Abstract
Two novel regulatory motifs, LDEVFL and C-terminal regulatory Glu (E)-rich motif (CREEM), are identified in the extreme C terminus of the ABC protein DrrA, which is involved in direct interaction with the N-terminal cytoplasmic tail of the membrane protein DrrB and in homodimerization of DrrA. Disulfide cross-linking analysis showed that the CREEM and the region immediately upstream of CREEM participate directly in forming an interaction interface with the N terminus of DrrB. A series of mutations created in the LDEVFL and CREEM motifs drastically affected overall function of the DrrAB transporter. Mutations in the LDEVFL motif also significantly impaired interaction between the C terminus of DrrA and the N terminus of DrrB as well as the ability of DrrA and DrrB to co-purify, therefore suggesting that the LDEVFL motif regulates CREEM-mediated interaction between DrrA and DrrB and plays a key role in biogenesis of the DrrAB complex. Modeling analysis indicated that the LDEVFL motif is critical for conformational integrity of the C-terminal domain of DrrA and confirmed that the C terminus of DrrA forms an independent domain. This is the first report which describes the presence of an assembly domain in an ABC protein and uncovers a novel mechanism whereby the ABC component facilitates the assembly of the membrane component. Homology sequence comparisons showed the presence of the LDEVFL and CREEM motifs in close prokaryotic and eukaryotic homologs of DrrA, suggesting that these motifs may play a similar role in other homologous drug and lipid export systems.
Keywords: ABC Transporter, ATPases, Drug Resistance, Drug Transport, Membrane Biogenesis, Membrane Proteins, DrrAB, Drug Efflux Pump, Membrane Protein Assembly
Introduction
Membrane proteins play vital roles in critical biological systems, which include solute transport, signal transduction, and energy conservation. Loss of function, or misassembly, of membrane proteins is frequently associated with severe medical conditions. However, due to the many hurdles involved in the study of these proteins, assembly and function of membrane protein complexes are still poorly understood. Moreover, diverse mechanisms and factors (both extrinsic and intrinsic) can contribute to localization and assembly of membrane proteins (1, 2), thus making the analysis of this process quite challenging. We are interested in elucidating the function of the DrrA and DrrB proteins of Streptomyces peucetius, which together form an ATP-driven efflux pump for doxorubicin and daunorubicin, two antibiotics used in the chemotherapy of cancer. DrrA belongs to the ABC family of proteins and forms the catalytic subunit (3, 4), whereas DrrB is an integral membrane protein containing eight transmembrane α-helices (5). Therefore, in this system, the catalytic function and the membrane transport function are present on separate subunits, which form a tetrameric complex and carry out doxorubicin efflux. Some other ABC transporters, such as P-glycoprotein and cystic fibrosis transmembrane conductance regulator, contain two nucleotide binding domains (NBDs)2 and two transmembrane domains fused together into a single, large polypeptide (6).
Previous studies have shown that DrrB is improperly assembled in the absence of DrrA. In this situation, DrrB is quickly degraded (7), or, if overexpressed, it severely inhibits growth (4), implying that interaction with DrrA is essential for stability and perhaps also for proper conformation of DrrB (7). Recent studies have shown that DrrA and DrrB proteins confer doxorubicin resistance only when the two proteins are expressed in cis in a translationally coupled manner (8). It is thus likely that the DrrA and DrrB proteins may be required to co-fold for the formation of a fully functional DrrAB complex, and translational coupling may facilitate this interaction. This raises the following question. Is a specific interaction between DrrA and DrrB involved in the assembly process? Here we show that the C terminus of DrrA indeed contains two novel motifs, which play a critical role in function and assembly of the DrrAB complex. One motif present at the extreme C terminus of DrrA is rich in glutamic acid residues and is termed the C-terminal regulatory Glu (E)-rich motif (CREEM) in this study. The second motif, termed LDEVFL, is located upstream of the CREEM. Interestingly, by homology sequence comparisons, these two motifs were found to be conserved in other prokaryotic and eukaryotic ABC proteins, the most noteworthy among these being the ABCA subfamily proteins 1, 2, 3, and 8, as well as Ced-7, which belong to the same family (DRA) of ABC proteins as DrrAB (9). ABCA proteins are involved in lipid efflux in mammalian cells, and defects in the function or assembly of proteins of this subfamily are associated with severe medical conditions, including Tangier and Alzheimer diseases (10, 11).
Recently, a spate of investigations has reported that the C-terminal domains of some ABC proteins may be associated with specialized functions. The amino acid sequence of the C-terminal domains is, however, only conserved among closely related proteins. For example, a vast majority of binding protein-dependent sugar transport systems harbor a ∼150-amino acid-long C-terminal extension containing three conserved regulatory motifs termed RDM1 to 3 (12, 13). In MalK of Escherichia coli, this region binds MalT and EIIglc and plays a key role in regulation of expression (14, 15) and in inducer exclusion (12, 16). Of the non-sugar ABC transporters, the C-terminal regulatory domain present in ModC of the molybdate/tungstate transporter (MaModBC) in Methanosarcina acetivorans binds molybdenum and is involved in trans-inhibition of the ATPase activity, which results in a decrease of the transport rate in response to an increase in concentration of the substrate in the cytoplasm (17). Similarly, C-terminal extensions present in MetN of the MetNI system (18) and in Wzt of the Wzt/Wzm system of E. coli are able to bind their respective pump substrates (19, 20). Crystal structure analysis suggests that the C-terminal domains of these proteins contain a similar β-sheet fold, although they contain diverse amino acid sequences and perform different functions in ABC transporters.
In the studies described here, we report that the CREEM and LDEVFL motifs present in the extreme C terminus of DrrA are critical for function of the DrrAB complex. We also show that this region of DrrA forms the point of contact with the N-terminal cytoplasmic tail of DrrB, thus leading to the proposal that the major role of the CREEM and LDEVFL motifs may be in assembly of the DrrAB complex. Interestingly, a 33-amino acid region in the C terminus of DrrA, encompassing residues in the LDEVFL motif, was also found to be involved in homodimerization of DrrA. The significance of these two interactions, both localized to the C-terminal end of DrrA, in protein assembly is discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Antibodies
The bacterial strains used in this study were E. coli TG1, N43, LE392ΔuncIC, HMS174, and XL1-Blue. The plasmids used in this study include pDx101 (drrAB in pSU2718) and pDx119 (drrAB in pET 16b). Various substitutions and deletions were created in the drrA and drrB genes in these plasmids. Rabbit polyclonal antibodies, generated against DrrA and DrrB previously (4), were used for Western blot analysis. Anti-SecY antibody was provided by the laboratory of Dr. P. C. Tai.
Media and Growth Conditions
For doxorubicin efflux experiments, cells were grown in TEA medium (50 mm triethanolamine HCl (pH 6.9), 15 mm KCl, 10 mm (NH4)2SO4, 1 mm MgSO4) supplemented with 0.5% (w/v) glycerol, 2.5 μg/ml thiamine, 0.5% (w/v) peptone, and 0.15% (w/v) succinate at 37 °C (21). For site-directed mutagenesis, XL1-Blue cells were grown at 37 °C in NYZ+ broth (pH 7.5; 1% (w/v) casein hydrolysate, 0.5% (w/v) yeast extract, 0.5% (w/v) NaCl) supplemented with 12.5 mm MgCl2, 12.5 mm MgSO4, and 0.4% (w/v) glucose (Stratagene, La Jolla, CA). For all other experiments, cells were grown in LB medium. Chloramphenicol was added to 20 μg/ml, and ampicillin was added to 75 μg/ml, where needed.
Site-directed Mutagenesis of DrrA
A QuikChange multisite-directed mutagenesis kit (Stratagene) was used to create various mutations in the drrA gene. The strategy involved the use of complementary primers that incorporated the change at the required position.
Mutagenesis of Residues in the LDEVFL and CREEM Motifs
pDx101 was used as a template. Primers were designed as described before (22). In the LDEVFL motif, conservative point mutations at positions 303 and 304 were created, which were named L303V and D304N, respectively. In addition, two sets of triple alanine substitutions were performed at 306, 307, and 308 and at 310, 311, and 312, resulting in V306A/F307A/L308A and L310A/T311A/G312A, respectively. Deletion of the LDEVFL motif was obtained by using a pair of primers consisting of 15 flanking bases on each side of the sequence to be deleted, and the resulting clone has been designated ΔLDEVFL. In the CREEM, 3, 4, or 5 glutamic acid residues, present within the last 10 amino acid region of DrrA, were altered to glutamine, aspartic acid, or glycine residues. The obtained mutants include E321D/E322D/E325D, E321G/E322G/E325G, E321Q/E322Q/E325Q/E326Q, and E321G/E322G/E325G/E326G/E327G, which are referred to as 3E-3D, 3E-3G, 4E-4Q, and 5E-5G, respectively.
