μm, final concentration). The samples were incubated for 30 min at 37 °C before 30 μl of the chromogenic substrate hippuryl-Arg (30 mm) was added. The reaction was allowed to continue for 60 min before 20 μl of 1 m HCl was added, followed by 20 μl of 1 m NaOH and 25 μl of 1 m NaH2PO4, pH 7.4. Upon addition of 60 μl of 6% cyanuric chloride in 1,4-dioxane, the samples were vortexed vigorously and centrifuged at 16,000 × g for 5 min. The supernatants were subsequently transferred to 96-well microtiter plates and the absorbance was measured at 405 nm in a FLUOStar Omega plate reader (BMG Labtech) using the end point mode. All data points were collected in triplicates.
Polyacrylamide Gel Electrophoresis
Proteins were separated by SDS-PAGE in 5–15% polyacrylamide gels (56). Samples were boiled for 5 min in the presence of 30 mm dithiothreitol and 1% SDS prior to electrophoresis. Some samples were transferred to polyvinylidene diflouride membranes (Immobilon-P, Millipore) (57) and processed for Western blotting using a TAFI polyclonal rabbit antiserum and enhanced chemiluminescence (GE Healthcare). For non-denaturing polyacrylamide gel electrophoresis, samples were analyzed in the same way as described for SDS-PAGE, except that SDS was omitted from all buffers and samples were not boiled nor treated with dithiothreitol. Coomassie Brilliant Blue G (25 mg/l) was added to the upper reservoir buffer to assist in the migration of basic proteins (58).
Preparation of TAFI-inhibitor Complexes
TAFI (2.5 μg) was incubated with 1 μg of PCI, LCI, or TCI for 1 h at 22 °C in a final volume of 25 μl of 20 mm Tris-HCl, 150 mm NaCl, pH 7.5. Complex formation between the enzyme and the inhibitors was examined by non-denaturing PAGE as described above.
MCP Inhibitor Affinity Chromatography
PCI, LCI, or TCI (2.5–3.5 mg) were coupled to 1 ml of HiTrap NHS-activated-Sepharose columns according to the manufacturer's instructions (GE Healthcare). Purified human TAFI (0.1 mg) in 50 mm Tris-HCl, 150 mm NaCl, pH 7.5 (binding buffer) was slowly applied onto each column at 22 °C. The flow was stopped for 15 min to allow the inhibitors and TAFI to interact. Additionally, 1 ml of pooled human plasma (from 10 individuals) diluted 5 times in binding buffer containing 5 mm EDTA was applied onto the TCI column in a separate experiment. In both cases the columns were washed extensively with binding buffer before a pH step gradient was applied, including (i) 50 mm Tris-HCl, pH 7.5, (ii) 50 mm Tris-HCl, pH 9.0, (iii) 50 mm phosphate buffer, pH 11.0, and (iv) 50 mm phosphate buffer, pH 12.0. Fractions were collected manually and subsequently analyzed by SDS-PAGE stained with Coomassie Blue or processed for Western blotting as described above.
Size-exclusion Chromatography of the TAFI-TCI Complex
TAFI (20 μg) and TCI (8 μg) were incubated in 50 mm Hepes, 150 mm NaCl, pH 7.5 for 1 h at 22 °C. The complex or TAFI alone (20 μg) were analyzed by size-exclusion chromatography by using a Superose 12 column (GE Healthcare) equilibrated in 50 mm Hepes, 150 mm NaCl, pH 7.5. The column was connected to an ÄKTAprime System (GE Healthcare). The elution was monitored at 280 nm using a flow rate of 0.3 ml/min, and fractions were collected every min for further analyses.
Surface Plasmon Resonance Spectroscopy
The kinetics of TAFI binding to TCI was examined by surface plasmon resonance analysis using a BIAcore 3000 instrument (Biacore, Sweden). A CM5 sensor chip was activated by N-ethyl-N'-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccimide as previously described (59), and purified TCI immobilized to a density corresponding to 32 fmol/mm2. An activated and blocked flow cell was used as control for nonspecific binding. The binding of TAFI was evaluated in 20 mm Tris-HCl, 100 mm NaCl, 0.005% (v/v) Tween 20, pH 7.5, at the indicated concentrations as determined by absorption at 280 nm. Kinetic parameters were determined by BIAevaluation 4.1 software using a Langmuir 1:1 binding model and simultaneous fitting of all curves in the concentration range.
Isothermal Titration Calorimetry
Binding studies were performed on a MicroCal VP isothermal titration calorimeter at 25 °C. The buffer used for both TAFI and TCI was 20 mm Tris-HCl, 50 mm NaCl, pH 7.5. TCI at 10 μm was titrated in 10-μl injections into 0.5 μm TAFI. Injection peaks were integrated using the Origin software provided by the calorimeter manufacturer. The integrated heats were progressively summed and fitted by nonlinear least-squares analysis to a one binding site model.