Single Cysteine Substitutions in DrrA
pDx101 containing a single cysteine substitution in drrB at position 23 (S23C) was used as the template (22). Single cysteine substitution mutants were created at amino acid position 325, 323, 319, 311, 302, 287, 253, or 232 in DrrA in this clone.
Deletion of the C Terminus of DrrA
This was achieved by removing 27 bases (positions 961–987) from the 3′-end of drrA while retaining the last 3 bases of the sequence in order to maintain translational stop/start overlap with drrB. An XhoI site was first introduced in the 3′-end of drrA by substituting 3 bases at positions 963, 966, and 968 in pDx101. These changes did not alter the coding sequence of drrA. The sequence of the primers used was as follows: UP-XhoI, 5′-GCCGATGACCGCTCGAGGGAAGAAGCG-3′; DN-XhoI, 5′-GCTTCTTCCCTCGAGCGGTCATCGGC-3′ This construct was designated pDx137. pDx137 DNA was then digested with XhoI (nucleotide 960 in drrA) and FseI (at nucleotide 84 in drrB) to remove a 139-bp fragment from the intergenic region of drrAB. A fragment corresponding to this region was then synthesized by mutually primed synthesis using single-stranded oligonucleotides. The oligonucleotides were designed so that 27 base pairs of drrA would be deleted from the synthesized fragment. The flanking regions contained XhoI and FseI restriction sites: Spacer27delUP, 5′-GCCGATGACCGCTCGAGGGCATGACGACGTCCCCCGGCACCGTGGAATCCACGACCCCTGTGAGCGGTCAGC-3′; Spacer27delDN, 5′-CCGTCGCGCGGGCCGGCCGTTCACCCGCGGACAGCACCGTCCGCAGCTGACCGCTCACAGGGGTCGTGGATT-3′.
The 112-bp fragment was digested with XhoI and FseI and then ligated to pDx137 DNA that had been digested with the same enzymes. The resulting clone has been designated ΔCREEM.
Doxorubicin Resistance Assay
Doxorubicin resistance assays were carried out as described earlier (23). Briefly, the indicated plasmids were transformed into E. coli N43 cells, which are doxorubicin-sensitive. A single colony was incubated in 5 ml of LB containing the desired antibiotic for 8 h. 1 μl of the above cells were streaked on M9 plates with a top layer containing 0, 4, 6, 8, or 10 μg/ml doxorubicin. Plates were covered with foil because doxorubicin is light-sensitive. Growth was recorded after incubation of plates at 37 °C for 24 h.
Doxorubicin Efflux Assay
The efflux assay was carried out according to the protocol previously developed in this laboratory.3 Briefly, E. coli LE392ΔuncIC cells (24) were transformed with the indicated plasmids; the cells were grown to mid-log phase (A600 nm = 0.6) and induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside for 1 h, harvested, washed twice with TEA, and resuspended in 1 ml of TEA. 10 μl of the cell suspension from above was incubated in 3 ml of TEA medium containing 10 μm doxorubicin and 5 mm 2,4-dinitrophenol for 11 h at 37 °C. The loaded cells were washed twice with 0.1 m MOPS buffer, pH 7.0, containing 2.0 mm MgSO4 and resuspended in 3 ml of MOPS buffer. The fluorescence spectra were recorded on an Alphascan-2 spectrofluorometer (Photon Technology International, London, Ontario, Canada). The excitation wavelength for doxorubicin was 480 nm, and emission was monitored at 590 nm. The excitation and emission slit widths were set at 0.75, and a time-based script was run. After an initial recording of fluorescence for 100 s at 37 °C, energy was provided in the form of 20 mm glucose, and recording was continued for an additional 400 s.
Preparation and Analysis of Cell Membranes
50 ml of E. coli TG1 cells containing the indicated plasmids were grown to mid-log phase and induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Growth was continued for an additional 3 h at 37 °C. The membrane fraction was prepared as follows. Cells were spun down and resuspended in 10 ml of buffer A (25 mm Tris-Cl (pH 7.5), 20% glycerol, 2 mm EDTA (pH 8.0), 1 mm DTT) and passed through a French press cell at 16,000 p.s.i., followed by centrifugation at 10,000 × g at 4 °C for 30 min to remove unbroken cells. The supernatant was centrifuged at 100,000 × g at 4 °C for 1 h. The pellet was resuspended in 10 ml of buffer A, washed twice, and finally resuspended in 100 μl of buffer A. The samples were heated at 55 °C for 10 min and loaded on a 10% SDS-polyacrylamide gel. The proteins were transferred onto the nitrocellulose membrane for 2 h at 55 V at room temperature, followed by Western blot analysis or autoradiography. Densitometric scanning of the bands in both the autoradiogram and the Western blots was performed by Multi Gauge Version 2.3 (FUJIFILM).
ATP Bindng Assay
Photolabeling of DrrA with [α-32P]ATP was carried out in membranes containing wild type DrrAB or DrrAB bearing mutations in DrrA (23). The ATP binding assay was carried out in a 100-μl reaction system containing buffer A, 0.1 mg of membrane protein, 10 μm ATP (pH7.5), 10 μCi of [α-32P]ATP, 35 μm doxorubicin, and 5 mm MgCl2. The reaction was exposed to UV light at 254 nm on ice for 30 min, followed by protein precipitation by 10% ice-cold TCA on ice for 30 min. Protein was recovered by centrifugation at 14,000 × g for 15 min and the pellet was resuspended into 20 μl of 4× Laemmli sample buffer and 5 μl of 1 m unbuffered Tris. The samples were resolved by polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane, as described above. The membrane was air-dried and exposed to x-ray film at −70 °C overnight, followed by autoradiography. The same blot was then examined by Western blot using anti-DrrA antibody.
Disulfide Cross-linking
A 100-μl reaction volume containing 250 μg of membrane protein in 0.1 m phosphate buffer pH 7.4 (0.1 m Na2HPO4 and 0.1 m NaH2PO4) was treated with thiospecific reagents, including copper phenanthroline (CuPhe; 3 mm CuSO4, 9 mm 1,10-phenanthroline) or 1 mm dithiobismaleimidoethane (DTME, Pierce). The cross-linking reaction was carried out at room temperature for 30 min and stopped by adding 4× Laemmli sample buffer. A 25-μl portion (50 μg of membrane protein) of the reaction mixture was then analyzed by 10% SDS-polyacrylamide gel, followed by Western blot analysis using anti-DrrA or anti-DrrB antibodies.
Co-purification of DrrA and DrrB
pDx119-derived clones bearing different mutations in DrrA were used for co-purification of DrrAB proteins. HMS174 cells containing the indicated plasmids were grown to mid-log phase at 37 °C. The expression of DrrAB was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C, and growth was continued for 16 h at 20 °C. DrrAB proteins were solubilized from a 5-mg membrane fraction with 5 ml of solubilization buffer (50 mm Tris-Cl (pH 7.5), 1% n-dodecyl-β-d-maltoside (DDM), 1 μl of 14.3 m β-mercaptoethanol, 20% glycerol, 200 mm NaCl) on ice for 1 h. After centrifugation at 100,000 × g at 4 °C for 1 h, the supernatant was loaded on a Ni2+-NTA column at 4 °C. The DrrAB proteins were eluted with the buffer containing 50 mm Tris-Cl (pH 7.5), 300 mm NaCl, 20% glycerol, 0.05% DDM, and 500 mm imidazole at 4 °C. The purified proteins were resolved on 12% SDS-polyacrylamide gels and analyzed by Western blot using anti-DrrA or anti-DrrB antibodies.
Modeling Analysis
AMMP protein structure modeling software (26) (available on the World Wide Web) was used for homology modeling of DrrA and its mutants using the crystal structure of MalK (Protein Data Bank entry 2R6G) (25) as a template. Modeling analysis of the membrane protein DrrB was carried out using the PHYRE protein fold recognition server (27) because DrrB is not homologous to the MalG or MalF protein or another crystallized membrane protein of the ABC family. The models of DrrA and DrrB complexes were then created by using the RosettaDock protein-protein docking server (28). Please note that docking analysis requires that the investigator provide a reasonable starting position for docking of two proteins. Based on the biochemical data presented in this paper, we provided the starting position where the relevant portion of the C terminus of DrrA and the N terminus of DrrB faced each other. Using this starting position, the RosettaDock server modeled the wild type and mutant DrrAB complexes. The PyMOL Molecular Graphics System (29) was used to view all of the predicted structures and for presentation of the models.