Analysis of TAFI-Protein Inhibitor Complexes by Matrix-assisted Laser Desorption Ionization Time-of-flight Mass Spectrometry (MALDI-TOF MS)
PCI, LCI, or TCI were added to 10 μl of human TAFI (0.1 mg/ml) dissolved in 50 mm Tris-HCl, 150 mm NaCl, pH 7.5, using a protease/inhibitor molar ratio of 1:1. After 20 min of incubation at 22 °C, samples were acidified and desalted using C18 Zip-tips (Millipore). All samples were mixed 1:1 (v/v) with matrix solution composed of 10 mg/ml 2,6-dihydroxyacetophenone (Sigma) in 30% acetonitrile containing 20 mm dibasic ammonium citrate, pH 5.5. Aliquots (0.5 μl) were spotted on the MALDI-TOF plate using the dried-droplet method and mass spectra were recorded in a Bruker Ultraflex mass spectrometer operated in linear mode. The instrument was calibrated using a protein mixture ranging from 4,000 to 20,000 Da (protein calibration standard I, Bruker Daltonics).
Crystallization of the TAFI-TCI Complex
Approximately 5 mg of bovine TAFI (1 mg/ml in 20 mm Tris-HCl, 200 mm NaCl, pH 7.5) was incubated with 2.5 mg of lyophilized TCI for 20 min at 22 °C. The resulting complex was immediately purified by using a HiLoad Superdex 75 26/60 column (GE Healthcare) equilibrated in 30 mm Tris-HCl, 150 mm NaCl, pH 7.5. The complex was concentrated and buffer-exchanged to 10 mm Tris-HCl, 50 mm NaCl, pH 7.5 with an Amicon Ultra-4 concentrator (10 kDa cut-off, Millipore) and a Microcon YM-10 concentrator (Millipore). Crystallization assays were performed following the sitting-drop vapor diffusion method by employing protein complex at 12.5 mg/ml. Reservoir solutions were prepared by a Tecan robot and 200 or 400 nl crystallization drops were dispensed on 96 × 3-well crystal Quick plates (Greiner) by a Cartesian nanodrop robot (Genomic Solutions) at the joint IBMB-CSIC/IRB/Barcelona Science Park High-Throughput Crystallography Platform (PAC). Best crystals appeared after 5 days in a Bruker steady-temperature crystal farm at 20 °C with 2.0 m (NH4)2SO4, 0.2 m Li2SO4, 0.1 m CAPS, pH 10.5 as reservoir solution. A complete diffraction dataset was collected at 100 K from a single N2 flash-cryo-cooled (Oxford Cryosystems) crystal on an ADSC Q315R CCD detector at beam line ID23-1 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) within the Block Allocation Group “BAG Barcelona.” The crystal had been previously harvested and cryo-protected with 2.5 m (NH4)2SO4, 0.2 m Li2SO4, 21% glycerol, 0.085 m CAPS, pH 10.5. It diffracted to 6 Å resolution, was cubic and harbored two complexes per asymmetric unit. Diffraction data were integrated, scaled, merged, and reduced with programs XDS (60) and SCALA within the CCP4 suite (61).
Structure Solution and Refinement
The structure of bovine TAFI-TCI was solved by Patterson-search methods with program PHASER using all diffraction data up to 6 Å resolution (62). The structure was solved by using the coordinates of the complex between bovine TAFIa and TCI (PDB 3D4U, (52)) and of the pro-domain of bovine TAFI (PDB 3DGV; (45)) as independent searching models. Two solutions were found for the complex at 88.4, 38.8, 302.6, 0.15, −0.15, 0.09, and 269.6, 136.4, 290.9, 0.17, 0.16, 0.44, respectively (α,β,γ, in Eulerian angles and x,y,z, as fractional unit-cell coordinates; final Z-score 33.9, log-likelihood gain 1450) (62), and confirmed P4132 as the correct space group. Rigid-body and TLS refinement was subsequently performed for the complex part with REFMAC5 (63).
Miscellaneous
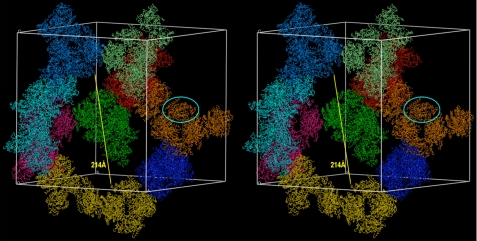
Fig. 6 was prepared with program TURBO-Frodo (64). The final coordinates of the bovine TAFI-TCI complex have been deposited with the PDB at www.pdb.org (access code 3OSL).
FIGURE 6.
Stereo animation displaying the crystal packing of the TAFI-TCI complex. Two complexes are present in the asymmetric unit of the cubic crystal form and one such complex is highlighted by a cyan ellipse. The respective glycosylated pro-domains are present in the crystals, but are disordered. The arrow pinpoints the width of the solvent channels in the crystal structure.
RESULTS
Inhibition of the Intrinsic TAFI Activity by Protein Inhibitors
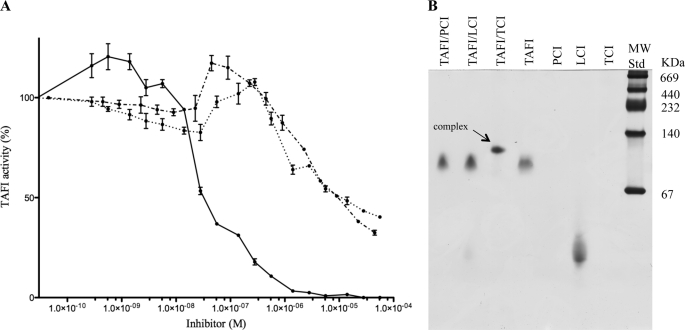
Initial studies revealed that the intrinsic proteolytic activity of TAFI was significantly decreased or abolished in the presence of PCI, LCI, or TCI (Fig. 1A). This finding prompted us to investigate the nature of the interaction between TAFI and the three protein MCP inhibitors. Even though they all decreased the intrinsic activity to a certain degree (Fig. 1A), only TCI formed a complex that resisted dissociation during non-denaturing PAGE (Fig. 1B).
FIGURE 1.
TAFI inhibition by protein inhibitors. TAFI (0.37 μm) cleaved the substrate Hippuryl-Arg at a constant rate, but its activity was strongly impaired upon incubation with increasing amounts of protein inhibitors, PCI (···), LCI (·−·) or TCI (−) (panel A). Complex formation between TAFI (2.5 μg) and the protein inhibitors (1 μg) was further studied by non-denaturing PAGE (panel B). The enzyme (2.5 μg) alone and each of the inhibitors (5 μg) were included as controls in the gel. PCI alone failed to enter the gel and TCI stained poorly.
Binding of TAFI to Immobilized Protein Inhibitors
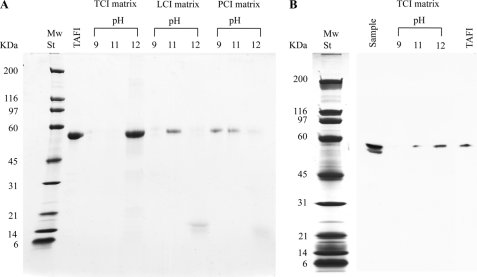
The enzyme-inhibitor interaction was further analyzed by affinity chromatography. Purified TAFI was applied to columns with immobilized PCI, LCI, or TCI and subsequently eluted by using a pH step gradient. SDS-PAGE analysis of the collected fractions revealed that TAFI bound to all three inhibitors (Fig. 2A). The highest affinity was observed for TCI (eluted at pH 12.0), followed by LCI (eluted at pH 11.0), and the lowest affinity was detected for PCI (eluted at pH 9.0). These results are consistent with the notion that both TCI and LCI may have evolved to inhibit TAFI, whereas PCI targets MCPs from the digestive tract of insects (3). The affinity of TCI for non-purified TAFI was investigated by TCI-affinity chromatography of pooled human plasma followed by Western blotting of the eluted fractions using a TAFI antibody (Fig. 2B). Similar to the purified protein, non-purified TAFI eluted at pH 12.0, suggesting that the interaction was specific. Additionally, no binding of TAFI to an “empty” column, i.e. without immobilized inhibitor, was detected (data not shown).
FIGURE 2.
TAFI binding to matrix-immobilized protein inhibitors. PCI, LCI, and TCI were immobilized separately on a Sepharose matrix and purified human TAFI was applied onto the different columns (panel A). Upon washing, the columns were eluted with a pH step gradient as described in “Experimental Procedures.” The eluted samples were then examined by SDS-PAGE and visualized by Coomassie staining. Purified TAFI (2 μg) was used as a control in the gel. Binding of non-purified human TAFI to TCI was verified by loading human plasma to the immobilized inhibitor and detecting TAFI in the eluate by ECL Western blotting (panel B). A portion of the sample (3 μl) and purified TAFI (0.03 μg) were included as controls on the gels.
Mass Spectrometry Analysis of the TAFI-Inhibitor Complexes
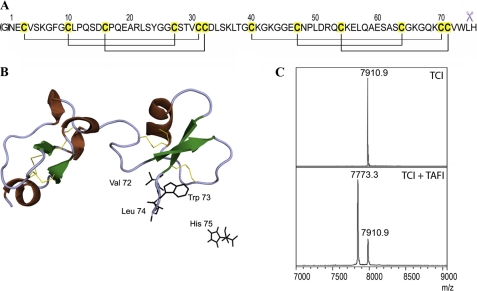
The TAFI-inhibitor complexes were dissociated by acidification and analyzed by MALDI-TOF MS. The analysis of released TCI revealed that the C-terminal residue His-75 had been removed, indicating that the interaction with TAFI mimics substrate binding, as observed for the CPA1/B1-TCI interactions (51) (Fig. 3, A and B). The molecular mass (MH+) of TCI was at m/z 7910.9 and it included an extra N-terminal Gly residue resulting from the recombinant production system used (46) (Fig. 3C, top spectrum). Upon interaction with TAFI, the majority of TCI displayed an m/z of 7773.7, corresponding to TCI without the C-terminal His-75 residue (Fig. 3B, bottom spectrum). Similarly, mass spectra of dissociated PCI and LCI revealed that a significant fraction of the C-terminal Gly-39 and Glu-66 residues, respectively, had been removed (data not shown). These results demonstrate unequivocally that TAFI is able to bind and cleave not only peptides, as previously shown (15), but also protein substrates.