RESULTS
A Glutamic Acid-rich Sequence at the Extreme C Terminus of DrrA Is Required for Function of the DrrAB Complex
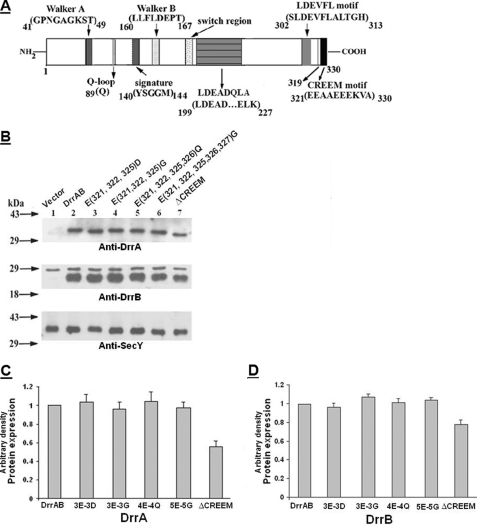
DrrA belongs to the ABC family of proteins and contains all the known conserved motifs (including Walker A, Walker B, Q-loop, signature, and switch motifs) required for the catalytic function of ABC proteins. These motifs are confined to the N-terminal domain of DrrA (residues 41–198) (23) (Fig. 1A), whereas the C-terminal domain (residues 199–330) is not known to contain any conserved motifs involved in function or assembly. In this study, we identified a glutamic acid-rich sequence (EEAAEEEKVA) at the extreme C terminus of DrrA, which suggested the possibility that the negatively charged residues present in this region may be involved in interaction between DrrA and DrrB. To determine the role of this region, the last 9 amino acids (residues 321–329) of DrrA were deleted, but the overlapping sequence between the translational stop of DrrA and the start of DrrB was retained. This truncation reduced the expression of DrrA and DrrB to roughly 55 and 78% of the wild type, respectively (Fig. 1, B (lane 7), C, and D). Doxorubicin resistance was severely compromised in this strain (Table 1). One explanation for these data could be that this region may play a role in stability of DrrA; however, when site-directed substitutions of 3, 4, or 5 glutamates with aspartates, glycines, or glutamines were carried out (resulting in 3E-3D, 3E-3G, 4E-4Q, and 5E-5G, as described under “Experimental Procedures”), the expression level of DrrA and DrrB remained about the same as in wild type cells (Fig. 1, B (lanes 3–6), C, and D). Doxorubicin resistance in the strain containing the 5E-5G mutation was found to be drastically affected, whereas it was only partially compromised in strains containing substitutions of 3 or 4 glutamates (Table 1). Together, the deletion and mutagenesis data indicate that the glutamic acid-rich region in the extreme C terminus of DrrA is important for function and suggest that it may play a role in DrrA-DrrB interaction and assembly of the complex. This region has been termed CREEM in this study. Further characterization of this motif is described in later sections.
FIGURE 1.
A, a schematic representation of the conserved motifs in DrrA. Previously identified motifs, located in the N-terminal domain of DrrA, include Walker A, Q-loop, signature, Walker B, and the switch motif (34). The motifs identified in this study, which include LDEADQLA, LDEVFL, and CREEM, are located in the C-terminal domain of DrrA. Numbers indicate the locations of specific amino acid residues. The position of residue 319 is also marked. B, effect of mutations in the CREEM on expression of DrrA and DrrB. Membranes were prepared as described under “Experimental Procedures.” 15 μg of membrane protein was analyzed on 12% SDS-polyacrylamide gels, followed by Western blot analysis with anti-DrrA (top), anti-DrrB (middle), and anti-SecY (bottom) antibodies. A nonspecific band of about 28 kDa was detected in the anti-DrrB blot in control membranes as well as membranes containing DrrAB proteins. Densitometric scanning of the bands in these blots was carried out as described under “Experimental Procedures.” Anti-SecY blot served as a loading control. C and D, histograms showing DrrA and DrrB expression, respectively, in various CREEM mutants. The wild type expression level was designated as 1. The data presented are averages of three independent experiments. Error bars, S.D.
TABLE 1.
Effect of mutations in the LDEVFL or CREEM motif on doxorubicin resistance
E. coli N43 cells carrying the indicated mutation on a plasmid were streaked on M9 plates containing different concentrations of doxorubicin. The growth was scored after incubation of the plates for 24 h at 37 °C. +++, very good growth; ++, good growth; +, some growth; +/−, very weak growth; −, no growth. This experiment was repeated four times. Dox, doxorubicin.
| Motif of DrrA | Mutation | 0 μg/ml Dox | 4 μg/ml Dox | 6 μg/ml Dox | 8 μg/ml Dox | 10 μg/ml Dox |
|---|---|---|---|---|---|---|
| Vector | +++ | − | − | − | − | |
| Wild type | +++ | +++ | +++ | +++ | ++ | |
| CREEM | E321D/E322D/E325D | +++ | +++ | ++ | + | − |
| CREEM | E321G/E322G/E325G | +++ | +++ | ++ | + | − |
| CREEM | E321Q/E322Q/E325Q/E326Q | +++ | +++ | ++ | + | − |
| CREEM | E321G/E322G/E325G/E326G/E327G | +++ | − | − | − | − |
| CREEM | CREEM deletion | +++ | − | − | − | − |
| LDEVFL | L303V | +++ | − | − | − | − |
| LDEVFL | D304N | +++ | ++ | + | +/− | − |
| LDEVFL | V306A/F307A/L308A | +++ | − | − | − | − |
| LDEVFL | L310A/T311A/G312A | +++ | − | − | − | − |
| LDEVFL | LDEVFL deletion | +++ | − | − | − | − |
The Extreme C Terminus of DrrA Interacts with the N-terminal Cytoplasmic Tail of DrrB
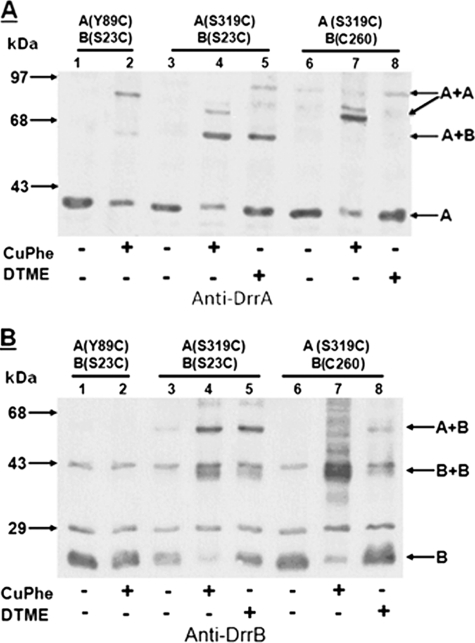
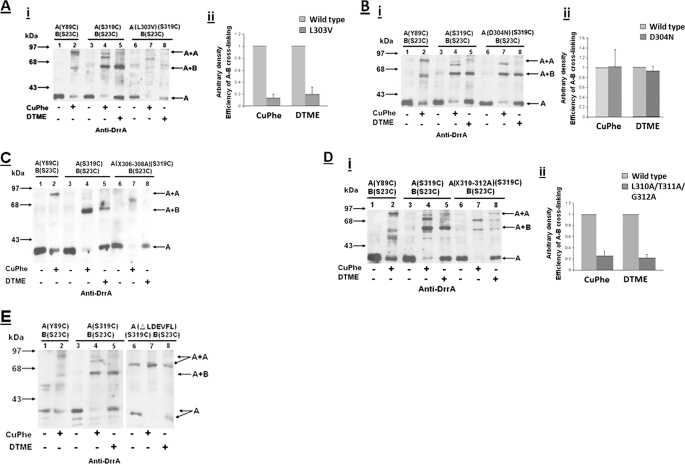
To determine if the extreme C terminus of DrrA interacts with DrrB, disulfide cross-linking experiments were performed. Previous studies, using a cysteine to amine cross-linker, N-(gammamaleimidobutyryloxy)succinimide esters, have shown that the N-terminal cytoplasmic tail of DrrB (residues 1–53) is the region that contacts DrrA (22). To determine if the extreme C terminus of DrrA is the region that makes the above contact with the N-terminal tail of DrrB, cysteine substitutions were created in these two domains. Residue Ser319 (immediately upstream of the CREEM) (Fig. 1A) was selected for the first cysteine substitution in DrrA, and it was tested in conjunction with a substitution S23C within the N-terminal tail of DrrB. This construct was termed A(S319C)B(S23C) to stress the location of the cysteines. (Note that all of the cysteine substitution mutants were named as above, unless mentioned otherwise). Strikingly, when membranes containing A(S319C)B(S23C) were treated with homobifunctional (disulfide) cross-linkers, CuPhe (arm length, 0 Å) or DTME (arm length, 13.3 Å), a species migrating at 65 kDa, which was previously identified as the size of DrrA-DrrB heterodimer (22), was detected by anti-DrrA antibody (Fig. 2A, lanes 4 and 5). The same species was also detected by anti-DrrB antibody (Fig. 2B, lanes 4 and 5), which implies the formation of a DrrA-DrrB heterodimer. These data suggest that the extreme C terminus of DrrA and the N-terminal tail of DrrB are in close proximity and interact with each other, highlighting the significance of this region of DrrA in mediating the association between the NBD and the transmembrane domain of this ABC transporter. To verify the specificity of this interaction, the same cross-linking experiments were carried out with A(S319C)B(C260), which contains the native cysteine at position 260 in DrrB. The DrrA-DrrB heterodimer was not formed in this situation, implying that residue 319 in DrrA specifically contacts the N-terminal tail of DrrB (Fig. 2, A and B, lanes 6–8).