FIGURE 3.
TAFI cleavage of TCI. TCI comprises 75 residues and six disulfide bonds (panel A). The cleavage of its C-terminal amino acid residue after interaction with A/B carboxypeptidases is indicated by scissors. The N-terminal glycine shown in brackets is the consequence of the heterologous expression system used. Richardson plot of TCI shows its two highly structural homologous domains and the secondary structure elements (ribbons for the concatenated 1,4-turns and the α-helix, arrows for the β-strands) (panel B). The disulfide bonds are shown as yellow sticks. The last residues of the C-terminal tail are shown as black sticks, with His-75 cleaved off after protease interaction. MALDI-TOF MS analysis of TCI shows a single peak in the spectrum, with a mass of 7910.9 Da corresponding to the intact inhibitor (panel C). When TAFI was incubated with TCI at equimolar concentration for 20 min, the products analyzed by MS clearly showed a C-terminally cleaved inhibitor with a mass of 7773.7 Da.
Characterization of the TAFI-TCI Complex by Size Exclusion Chromatography, Surface Plasmon Resonance, and Isothermal Titration Calorimetry
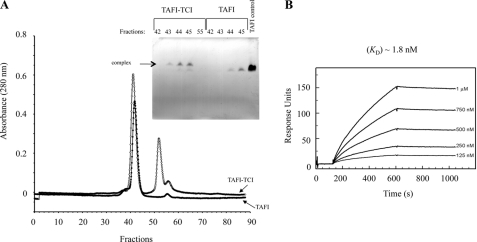
The TAFI-TCI complex eluted a few fractions ahead of free TAFI during size-exclusion chromatography, as confirmed by non-denaturing PAGE (Fig. 4A). Surface plasmon resonance (SPR) analysis revealed that TAFI bound to TCI in a concentration-dependent manner (Fig. 4B) with very high affinity (Kd 1.8 nm). Interestingly, high-affinity binding was the result of a very low dissociation rate, as evident from the dissociation curve (Fig. 4B). The nm affinity was confirmed by isothermal titration calorimetry (ITC), which revealed a Kd of 19 nm (data not shown). The difference between the two values may be attributed to the effect of immobilization and/or the small ITC signals arising from the sub-μm protein concentrations required for saturation data (65). The values compare well with Kd values determined for the interaction of TCI with other A/B-type MCPs (49).
FIGURE 4.
TAFI-TCI complex stability. TAFI (20 μg) was incubated with TCI (8 μg) and applied onto a Superose 12 size-exclusion column (panel A). The elution profile of the TAFI-TCI complex (●) and TAFI alone (○) was examined by non-denaturing PAGE (panel A, inset). Purified TAFI (5 μg) was included as a control on the gel. The dissociation constant of the TAFI-TCI interaction was further investigated by surface plasmon resonance analysis (panel B). Human purified TAFI was applied at the indicated concentrations onto a TCI-coated flow cell, resulting in high binding affinity (Kd 1.8 nm).
Crystallization of the TAFI-TCI Complex
Because of the heterogeneity of the carbohydrates attached to the pro-domain of human TAFI purified from pooled plasma (18), bovine TAFI purified from one single cow was used for the structural studies with TCI. The presence of intact complex, i.e. including the pro-domain and the catalytic moiety of TAFI, as well as the inhibitor, was confirmed by both SDS-PAGE and mass spectrometry analyses of carefully washed and dissolved protein crystals (Fig. 5). These crystals belonged to a highly-symmetric cubic crystallographic space group and diffracted to low resolution, suggesting high solvent content and/or large disordered polypeptide regions. This is reminiscent of the previously obtained structure of domain II of duck carboxypeptidase D, which contained 80 extra disordered residues at the C terminus (66). These crystals likewise belonged to a cubic space group (P213) and displayed large volumes occupied by solvent or disordered polypeptide. However, they diffracted to significantly higher resolution (2.7 Å) than initially expected.
FIGURE 5.
Protein analysis of the TAFI-TCI crystals. Crystals of the complex were washed extensively with reservoir solution, dissolved in water and subjected to SDS-PAGE stained with Coomassie (panel A). The pinpointed 60-kDa band was analyzed by peptide mass fingerprint at the IBB-UAB Proteomics Facility. The tryptic peptides detected by MALDI-TOF MS are shown in red in the TAFI sequence (panel B) and include peptides derived from the pro-domain. The catalytic domain is shown in bold.