FIGURE 2.
Disulfide cross-linking between S319C in DrrA and S23C or Cys260 in DrrB. Two different cross-linkers, CuPhe and DTME, were used. Anti-DrrA and anti-DrrB antibodies were used to identify the cross-linked species. A and B, lanes 1 and 2, A(Y89C)B(S23C). This strain contains one cysteine in the Q-loop at residue 89 of DrrA and another cysteine in the N-terminal tail of DrrB at residue 23, and it shows the previously characterized DrrA homodimer (23). Lanes 3–5, A(S319C)B(S23C). Lanes 6–8, A(S319C)B(C260). The plus or minus at the bottom of the gels indicates the presence or absence of the cross-linker. The position of the two DrrA homodimeric species, present in lanes 4 and 7, is marked with an oblique arrow. This experiment was repeated three times.
Two minor species of higher molecular mass (roughly 72 kDa, marked with an oblique arrow in Fig. 2A) were also identified by anti-DrrA antibody in CuPhe-treated A(S319C)B(S23C) membranes (Fig. 2A, lane 4). Because these species were not detected by anti-DrrB antibody, they are likely to be DrrA homodimers, and they correspond in size to the expected size of the DrrA dimer. Interestingly, these potential DrrA homodimeric species become the major cross-linked species in membranes containing A(S319C)B(C260) (Fig. 2A, lane 7), when no cysteine is present in the N-terminal tail of DrrB. We have previously shown that DrrA is trapped in a homodimeric conformation when a cysteine (Y89C) is introduced into the Q-loop of DrrA (23) (also shown in Fig. 2A, lane 2, marked with a horizontal arrow). This event reflects the head-to-tail dimerization of the N-terminal catalytic domain of DrrA as well as other ABC proteins (30–32). The DrrA homodimer produced by Y89C-Y89C cross-linking is slightly bigger in size (78 kDa) than the S319C-S319C species, which is probably due to different conformations of the DrrA dimer produced in these two situations. Furthermore, these two dimerization events are likely to be distinct; previous studies showed that Y89C-mediated dimerization of DrrA is affected by ATP (23), whereas S319C-mediated dimerization is not influenced by ATP (data not shown). A species marked as B+B, which corresponds to the size of DrrB homodimer, was also produced in membranes containing either S23C or Cys260 (Fig. 2B, lanes 4, 5, 7, and 8). Because this species is produced in all cysteine-containing DrrB variants (22), it is likely to be the result of nonspecific association between cysteines in DrrB.
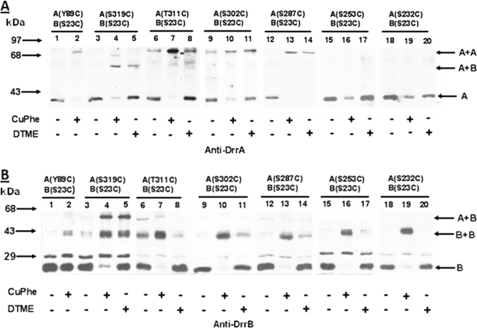
To further define the DrrA-DrrB contact region within the C terminus of DrrA, single cysteine substitutions were introduced both upstream and downstream of residue Ser319 in DrrA in a strain already containing S23C as the only cysteine in DrrB. Two downstream cysteines introduced within the CREEM, A323C and E325C, showed high efficiency of disulfide cross-linking with S23C (supplemental Fig. S1), as seen previously between S319C and S23C (Fig. 2). Analysis of the cysteine substitutions upstream of Ser319 (at residue 311, 302, 287, 253, or 232) provided different results. In membranes containing A(T311C)B(S23C), two cross-linked species were detected: a minor species of DrrA-DrrB heterodimer identified by both anti-DrrA and anti-DrrB antibodies and a major DrrA dimeric species detected by only anti-DrrA antibody (Figs. 3, A and B, lanes 7 and 8). The major species is similar to the 78-kDa DrrA homodimer seen in A(Y89C)B(S23C) (Fig. 3A, lane 2). As the location of cysteine substitution moved further away from the C-terminal end of DrrA (e.g. in A(S302C)B(S23C) or A(S287C)B(S23C)), the DrrA-DrrB heterodimer was undetectable, and only DrrA homodimer formation was observed (Fig. 3, A and B, lanes 9–14). In A(S253C)B(S23C) or A(S232C)B(S23C), neither DrrA-DrrB heterodimer nor the DrrA homodimer was seen (Fig. 3, A and B, lanes 15–20). Taken together, the results discussed above suggest that the extreme C terminus of DrrA participates in two interactions; the region containing the CREEM, up to residue Ser319, is involved in DrrA-DrrB heterodimerization, whereas the 33-residue region upstream of Ser319 (residues 287–319) is involved in DrrA homodimerization. Interestingly, residue 319 is involved in both DrrA-DrrA and DrrA-DrrB interactions, although the DrrA-DrrB interaction between S319C and S23C is preferred, as seen in Fig. 2, A and B.
FIGURE 3.
Disulfide cross-linking between T311C, S302C, S287C, S253C, or S232C in DrrA and S23C in DrrB. Cysteine scan of the C terminus of DrrA (residues 232–311) was performed by introducing cysteines at the indicated positions, followed by a disulfide cross-linking assay for each of these cysteine substitution mutants. A and B, lanes 1 and 2, A(Y89C)B(S23C); lanes 3–5, A(S319C)B(S23C); lanes 6–8, A(T311C)B(S23C); lanes 9–11, A(S302C)B(S23C); lanes 12–14, A(S287C)B(S23C); lanes 15–17, A(S253C)B(S23C); lanes 18–20, A(S232C)B(S23C). This experiment was repeated three times.
The C-terminal Domain of DrrA Contains Conserved Motifs
To determine if the sequence of the C-terminal domain of DrrA is conserved, the last 132 amino acids (residues 199–330), located right after the switch motif, were subjected to an NCBI BLAST search. Surprisingly, this search picked up 99 bacterial and archaeal ABC members. All of these proteins are ABC components of putative doxorubicin or multidrug resistance ABC transporters from various phyla. Although most of these proteins remain uncharacterized, based on their homology with DrrA, it is likely that they belong to the DRA family of ABC proteins. A multiple sequence alignment of all 99 homologs using ClustalW strongly pointed out two highly conserved regions, which we have named LDEADQLA and LDEVFL in this study (supplemental Fig. S2). The LDEADQLA motif covers 29 residues (positions 199–227), whereas the LDEVFL motif contains 12 residues (positions 302–313). These motifs are named according to the sequence of the amino acid residues in these motifs, and their location in DrrA is marked in the schematic presented in Fig. 1A. Eight of the 99 homologs also exhibited a glutamic acid-rich sequence in their extreme C termini, which was defined as CREEM in DrrA (see above). These members were therefore chosen for a separate multiple sequence alignment using the last 30-amino acid region of these proteins. This alignment highlights both LDEVFL and CREEM motifs in this group of proteins (Fig. 4A). In summary, we find that the LDEADQLA and LDEVFL motifs are highly conserved in DrrA and all members identified by the BLAST search. However, the CREEM is conserved in only a few members, which might indicate that CREEM plays a more specific role in the function of only some of these transporters.
FIGURE 4.
Sequence alignment of the C terminus of DrrA and its prokaryotic or eukaryotic homologs. A, sequence alignment by ClustalW of the last 30 amino acids of DrrA with C-terminal sequences of bacterial homologs identified by an NCBI BLAST search. The conserved sequences are termed the LDEVFL motif and CREEM. B, sequence alignment of the last 29 amino acids of DrrA with 25 amino acids of the C terminus of NBDI (top) or 26 amino acids of the C terminus of NBDII (bottom) of ABCA2 proteins from different species. Both LDEVFL and CREEM motifs are shown by this alignment.
Interestingly, when a TC-BLAST analysis (based on the Transport Classification Database, available on the World Wide Web) of the last 132-amino acid sequence of DrrA was performed, it identified mostly eukaryotic homologs, which included ABCA1, ABCA2, ABCA3, and ABCA8 from Homo sapiens (and other mammals) and Ced-7 (cell death protein 7) from Caenorhabditis elegans. These eukaryotic proteins are evolutionarily more closely related to bacterial proteins, DrrA and NodI, than to the eukaryotic ABC transporters (33) and are assigned to the same family (DRA) but to different subfamilies (DRR and ABCA, respectively) of ABC exporters (9, 34). Like most other eukaryotic members of the ABC superfamily, ABCA proteins contain two NBDs and two transmembrane domains within a large molecule (35). The DrrA sequence was, therefore, aligned with the corresponding region in NBDI or NBDII of these homologs. It was found that both the NBDI and NBDII of these proteins contain the conserved LDEADQLA and LDEVFL motifs (supplemental Figs. S3 and S4). Only ABCA2 was, in addition, also seen to contain the CREEM, which is highlighted by a manual alignment of the last 29 residues of DrrA with the corresponding region of three ABCA2 isoforms from different species (Fig. 4B). Taken together, the above analyses allowed the identification of three conserved motifs, LDEADQLA, LDEVFL, and CREEM, within the C-terminal domain of DrrA. Two motifs, LDEVFL and CREEM, at the extreme C terminus of DrrA, were selected for further characterization, as described below. (For clarity, the usage of term “extreme C terminus” in this paper refers to the last 50 amino acids and includes both the CREEM and LDEVFL motifs of DrrA).
The Conserved Motifs in the Extreme C Terminus of DrrA Are Involved in Function and Assembly
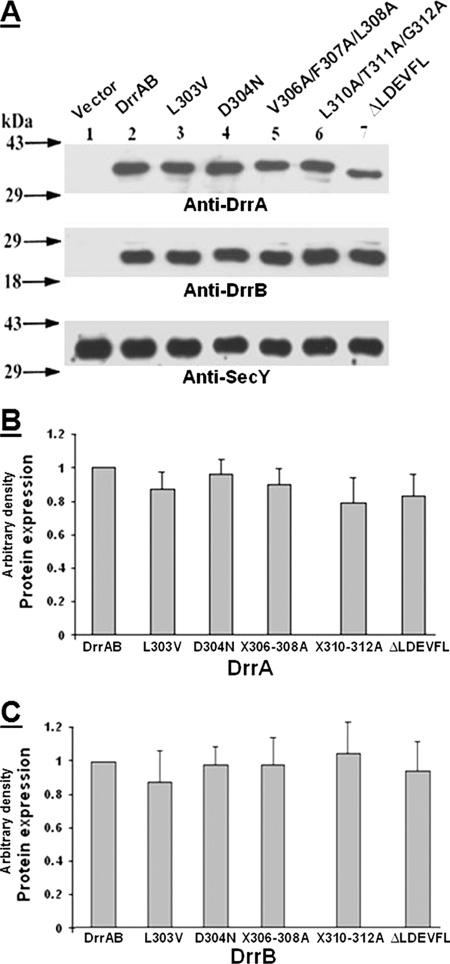
Point mutations and deletions of the conserved residues in the LDEVFL and CREEM motifs were created through site-directed mutagenesis. Mutations created in the LDEVFL motif include L303V, D304N, V306A/F307A/L308A (three residues simultaneously mutated to alanine), L310A/T311A/G312A, and ΔLDEVFL (residues 302–313 deleted). Mutations in the CREEM, as described earlier, include 3E-3G, 3E-3D, 4E-4Q, 5E-5G, and ΔCREEM (residues 321–329 deleted). The effect of these mutations on expression of the DrrA and DrrB proteins, ATP binding to DrrA, doxorubicin resistance, and doxorubicin efflux was studied. Furthermore, the effect of these mutations on interaction between DrrA and DrrB was examined by disulfide cross-linking experiments. Co-purification of DrrA and DrrB on Ni2+-NTA resin was used as an indicator of assembly of native DrrAB complexes. These studies are described below.
Effect of Mutations in LDEVFL and CREEM on Expression of DrrA and DrrB
Western blot analysis of the membrane fractions generated from the mutants described above showed that the average DrrA and DrrB expression varied between 80 and 100% in the different point mutants in the CREEM and LDEVFL motifs (Figs. 1, B–D, and 5, A–C), therefore suggesting that these motifs do not play a significant role in stable maintenance of DrrB in the membrane. The strain containing ΔCREEM, however, showed an average expression of 55 and 78%, respectively, for DrrA and DrrB (Fig. 1, B–D), which is most likely due to deletion of multiple residues in this motif.
FIGURE 5.
A, effect of mutations in the LDEVFL motif on expression of DrrA and DrrB. Conditions described in the legend to Fig. 1B were used. Western blot analysis was carried out with anti-DrrA (top), anti-DrrB (middle), or anti-SecY (bottom) antibody. The anti-SecY blot served as a loading control. B and C, histograms showing the relative amounts of DrrA and DrrB expression, respectively, in various LDEVFL mutants. The wild type expression level was designated as 1. The data presented are averages of three independent experiments. Error bars, S.D.
Effect on ATP Binding to DrrA
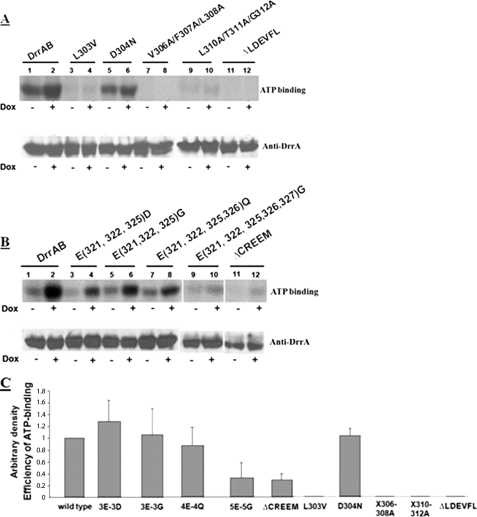
UV-induced [α-32P]ATP binding to DrrA was analyzed, as described under “Experimental Procedures.” The data in Fig. 6, A and B, upper gels, represent autoradiograms showing ATP binding, whereas the lower gels show Western blot analysis of the same blots with anti-DrrA antibodies. The ATP binding efficiency of the wild type DrrA and each mutant was calculated as the ratio of ATP bound (in the presence of doxorubicin) to the amount of DrrA in the sample. The efficiency of ATP binding was then plotted in a histogram designating the efficiency of wild type as 1 (Fig. 6C). (Please note that the gels shown in Fig. 6, A and B, are representative, and the data in Fig. 6C reflect the average of three independent experiments.) Surprisingly, ATP binding to L303V, V306A/F307A/L308A, L310A/T311A/G312A, or ΔLDEVFL was found to be abolished both in the absence and presence of doxorubicin (Figs. 6, A and C), indicating that the residues in the LDEVFL motif are critical for both basal and doxorubicin-stimulated nucleotide binding to DrrA. The D304N mutant was, however, unaffected (Figs. 6, A and C). In the CREEM, 3E-3G, 3E-3D, and 4E-4Q showed normal ATP binding, whereas ATP binding to 5E-5G or ΔCREEM was significantly reduced (Fig. 6, B and C). Together, these results indicate that the conserved motifs in the C terminus of DrrA influence ATP binding in a significant manner.
FIGURE 6.
Effect of mutations in the LDEVFL or CREEM motif on ATP binding. UV-induced [α-32P]ATP adduct formation was analyzed in membrane fractions, as described under “Experimental Procedures.” The reaction was performed both in the presence (marked by a plus sign) or absence (marked by minus sign) of 35 μm doxorubicin. A and B, upper gels, autoradiograms showing [α-32P]ATP binding to wild type DrrAB and various LDEVFL and CREEM mutants. A and B, lower gels, Western blot analysis of the nitrocellulose membranes from the upper gels in A and B, respectively, using anti-DrrA antibodies. C, histogram showing the efficiency of ATP binding to wild type DrrA and mutants. The wild type efficiency was designated as 1. The data presented are averages of three independent experiments. Error bars, S.D.
Effect on Doxorubicin Resistance
The mutations described above were also found to have a significant effect on the overall function of the efflux pump (Table 1). It was found that L303V, V306A/F307A/L308A, L310A/T311A/G312A, and ΔLDEVFL mutations confer doxorubicin sensitivity on the cells, whereas the D304N mutation has only a minor effect on doxorubicin resistance. Mutations in the CREEM affected doxorubicin resistance to varying degrees, with 5E-5G and ΔCREEM having the most drastic effect (Table 1).