Crystal Structure Solution of the TAFI-TCI Complex
The structure of TAFI-TCI was solved by Patterson search techniques, assaying several strategies and combination of searching models, which corresponded to 100% identical chemical species: bovine TAFIa, TCI, and the pro-domain of bovine TAFI (45, 52). Unambiguous solutions were found for the mature enzyme moiety and the inhibitor, but not for the pro-domain. These calculations confirmed the presence of just two complexes in the asymmetric unit and entailed that the value of the associated Matthews-coefficient (VM = 7.1Å3/Da) and the solvent content (83%) were at the very far end of the distribution usually found in protein crystals. In general, such high values correlate with poorly diffracting crystals and high-symmetry space groups, as is the present case (67, 68).
Given the low resolution of the diffraction data, only rigid body refinement of the correctly placed molecules and TLS-refinement were carried out. These calculations rendered final Rfactor and Rfree values of 31.3% and 32.0%, respectively, which account for an essentially correct solution (Table 1). All crystallographic contacts required for the integrity of the crystals were performed by the catalytic moiety (through a local non-crystallographic 2-fold axis) or the latter plus the inhibitor moiety (crystallographic contacts), leaving large cavities spanning over 200 Å in diameter left to bulk solvent (Fig. 6).
TABLE 1.
Crystallographic data
| Space group/cell constants (a in Å) | P4132/279.1 |
| Wavelength (Å) | 1.0723 |
| No. of measurements/unique reflections | 219,812/9,806 |
| Resolution range (Å) (outermost shell)a | 49.3–6.00 (6.32–6.00) |
| Completeness (%) | 99.8 (100) |
| Rr.i.m. (= Rmeas)b/ Rp.i.m.b | 0.143 (0.365)/0.030 (0.075) |
| Average intensity (<[<I>/σ(<I>)]>) | 20.7 (10.4) |
| Average multiplicity | 22.4 (23.6) |
| Resolution range used for refinement (Å) | 49.3–6.00 |
| No. of reflections used (test set) | 9,014 (739) |
| Crystallographic Rfactor (free Rfactor)b | 0.313 (0.320) |
| No. of protein atoms/ions in model | 5,980/2 (Zn2+) |
| Rmsd from target values | |
| Bonds (Å)/angles (°) | 0.014/1.34 |
| Average B-factors for all atoms/ions (Å2) | 37.7 |
a Values in parentheses refer to the outermost resolution shell if not otherwise stated.
b See Ref. 71 for definitions.
The final structure shows that the complex between the catalytic moiety of TAFI and TCI is indistinguishable from the one found in the TAFIa-TCI complex (52, 53). Briefly, TAFI conforms to the α/β-hydrolase fold of zymogens of the M14 family of MCPs (2, 3). The protein inhibitor contacts the active site of the mature enzyme moiety through its C-terminal domain and an exosite through its N-terminal domain, thus explaining its high inhibitory efficiency (49). The N terminus of the catalytic moiety points into the solvent area and this provides a reasonable explanation for the absence of a Patterson-search solution for the pro-domain: the latter, connected by a flexible linker with the catalytic moiety, would point toward the bulk solvent and would not participate in any crystal contact, thus potentially adopting a number of possible positions (Fig. 7).
FIGURE 7.
Schematics depicting the suggested conformation of TAFI-TCI complex. The flexible pro-domain (in cyan, linker in dark blue) moves away from the catalytic moiety in TAFI (in blue, catalytic zinc ion in magenta) and is replaced by TCI (N-terminal domain in orange, flexible linker in yellow, C-terminal domain in red).
DISCUSSION
The intrinsic proteolytic activity of TAFI was inhibited by three different protein MCP inhibitors, PCI, LCI, and TCI. In addition, TAFI bound to these inhibitors when immobilized on NHS-activated Sepharose. The interaction between TAFI and TCI appeared to be the strongest based on the late elution during the pH step gradient, and the resistance of the complex to both non-denaturing PAGE and size-exclusion chromatography. It is worth noting that PCI, LCI, and TCI exert the same inhibitory mechanism toward their target MCPs, with the C-terminal tail being the primary binding site of the inhibitors. The latter docks into the active site of the enzyme in a substrate-like manner, forming a productive enzyme-inhibitor complex (3). This interaction results in the release of the C-terminal amino acid residue upon complex formation (3). In the present work, the C-terminal residues of PCI, LCI, and TCI had all been trimmed upon dissociation from TAFI, substantiating the authenticity of complex formation. Such binding might only be feasible for high-affinity protein inhibitors and/or substrates, which give rise to productive complexes. However, further studies are required to decipher this in detail.