Effect on Doxorubicin Efflux
To further confirm the importance of LDEVFL and CREEM motifs in function, doxorubicin efflux by the DrrAB proteins bearing the above mutations was tested. Doxorubicin is fluorescent in solution; however, its fluorescence is quenched upon binding to DNA inside the cells. This property was used to measure doxorubicin efflux in E. coli cells expressing DrrA and DrrB. Efflux was initiated by providing glucose to preloaded E. coli cells, and the resulting increase in doxorubicin fluorescence was recorded. The rate of efflux was determined by calculating the slope of the linear portion of each curve shown in supplemental Fig. S5, A–C. The efficiency of efflux in each LDEVFL or CREEM mutant was then calculated as the percentage of the mutant slope/wild type slope. Control cells containing just the vector showed about 30% efflux of the strain containing wild type DrrAB proteins (Table 2). This basal efflux seen in control E. coli cells is most likely contributed by the AcrAB pump, which is known to carry out efflux of many different antibiotics by coupling it to the energy of proton gradients (36). Mutations in the LDEVFL motif resulted in varying degrees of effect on doxorubicin efflux. The most severe effect was seen in the L303V, V306A/F307A/L308A, and ΔLDEVFL mutants, whereas D304N remained relatively unaffected (Table 2). Of the CREEM mutants, only the 5E-5G mutation and ΔCREEM showed a reduction in efflux, whereas 3E-3G, 3E-3D, and 4E-4Q exhibited normal doxorubicin efflux (supplemental Fig. S5C and Table 2; note that the efflux curves for 3E-3D and 4E-4Q are similar to that of 3E-3G and are not shown in supplemental Fig. S5C). Overall, the results of the doxorubicin efflux assay are consistent with the doxorubicin resistance results. Taken together, these data show that both LDEVFL and CREEM motifs are essential for the function of the DrrAB transporter. Specifically, residues at positions 303, 306, 307, 308, 310, 311, and 312 in LDEVFL are key residues that are indispensable for the overall function. Glutamic acid residues in the CREEM also play an important role, although a significant reduction in doxorubicin resistance, doxorubicin efflux, or ATP binding is seen only when all 5 glutamic acid residues are mutated.
TABLE 2.
Effect of mutations in the LDEVFL or CREEM motif on doxorubicin efflux
Doxorubicin efflux was measured as described under “Experimental Procedures.” Based on the data shown in supplemental Fig. S4, the efficiency of efflux in each LDEVFL or CREEM mutant was calculated, as described under “Results.” The values shown are the average of the data obtained from three independent experiments. The top shows the percentage efflux in various mutations in the LDEVFL motif. The bottom shows the percentage efflux in mutations in the CREEM.
| Wild type | Vector | L303V | D304N | V306A/F307A/L308A | L310A/T311A/G312A | Δ LDEVFL |
|---|---|---|---|---|---|---|
| % | % | % | % | % | % | % |
| 100 | 30.6 | 31.5 | 78 | 26.9 | 43.5 | 35.8 |
| Wild type | Vector | 3E-3D | 3E-3G | 4E-4Q | 5E-5G | Δ CREEM |
| 100 | 36.5 | 100 | 97 | 109 | 54.5 | 55 |
Effect on DrrA-DrrB Interaction
To determine the role of the LDEVFL and CREEM motifs in DrrA-DrrB interaction, each of the above mutations was introduced into a strain containing A(S319C)B(S23C). The effect of each mutation on DrrA-DrrB heterodimer formation was studied by disulfide cross-linking between S319C and S23C, as described above. The intensity of the DrrA-DrrB cross-linked species produced by CuPhe or DTME was determined by densitometric scanning of the Western blots. The efficiency of cross-linking in each mutant was then calculated as the ratio of the DrrA-DrrB heterodimer (e.g. Fig. 7A, i, lane 7) to the DrrA monomer (from the untreated sample) in the same set (Fig. 7A, i, lane 6). A similar calculation was done for the wild type sample containing A(S319C)B(S23C) from the same blot (Fig. 7A, i, lanes 3–5). The efficiency of cross-linking was plotted in a histogram designating the wild type efficiency as 1 (Fig. 7A, ii). The data in Fig. 7A, i and ii, strongly indicate that the L303V mutation drastically reduces the efficiency of S319C-S23C cross-linking by both CuPhe and DTME. On the other hand, the D304N was found to have no effect on the cross-linking efficiency (Fig. 7B, i (lanes 6–8) and ii). Strikingly, the formation of the DrrA-DrrB heterodimer was completely abolished in V306A/F307A/L308A and ΔLDEVFL mutants (Fig. 7, C and E (lanes 6–8)). Due to the absence of the detectable DrrA-DrrB heterodimeric species in these two mutants, the efficiency of cross-linking could not be determined. (Note that in the strain containing ΔLDEVFL, the size of the DrrA monomer as well as the DrrA homodimer is smaller than the corresponding species in wild type, as expected) Finally, the L310A/T311A/G312A mutant also showed significantly reduced cross-linking efficiency (Fig. 7D, i (lanes 6–8) and ii). Together, these data strongly suggest that the LDEVFL motif is crucial for mediating DrrA and DrrB interaction. Once again, residues 303, 306, 307, 308, 310, 311, and 312 were found to be essential. Because mutations in these residues do not significantly affect stable expression of DrrA and DrrB but drastically affect function of the transporter and association of subunits, it can be concluded that interaction between the C terminus of DrrA and the N-terminal tail of DrrB plays a specific role in a higher order process, such as assembly. In the ΔCREEM mutant, the cross-linking efficiency was reduced to between 60 and 80% of wild type. However, all other CREEM mutations (including 5E-5G) exhibited wild type-like cross-linking between S319C and S23C (data not shown), indicating that the specific interaction between Ser319 and Ser23 in DrrB can occur independently.
FIGURE 7.
Effect of LDEVFL mutations on disulfide cross-linking between DrrA(S319C) and DrrB(S23C). Each LDEVFL mutation was introduced into a strain containing A(S319C)B(S23C) for disulfide cross-linking experiments. A, i, effect of L303V mutation on disulfide cross-linking. Lanes 1 and 2, A(Y89C)B(S23C); lanes 3–5, A(319C)B(S23C); lanes 6–8, A(S319C)B(S23C) containing the L303V mutation. Western blot analysis was carried out with anti-DrrA antibody. A, ii, histogram showing the efficiency of cross-linking in wild type and mutant. The wild type efficiency was designated as 1. The data presented are averages of three independent experiments. Error bars, S.D. B and D, effect of D304N and X310–312A (L310A/T311A/G312A) on disulfide cross-linking, respectively. Data are organized as in A. C and E, effect of X306–308A and ΔLDEVFL on disulfide cross-linking, respectively.
Effect on Co-purification of DrrA and DrrB
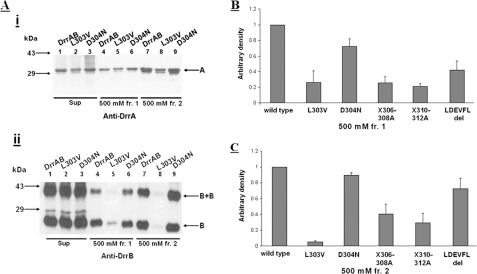
Co-purification is commonly used as an indicator of association and assembly of protein complexes (37, 38). To confirm the role of the C terminus of DrrA in assembly of the complex, drrAB genes bearing the mutations in the LDEVFL or CREEM motif were subcloned into pET16b vector, which places a His6 tag at the N terminus of DrrA. Co-elution of DrrA and DrrB in the same fraction during the purification process would suggest that these two proteins are associated with each other. The DrrAB complex was solubilized from the membrane fraction with 1% DDM, followed by standard nickel affinity chromatography. The elution profiles of several LDEVFL and CREEM mutants were determined; however, only the elution profiles of wild type DrrAB and the LDEVFL mutants, L303V and D304N, are shown in Fig. 8A. The data in lanes 4 and 7, i and ii, show that wild type DrrA and DrrB co-elute in two successive elution fractions, each containing 500 mm imidazole (labeled as fr.1 and fr.2). The L303V mutation, however, drastically affected co-purification of DrrA with DrrB (Fig. 8A, i and ii, lanes 5 and 8). The D304N mutation showed only a minor effect (Fig. 8A, i and ii, lanes 6 and 9). The efficiency of co-purification of DrrA and DrrB was determined by densitometric scanning of the Western blots, and the ratio of DrrB to DrrA purified in each fraction was calculated. Note that the homodimer of DrrB was also observed in both 500 mm fraction 1 and 500 mm fraction 2, and this species was also scanned and included in the calculation of the amount of DrrB eluted. When plotted in a histogram, the ratio of DrrB to DrrA for the wild type sample was set as 1. The data in Fig. 8, B and C, show that in addition to the drastic effect of L303V, other LDEVFL mutants, including V306A/F307A/L308A, L310A/T311A/G312A, and ΔLDEVFL, also affected the ability of DrrA and DrrB to co-purify. All CREEM mutants tested (including 5E-5G and ΔCREEM) exhibited wild type-like co-purification efficiency (data not shown), showing once again that the region immediately upstream of CREEM (especially residue Ser319) is able to associate with DrrB independently.
FIGURE 8.
Effect of mutations in the LDEVFL motif on co-purification of DrrA and DrrB. A, Western blot analysis using anti-DrrA and anti-DrrB antibodies. Lanes 1–3, Sup, DrrAB proteins solubilized from membrane fraction with 1% DDM; lanes 4–6, 500 mm fraction 1 (fr.1), the first 500 mm imidazole elution fraction; lanes 7–9, 500 mm fraction 2 (fr.2), the second 500 mm imidazole elution fraction. The intensity of bands in Western blots was determined by densitometric scanning, and co-purification efficiencies were plotted in histograms shown in B and C. This experiment was repeated three times. Error bars in B and C, S.D.