The TAFI-TCI dissociation constant Kd was determined to be between 1.8 nm (SPR data) and 19 nm (ITC), confirming a tight binding and thus motivating further evaluation of the interaction by structural analysis. The crystallized complex resembled the previously published complex between the proteolytically activated TAFIa variant and TCI (52), but diffracted at much lower resolution. Inhibition of TAFI by TCI was accomplished through tight binding of the C-terminal domain of the inhibitor to the active-site cleft, thus abolishing any residual enzymatic activity. Additional binding of the N-terminal inhibitor domain to an exosite contributed to tight binding. Accordingly, we can conclude that TCI forms a strong complex with TAFI, equivalent to the complex formed between TCI and TAFIa (52, 53). The binding similarly involves a double-headed interaction that anchors the inhibitor to the surface of TAFI in a successful competition with the pro-domain. The interaction displaces the pro-peptide from its position, allowing it to be loosely linked to the enzyme-inhibitor complex (see Fig. 7). It is conceivable that interactions between the pro-domain and other proteins may influence its position relative to the mature enzyme moiety and therewith the proteolytic activity of TAFI, particularly in light of the unusual structural flexibility of the pro-domain demonstrated in the current study. Because protein MCP inhibitors eliminate the activity of both TAFI and TAFIa, it is difficult to design experiments that distinguish between the activities of the two forms. Consequently, studies with MCP inhibitors against TAFIa activity to argue that TAFI activity is irrelevant will have to be reconsidered (69, 70).
In summary, we show that protein MCP inhibitors form authentic complexes with TAFI, inhibiting its intrinsic activity. TAFI cleaves the C-terminal residues of such inhibitors once bound. However, its action toward endogenous substrates and the derived physiological implications remain to be ascertained. The structure analysis of the TAFI-TCI complex reveals that the pro-domain is displaced by the inhibitor. The pro-domain has in this way the capacity to flip away from its position as a gatekeeper of the active site, unlocking the access to the active site residues. In this context, it should be considered whether interactions with endogenous factors may aid in this relocation, regulating the proteolytic activity of TAFI itself in vivo.
Acknowledgments
We thank Prof. Peter Højrup for performing amino acid analysis and acknowledge the help provided by EMBL and ESRF synchrotron local contacts. Funding for data collection was provided by ESRF.
This work was supported by grants from the Danish Natural Science Research Council and by Grants BIO2007-68046, BIO2008-04080-E, PSE-010000-2009-8, 2009SGR1036, BIO2009-10334, and the CONSOLIDER-INGENIO 2010 Project “La Factoría de Cristalización” (CSD2006-00015) from Spanish and Catalan agencies, as well as FP7-HEALTH-F3-2009-223101 “AntiPathoGN” and FP7-HEALTH-2010-261460 “Gums&Joints” from the European Union.
The atomic coordinates and structure factors (code 3OSL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MCP
- metallocarboxypeptidase
- TAFIa
- (activated) thrombin-activatable fibrinolysis inhibitor
- ITC
- isothermal titration calorimetry
- MALDI-TOF MS
- matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- LCI
- leech carboxypeptidase inhibitor
- PCI
- potato carboxypeptidase inhibitor
- (P)CPA/B
- (pro-)carboxypeptidase A/B
- SPR
- surface plasmon resonance
- TCI
- tick carboxypeptidase inhibitor.
REFERENCES
- 1.Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomis-Rüth F. X. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 319–345 [DOI] [PubMed] [Google Scholar]
- 3.Arolas J. L., Vendrell J., Aviles F. X., Fricker L. D. (2007) Curr. Pharm. Des. 13, 349–366 [DOI] [PubMed] [Google Scholar]
- 4.Skidgel R. A., Erdös E. G. (2004), pp. 837–840, Academic Press, London [Google Scholar]
- 5.Eaton D. L., Malloy B. E., Tsai S. P., Henzel W., Drayna D. (1991) J. Biol. Chem. 266, 21833–21838 [PubMed] [Google Scholar]
- 6.Hendriks D., Scharpe S., van Sande M., Lommaert M. P. (1989) Clin. Chem. 35, 177. [PubMed] [Google Scholar]
- 7.Hendriks D., Wang W., Scharpé S., Lommaert M. P., van Sande M. (1990) Biochim. Biophys. Acta 1034, 86–92 [DOI] [PubMed] [Google Scholar]