DISCUSSION
This paper describes the identification of two unique motifs, CREEM and LDEVFL, in the extreme C terminus of DrrA that may constitute a novel assembly domain for the DrrAB complex in the membrane. Cysteine substitution and cross-linking experiments showed that the amino acid residues Ser319–Glu325 (comprising the CREEM and the upstream region up to Ser319) interact specifically with the N-terminal cytoplasmic tail of DrrB. Interestingly, mutations and deletions created in the LDEVFL and CREEM motifs drastically affected doxorubicin efflux and resistance. Some mutations, in addition, also abolished ATP binding to DrrA. Because these motifs lie far away from the motifs known to be critical for ATP binding to the ABC proteins, a drastic effect of these mutations on ATP binding was unexpected. Moreover, several ABC proteins, including KpsT (E. coli) (39) and MJ0796 (Methanococcus jannaschii) (40, 41) do not contain C-terminal extensions, yet they carry out ATP binding successfully. The results obtained in this study can be explained, however, if we consider the fact that ATP binding to DrrA occurs only when DrrA is in complex with DrrB (7). Therefore, if interaction between DrrA and DrrB is disrupted by mutations in either of these motifs, it could affect ATP binding to DrrA. Interestingly, we also found that not all mutations in these motifs compromise ATP binding equally. In some situations, mutations in the neighboring residues resulted in strikingly different effects; for example, the L303V mutation abolished doxorubicin resistance as well as ATP binding, whereas the D304N mutation showed no effect. Much more extensive mutagenesis analysis is therefore needed to better understand the functions of individual residues in these motifs. The most significant finding emerging from the mutagenesis analysis conducted so far is that some mutations in the LDEVFL motif also prevent DrrA and DrrB interaction (judged by disulfide cross-linking) as well as co-purification, which are two indicators of the ability of DrrA and DrrB to form a complex. Therefore, based on the data discussed above, we propose that the CREEM of DrrA, including residues up to Ser319, forms an interaction module for a specific interaction between DrrA and DrrB, whereas the LDEVFL motif plays a crucial role in regulating this interaction and in assembly of the DrrAB complex. This is a novel and previously uninvestigated aspect of ABC protein function and biogenesis. A number of reports have recently appeared suggesting that the C-terminal extensions of ABC proteins may be associated with specialized functions (12–20); the possibility that the ABC proteins may play a critical role in membrane protein biogenesis has never been suggested or explored before. Because the LDEVFL and CREEM motifs are conserved in other prokaryotic and eukaryotic homologs belonging to the DRA family, it is likely that these motifs will play a similar role in these homologous systems.
Another interesting aspect of the work presented here is the finding that the C terminus of DrrA also participates in DrrA homodimerization. This function is localized in a 33-amino acid region upstream of Ser319, and it encompasses the LDEVFL motif. Thus, two interaction interfaces, DrrA-DrrB and DrrA-DrrA, were found to exist in the C-terminal end of DrrA. These studies also indicate that residue Ser319 exists in equilibrium between A-A ↔ A-B, which raises a number of important questions. For example, under what conditions does the equilibrium shift to the AB species, and vice versa? Further, what is the relationship between DrrA dimerization localized in the C terminus and the head-to-tail dimerization of the N-terminal catalytic domain of DrrA, described by us earlier (23). In MalK of E. coli, the regulatory C-terminal domains of MalK were shown to remain in contact throughout the catalytic cycle, thus suggesting that the C terminus plays a role in stabilization of the MalK dimer (42). Dimerization of the C terminus of DrrA could similarly be involved in stabilization of the DrrA dimer; however, we are leaning toward the possibility that this event may be transient, and it may be an important prerequisite for DrrA and DrrB interaction. In previous studies using general cross-linkers (such as DTSSP or DSP) DrrA-DrrB heterodimer was the only species detected, whereas DrrA dimer was never isolated (7), suggesting the transient nature of the DrrA dimer. Furthermore, we find that residue 319 can participate in both A-B and A-A interactions, implying that A-B and A-A species involving the C terminus of DrrA are formed in a mutually exclusive manner.
Several ABC proteins, including MalK, ModC, and MetN, which contain regulatory C-terminal domains, have been crystallized recently (17, 18, 25). Although the C-terminal domains of these proteins do not share significant amino acid identity, they all show the presence of β-sheet-rich folds characteristic of β-barrel or β-sandwich structures. In ModC and MetN, these domains are critical for binding of the specific pump substrate, resulting in trans-inhibition of ATPase activity and further substrate uptake (17, 18). In the case of MalK, the C-terminal domain binds cytoplasmic proteins MalT and EIIglc, which play a role in gene expression and inducer exclusion, respectively (13). We used the closely related bacterial homolog MalK as a template to model the structure of wild type DrrA as well as several mutants in the conserved LDEVFL motif, which are shown through biochemical analysis in this study to negatively influence interaction of the C terminus of DrrA with the N terminus of DrrB and drastically affect function of the DrrAB complex. The model of wild type DrrA, (generated by AMMP modeling software (26)), indicates that the structure of the N-terminal domain of DrrA (containing the ABC cassette) is almost identical to the N terminus of MalK (Fig. 9, A and B). Such conservation of structure is not unexpected, given the high homology between the N-terminal domains of DrrA and MalK. The C-terminal domain of DrrA exhibits a β-sheet-rich structure that is also similar, but not identical, to the structure of the C-terminal domain of MalK. In the model of DrrA shown in Fig. 9B, the LDEVFL motif (shown in yellow) is partially present in beta strand 6, whereas most of this motif is present in a loop region. Similarly, the CREEM (shown in purple) is also seen in a coil or loop region in this model.
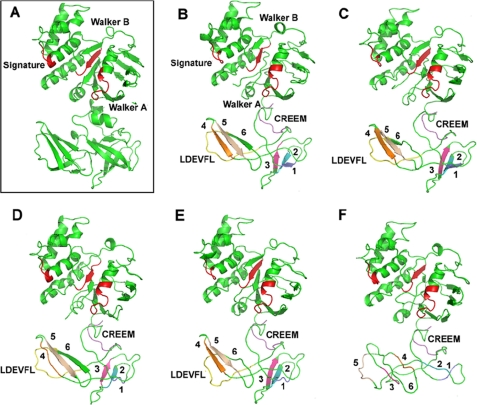
FIGURE 9.
Structure modeling of DrrA. Wild type DrrA and mutants in the LDEVFL motif of DrrA were modeled based on the known crystal structure of MalK (Protein Data Bank entry 2R6G) using AMMP protein structure modeling software (26), as described under “Experimental Procedures.” A, crystal structure of MalK (Protein Data Bank entry 2R6G) is shown in a rectangle. B–F, predicted structures of wild type DrrA and mutants in the LDEVFL motif of DrrA. B, wild type DrrA; C, D304N; D, L303V; E, X306–308A; F, ΔLDEVFL. In all panels, the Walker A, signature, and Walker B motifs in the N-terminal domain of DrrA are shown in red. The LDEVFL and CREEM motifs in the C-terminal domain are shown in yellow and purple, respectively. The β-strands in the C-terminal domain of DrrA are shown in different colors and are labeled 1–6.
Modeling analysis of mutations in the LDEVFL motif, which include L303V, X306–308A, and ΔLDEVFL, showed varying degrees of structural alterations in the C-terminal domain of DrrA (Fig. 9, C–F). Importantly, however, the structure of the N-terminal domain of DrrA remained largely unaltered in all mutants. Overall, the severity of structural changes observed in the C-terminal domain correlated well with the biochemical effects of mutations reported earlier in this paper. For example, the D304N mutation, which showed minimal effects on DrrAB interaction and function, showed no significant change in the structure of the N- or the C-terminal domain of DrrA (Fig. 9C). In the L303V mutant, which significantly affected interaction and function, β-strands 1 (shown in slate blue) and 4 (shown in orange) present 25 amino acids and 60 amino acids upstream of Leu303, respectively, were no longer formed (Fig. 9D), therefore suggesting that even a conservative mutation in the LDEVFL motif has significant effects on the conformation of the C-terminal domain. Structural changes were also observed in the X306–308A mutation; specifically, β-strands 1 and 6 were not seen. Finally, the most drastic changes, as expected, were observed when the 13-amino acid-long LDEVFL motif was deleted. This deletion resulted in a complete loss of the β-sheet structure (strands 1–6) of the C-terminal domain without altering the structure of the N-terminal domain of DrrA (Fig. 9F). These analyses not only confirm that the C terminus of DrrA forms an independent domain; they also indicate that the conserved LDEVFL motif plays a critical role in maintaining the conformational integrity of the C-terminal domain of DrrA, which would be essential for the ability of this domain to interact with the DrrB protein and for overall function of the complex. This conclusion is supported by DrrA and DrrB docking analysis performed by the Rosetta dock server (28), as described under “Experimental Procedures.” Results of the docking analysis (shown in supplemental Fig. S6) indicate that L303V and X306–308A point mutations in the LDEVFL motif significantly alter the orientation of DrrB relative to DrrA. The D304N mutation, however, showed no significant effect. Furthermore, the distance between residue Ser319 in DrrA and Ser23 in DrrB was also significantly increased in L303V (31.3 Å; supplemental Fig. S6C) and X306–308A (37.8 Å; supplemental Fig. S6D) as compared with in the wild type (15.5 Å; supplemental Fig. S6A). In D304N, as expected, the distance (14.3 Å) between these residues remained the same as in the wild type complex (supplemental Fig. S6B). The modeling analysis of DrrA is therefore consistent with the biochemical studies; both kinds of analyses highlight the importance of the C-terminal domain of DrrA in the interaction and function of the DrrAB complex. Further understanding of the roles of the conserved motifs identified in this study will be obtained by genetic analysis, especially by isolation of suppressors of the mutations shown to be important, as well as by crystal structural analysis of the DrrA and DrrB complex.
In summary, we propose that the interaction between the extreme C terminus of DrrA and the N terminus of DrrB, identified in this study, represents an initial interaction important for localization and biogenesis of the complex in the membrane. This is shown as conformation I (Confo. I) in the working model presented in supplemental Fig. S7, where the N-terminal ATP-binding domains of DrrA are in the “open” conformation, whereas sequences in the extreme C terminus of DrrA (residues 319–325, including part of CREEM) form an interface with the N-terminal tail of DrrB. It is highly likely that this interaction defines the contact points in the DrrAB heterodimer isolated previously by the use of general cross-linkers (7). Conformation II (Confo. II) results from binding of doxorubicin to DrrB. In this state, the N-terminal tail of DrrB disengages from the C terminus of DrrA. Based on studies reported earlier (23), we suggest that the N-terminal tail of DrrB is now involved in communicating conformational changes to the Q-loop region (represented by residue 89) in the N terminus of DrrA. Simultaneously, the C terminus of DrrA, which is disengaged from DrrB, undergoes homodimerization through the 33-residue region (residues 287–319, including the LDEVFL motif). This is followed by binding of ATP to the N- terminal nucleotide binding domains of DrrA, resulting in the “closed” state (Confo. III) produced by head-to-tail dimerization of the NBDs (23). Hydrolysis of ATP returns the complex to the resting state (Confo. I). This study defines novel interactions between the ABC component and the transmembrane component of the DrrAB transporter. It raises many interesting questions and opens new avenues for understanding the assembly of membrane proteins.
Supplementary Material
Acknowledgments
We thank Drs. P. C. Tai and John Houghton for valuable comments and suggestions during preparation of the manuscript. We also thank Guoxing Fu and Xue Wang for help with modeling analysis of DrrA and DrrB.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM51981-09 (to P. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
M. Sharma and P. Kaur, unpublished data.
- NBD
- nucleotide binding domain
- CREEM
- C-terminal regulatory Glu (E)-rich motif
- 3E-3D
- E321D/E322D/E325D
- 3E-3G
- E321G/E322G/E325G
- 4E-4Q
- E321Q/E322Q/E325Q/E326Q
- 5E-5G
- E321G/E322G/E325G/E326G/E327G
- X306–308A
- V306A/F307A/L308A
- X310–312A
- L310A/T311A/G312A
- CuPhe
- copper phenanthroline
- DTME
- dithiobismaleimidoethane
- DDM
- n-dodecyl-β-d-maltoside.
REFERENCES
- 1.de Gier J. W., Luirink J. (2001) Mol. Microbiol. 40, 314–322 [DOI] [PubMed] [Google Scholar]
- 2.Riordan J. R. (2005) Annu. Rev. Physiol. 67, 701–718 [DOI] [PubMed] [Google Scholar]
- 3.Guilfoile P. G., Hutchinson C. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8553–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur P. (1997) J. Bacteriol. 179, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandlur S. M., Wei L., Levine J., Russell J., Kaur P. (2004) J. Biol. Chem. 279, 27799–27806 [DOI] [PubMed] [Google Scholar]
- 6.Borges-Walmsley M. I., McKeegan K. S., Walmsley A. R. (2003) Biochem. J. 376, 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaur P., Russell J. (1998) J. Biol. Chem. 273, 17933–17939 [DOI] [PubMed] [Google Scholar]
- 8.Pradhan P., Li W., Kaur P. (2009) J. Mol. Biol. 385, 831–842 [DOI] [PubMed] [Google Scholar]
- 9.Holland I. B. (2003) ABC Proteins: From Bacteria to Man, pp. 3–35, Academic Press, Inc., London [Google Scholar]
- 10.Albrecht C., Viturro E. (2007) Pflugers Arch. 453, 581–589 [DOI] [PubMed] [Google Scholar]
- 11.Macé S., Cousin E., Ricard S., Génin E., Spanakis E., Lafargue-Soubigou C., Génin B., Fournel R., Roche S., Haussy G., Massey F., Soubigou S., Bréfort G., Benoit P., Brice A., Campion D., Hollis M., Pradier L., Benavides J., Deleuze J. F. (2005) Neurobiol. Dis. 18, 119–125 [DOI] [PubMed] [Google Scholar]
- 12.Böhm A., Diez J., Diederichs K., Welte W., Boos W. (2002) J. Biol. Chem. 277, 3708–3717 [DOI] [PubMed] [Google Scholar]
- 13.Lee S. J., Böhm A., Krug M., Boos W. (2007) Trends Microbiol. 15, 389–397 [DOI] [PubMed] [Google Scholar]
- 14.Panagiotidis C. H., Boos W., Shuman H. A. (1998) Mol. Microbiol. 30, 535–546 [DOI] [PubMed] [Google Scholar]
- 15.Bukau B., Ehrmann M., Boos W. (1986) J. Bacteriol. 166, 884–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kühnau S., Reyes M., Sievertsen A., Shuman H. A., Boos W. (1991) J. Bacteriol. 173, 2180–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber S., Comellas-Bigler M., Goetz B. A., Locher K. P. (2008) Science 321, 246–250 [DOI] [PubMed] [Google Scholar]
- 18.Kadaba N. S., Kaiser J. T., Johnson E., Lee A., Rees D. C. (2008) Science 321, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuthbertson L., Kimber M. S., Whitfield C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuthbertson L., Powers J., Whitfield C. (2005) J. Biol. Chem. 280, 30310–30319 [DOI] [PubMed] [Google Scholar]
- 21.Dey S., Rosen B. P. (1995) J. Bacteriol. 177, 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur P., Rao D. K., Gandlur S. M. (2005) Biochemistry 44, 2661–2670 [DOI] [PubMed] [Google Scholar]
- 23.Rao D. K., Kaur P. (2008) Biochemistry 47, 3038–3050 [DOI] [PubMed] [Google Scholar]
- 24.Angov E., Brusilow W. S. (1988) J. Bacteriol. 170, 459–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J. (2007) Nature 450, 515–521 [DOI] [PubMed] [Google Scholar]
- 26.Harrison R. W., Chatterjee D., Weber I. T. (1995) Proteins 23, 463–471 [DOI] [PubMed] [Google Scholar]
- 27.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 28.Lyskov S., Gray J. J. (2008) Nucleic Acids Res. 36, W233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 30.Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 31.Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. (2002) Mol. Cell. 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Lu G., Lin J., Davidson A. L., Quiocho F. A. (2003) Mol. Cell. 12, 651–661 [DOI] [PubMed] [Google Scholar]
- 33.Luciani M. F., Denizot F., Savary S., Mattei M. G., Chimini G. (1994) Genomics 21, 150–159 [DOI] [PubMed] [Google Scholar]
- 34.Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Microbiol. Mol. Biol. Rev. 72, 317–364, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dean M., Rzhetsky A., Allikmets R. (2001) Genome Res. 11, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 36.Zgurskaya H. I., Nikaido H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nehme D., Li X. Z., Elliot R., Poole K. (2004) J. Bacteriol. 186, 2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song K. S., Li S., Okamoto T., Quilliam L. A., Sargiacomo M., Lisanti M. P. (1996) J. Biol. Chem. 271, 9690–9697 [DOI] [PubMed] [Google Scholar]
- 39.Nsahlai C. J., Silver R. P. (2003) FEMS Microbiol. Lett. 224, 113–118 [DOI] [PubMed] [Google Scholar]
- 40.Moody J. E., Thomas P. J. (2005) J. Bioenerg. Biomembr. 37, 475–479 [DOI] [PubMed] [Google Scholar]
- 41.Yuan Y. R., Blecker S., Martsinkevich O., Millen L., Thomas P. J., Hunt J. F. (2001) J. Biol. Chem. 276, 32313–32321 [DOI] [PubMed] [Google Scholar]
- 42.Samanta S., Ayvaz T., Reyes M., Shuman H. A., Chen J., Davidson A. L. (2003) J. Biol. Chem. 278, 35265–35271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.