- 8.Bajzar L., Manuel R., Nesheim M. E. (1995) J. Biol. Chem. 270, 14477–14484 [DOI] [PubMed] [Google Scholar]
- 9.Wang W., Hendriks D. F., Scharpé S. S. (1994) J. Biol. Chem. 269, 15937–15944 [PubMed] [Google Scholar]
- 10.Avilés F. X., Vendrell J., Guasch A., Coll M., Huber R. (1993) Eur. J. Biochem. 211, 381–389 [DOI] [PubMed] [Google Scholar]
- 11.Reverter D., García-Sáez I., Catasús L., Vendrell J., Coll M., Avilés F. X. (1997) FEBS Lett. 420, 7–10 [DOI] [PubMed] [Google Scholar]
- 12.Lacko A. G., Neurath H. (1970) Biochemistry 9, 4680–4690 [DOI] [PubMed] [Google Scholar]
- 13.Reeck G. R., Neurath H. (1972) Biochemistry 11, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 14.Willemse J. L., Polla M., Hendriks D. F. (2006) Anal. Biochem. 356, 157–159 [DOI] [PubMed] [Google Scholar]
- 15.Valnickova Z., Thogersen I. B., Potempa J., Enghild J. J. (2007) J. Biol. Chem. 282, 3066–3076 [DOI] [PubMed] [Google Scholar]
- 16.García-Sáez I., Reverter D., Vendrell J., Avilés F. X., Coll M. (1997) EMBO J. 16, 6906–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coll M., Guasch A., Avilés F. X., Huber R. (1991) EMBO J. 10, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valnickova Z., Christensen T., Skottrup P., Thøgersen I. B., Højrup P., Enghild J. J. (2006) Biochemistry 45, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 19.Buelens K., Hillmayer K., Compernolle G., Declerck P. J., Gils A. (2008) Circ. Res. 102, 295–301 [DOI] [PubMed] [Google Scholar]
- 20.Valnickova Z., Enghild J. J. (1998) J. Biol. Chem. 273, 27220–27224 [DOI] [PubMed] [Google Scholar]
- 21.Valnickova Z., Thaysen-Andersen M., Højrup P., Christensen T., Sanggaard K. W., Kristensen T., Enghild J. J. (2009) BMC Biochem. 10, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajzar L., Morser J., Nesheim M. (1996) J. Biol. Chem. 271, 16603–16608 [DOI] [PubMed] [Google Scholar]
- 23.Tan A. K., Eaton D. L. (1995) Biochemistry 34, 5811–5816 [DOI] [PubMed] [Google Scholar]
- 24.Mao S. S., Cooper C. M., Wood T., Shafer J. A., Gardell S. J. (1999) J. Biol. Chem. 274, 35046–35052 [DOI] [PubMed] [Google Scholar]
- 25.Marx P. F., Dawson P. E., Bouma B. N., Meijers J. C. (2002) Biochemistry 41, 6688–6696 [DOI] [PubMed] [Google Scholar]
- 26.Valnickova Z., Thogersen I. B., Christensen S., Chu C. T., Pizzo S. V., Enghild J. J. (1996) J. Biol. Chem. 271, 12937–12943 [DOI] [PubMed] [Google Scholar]
- 27.Wang W., Boffa M. B., Bajzar L., Walker J. B., Nesheim M. E. (1998) J. Biol. Chem. 273, 27176–27181 [DOI] [PubMed] [Google Scholar]
- 28.Boffa M. B., Bell R., Stevens W. K., Nesheim M. E. (2000) J. Biol. Chem. 275, 12868–12878 [DOI] [PubMed] [Google Scholar]
- 29.Marx P. F., Hackeng T. M., Dawson P. E., Griffin J. H., Meijers J. C., Bouma B. N. (2000) J. Biol. Chem. 275, 12410–12415 [DOI] [PubMed] [Google Scholar]
- 30.Boffa M. B., Wang W., Bajzar L., Nesheim M. E. (1998) J. Biol. Chem. 273, 2127–2135 [DOI] [PubMed] [Google Scholar]
- 31.Sakharov D. V., Plow E. F., Rijken D. C. (1997) J. Biol. Chem. 272, 14477–14482 [DOI] [PubMed] [Google Scholar]
- 32.Mosnier L. O., Meijers J. C., Bouma B. N. (2001) Thromb. Haemost. 85, 5–11 [PubMed] [Google Scholar]
- 33.Bouma B. N., Meijers J. C. (2003) J. Thromb. Haemost. 1, 1566–1574 [DOI] [PubMed] [Google Scholar]
- 34.Redlitz A., Nicolini F. A., Malycky J. L., Topol E. J., Plow E. F. (1996) Circulation 93, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 35.te Velde E. A., Wagenaar G. T., Reijerkerk A., Roose-Girma M., Borel Rinkes I. H., Voest E. E., Bouma B. N., Gebbink M. F., Meijers J. C. (2003) J. Thromb. Haemost. 1, 2087–2096 [DOI] [PubMed] [Google Scholar]
- 36.Campbell W., Okada H. (1989) Biochem. Biophys. Res. Commun. 162, 933–939 [DOI] [PubMed] [Google Scholar]
- 37.Shinohara T., Sakurada C., Suzuki T., Takeuchi O., Campbell W., Ikeda S., Okada N., Okada H. (1994) Int. Arch. Allergy Immunol. 103, 400–404 [DOI] [PubMed] [Google Scholar]
- 38.Myles T., Nishimura T., Yun T. H., Nagashima M., Morser J., Patterson A. J., Pearl R. G., Leung L. L. (2003) J. Biol. Chem. 278, 51059–51067 [DOI] [PubMed] [Google Scholar]
- 39.Swaisgood C. M., Schmitt D., Eaton D., Plow E. F. (2002) J. Clin. Invest. 110, 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung L. L., Myles T., Nishimura T., Song J. J., Robinson W. H. (2008) Mol. Immunol. 45, 4080–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renckens R., Roelofs J. J., ter Horst S. A., van 't Veer C., Havik S. R., Florquin S., Wagenaar G. T., Meijers J. C., van der Poll T. (2005) J. Immunol. 175, 6764–6771 [DOI] [PubMed] [Google Scholar]
- 42.Bedaiwy M. A., Falcone T., Mascha E. J., Casper R. F. (2006) Obstet. Gynecol. 108, 162–168 [DOI] [PubMed] [Google Scholar]
- 43.Fujimoto H., Gabazza E. C., Taguchi O., Nishii Y., Nakahara H., Bruno N. E., D'Alessandro-Gabazza C. N., Kasper M., Yano Y., Nagashima M., Morser J., Broze G. J., Suzuki K., Adachi Y. (2006) Am. J. Pathol. 168, 1086–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marx P. F., Brondijk T. H., Plug T., Romijn R. A., Hemrika W., Meijers J. C., Huizinga E. G. (2008) Blood 112, 2803–2809 [DOI] [PubMed] [Google Scholar]
- 45.Anand K., Pallares I., Valnickova Z., Christensen T., Vendrell J., Wendt K. U., Schreuder H. A., Enghild J. J., Avilés F. X. (2008) J. Biol. Chem. 283, 29416–29423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanglas L., Aviles F. X., Huber R., Gomis-Rüth F. X., Arolas J. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villanueva J., Canals F., Prat S., Ludevid D., Querol E., Avilés F. X. (1998) FEBS Lett. 440, 175–182 [DOI] [PubMed] [Google Scholar]
- 48.Reverter D., Vendrell J., Canals F., Horstmann J., Avilés F. X., Fritz H., Sommerhoff C. P. (1998) J. Biol. Chem. 273, 32927–32933 [DOI] [PubMed] [Google Scholar]
- 49.Arolas J. L., Lorenzo J., Rovira A., Castellà J., Aviles F. X., Sommerhoff C. P. (2005) J. Biol. Chem. 280, 3441–3448 [DOI] [PubMed] [Google Scholar]
- 50.Pallarès I., Bonet R., García-Castellanos R., Ventura S., Avilés F. X., Vendrell J., Gomis-Rüth F. X. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3978–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arolas J. L., Popowicz G. M., Lorenzo J., Sommerhoff C. P., Huber R., Aviles F. X., Holak T. A. (2005) J. Mol. Biol. 350, 489–498 [DOI] [PubMed] [Google Scholar]
- 52.Sanglas L., Valnickova Z., Arolas J. L., Pallarés I., Guevara T., Solà M., Kristensen T., Enghild J. J., Aviles F. X., Gomis-Rüth F. X. (2008) Mol. Cell 31, 598–606 [DOI] [PubMed] [Google Scholar]
- 53.Sanglas L., Arolas J. L., Valnickova Z., Aviles F. X., Enghild J. J., Gomis-Ruth F. X. (2010) J. Thromb. Haemost. 1056–1065 [DOI] [PubMed] [Google Scholar]
- 54.Deutsch D. G., Mertz E. T. (1970) Science 170, 1095–1096 [DOI] [PubMed] [Google Scholar]
- 55.Ozols J. (1990) Methods Enzymol. 182, 587–601 [DOI] [PubMed] [Google Scholar]
- 56.Bury A. (1981) J. Chromatogr. 491–450 [Google Scholar]
- 57.Matsudaira P. (1987) J. Biol. Chem. 262, 10035–10038 [PubMed] [Google Scholar]
- 58.Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 59.Birn H., Verroust P. J., Nexo E., Hager H., Jacobsen C., Christensen E. I., Moestrup S. K. (1997) J. Biol. Chem. 272, 26497–26504 [DOI] [PubMed] [Google Scholar]
- 60.Kabsch W. (2001) in International Tables for Crystallography. Volume F: Crystallography of Biological Macromolecules. (Rossmann M. G., Arnold E. eds), 1st Ed., pp. 730–734, Kluwer Academic Publishers (for The International Union of Crystallography), Dordrecht, The Netherlands [Google Scholar]
- 61.CCP4 (1994) Acta Crystallogr. Sect. D. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 62.Read R. J. (2001) Acta. Crystallogr. D. Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 63.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 64.Carranza C., Inisan A.-G., Mouthuy-Knoops E., Cambillau C., Roussel A. (1999) Turbo-Frodo in AFMB Activity Report 1996–1999, pp. 89–90, CNRS-UPR 9039, Marseille [Google Scholar]
- 65.Perozzo R., Folkers G., Scapozza L. (2004) J. Recept. Signal Transduct. Res. 24, 1–52 [DOI] [PubMed] [Google Scholar]
- 66.Gomis-Rüth F. X., Companys V., Qian Y., Fricker L. D., Vendrell J., Avilés F. X., Coll M. (1999) EMBO J. 18, 5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kantardjieff K. A., Rupp B. (2003) Protein Sci. 12, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 69.Willemse J. L., Heylen E., Hendriks D. F. (2007) J. Thromb. Haemost. 5, 1334–1336 [DOI] [PubMed] [Google Scholar]
- 70.Foley J. H., Kim P., Nesheim M. E. (2008) J. Biol. Chem. 283, 8863–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mallorquí-Fernández N., Manandhar S. P., Mallorquí-Fernández G., Usón I., Wawrzonek K., Kantyka T., Solà M., Thøgersen I. B., Enghild J. J., Potempa J., Gomis-Rüth F. X. (2008) J. Biol. Chem. 283, 2871–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]