Abstract
IRF-1 is a tumor suppressor protein that activates gene expression from a range of promoters in response to stimuli spanning viral infection to DNA damage. Studies on the post-translational regulation of IRF-1 have been hampered by a lack of suitable biochemical tools capable of targeting the endogenous protein. In this study, phage display technology was used to develop a monoclonal nanobody targeting the C-terminal Mf1 domain (residues 301–325) of IRF-1. Intracellular expression of the nanobody demonstrated that the transcriptional activity of IRF-1 is constrained by the Mf1 domain as nanobody binding gave an increase in expression from IRF-1-responsive promoters of up to 8-fold. Furthermore, Mf1-directed nanobodies have revealed an unexpected function for this domain in limiting the rate at which the IRF-1 protein is degraded. Thus, the increase in IRF-1 transcriptional activity observed on nanobody binding is accompanied by a significant reduction in the half-life of the protein. In support of the data obtained using nanobodies, a single point mutation (P325A) involving the C-terminal residue of IRF-1 has been identified, which results in greater transcriptional activity and a significant increase in the rate of degradation. The results presented here support a role for the Mf1 domain in limiting both IRF-1-dependent transcription and the rate of IRF-1 turnover. In addition, the data highlight a route for activation of downstream genes in the IRF-1 tumor suppressor pathway using biologics.
Keywords: Antibodies, Interferon, Protein Domains, Transcription Factors, Tumor Suppressor, IRF-1, Intracellular Activation, Nanobody, Negative Regulation, Epitope
Introduction
The skipping of exon 3 and/or 2 seen in transcripts for the IRF-1 tumor suppressor has been linked to the development of human hemopoietic malignancies, such as leukemia and myelodysplastic syndrome (1–3). More recently, the loss of exons 7–9 in various combinations has been reported in cervical cancer, and this leads to the generation of C-terminally truncated IRF-1 that can compete with the wild-type protein for DNA binding. The C-terminal mutants have longer half-lives and, unlike wild-type IRF-1, can be expressed in a cell cycle independent manner (4). Understanding the function of the Mf1 domain of IRF-1, a regulatory subdomain that is located at the C terminus of the protein, could therefore increase our current knowledge of the role that truncated proteins play in tumor progression.
The extreme C-terminal region of IRF-1 (Mf1 domain; amino acids 301–325) is a regulatory domain that plays a role in both positive and negative modulation of target gene expression (5). In addition, the Mf1 domain plays a role in determining the rate of IRF-1 protein degradation, with removal of the C terminus leading to a significant increase in the half-life of the protein (6, 7). Furthermore, binding of members of the Hsp70 family of molecular chaperones to the Mf1 domain appears to be critical for the normal function and regulation of IRF-1 (8). Thus previous studies have pointed to a potentially important role for the Mf1 domain of IRF-1 in its homeostatic regulation.
The studies described above, by necessity, relied on the use of exogenous IRF-1 mutant proteins; therefore, we do not know whether the Mf1 domain is rate-limiting for IRF-1-mediated gene activation within the context of the endogenous protein. To address this issue, we have used antibody phage display to screen for scFv3 antibody fragments (9, 10) to the Mf1 domain. The introduction of scFv nanobodies into a cell system showed that the Mf1 domain was rate-limiting for IRF-1-mediated transcription under normal cellular conditions, providing good evidence that IRF-1 transcriptional function is subject to post-translational regulation. The data also highlight an unexpected role for the extreme C terminus in the negative regulation of IRF-1 turnover.
MATERIALS AND METHODS
Reagents and Cells
Anti-IRF-1 (BD Biosciences), anti-IRF-1 C20 (Santa Cruz Biotechnology), anti-IRF-2 (Abcam), anti-His (Novagen), anti-HP1α (Millipore), anti-caspase3 (Santa Cruz Biotechnology), anti-ISG20 (Abnova), anti-PKR (Santa Cruz Biotechnology), anti-CDK2 (Santa Cruz Biotechnology), anti-GAPDH (Abcam), and anti-GFP (Clontech) were used at a 1:1000 dilution; and HRP-protein A (BD Biosciences) was used at 1 μg/ml. Cycloheximide (Supelco) was dissolved in H2O to 5 mg/ml and used at a concentration of 30 μg/ml. MG132 (Calbiochem) was dissolved in DMSO to 10 mm and used at 50 μm. pcDNA3-IRF-1 and IRF-1 ΔC25 were as described previously (5); pcDNA3 IRF-1 P325A was made using a QuikChange site-directed mutagenesis kit. A375 and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin at 37 °C and 10% CO2. Cells were transfected at 80% confluence using Attractene (Qiagen).
Generation of Anti-IRF-1 scFv Antibodies
Biotin-SGSG-linked peptide (LDSLLTPVRLPSIQAIPCAP) was immobilized on an immunotube (Nunc) pre-coated with 20 μg/ml streptavidin. The Tomlinson I and J Human Single Fold scFv Libraries (Geneservice) were used for three rounds of selection following the manufacturer's protocol. scFv were expressed from HB2151 bacteria infected with scFv displaying phage. Expression was induced with 9 mm isopropyl 1-thio-β-d-galactopyranoside, shaking at 30 °C overnight. A nitrocellulose membrane was incubated with 0.3 μg/μl GST-IRF-1 in PBS and blocked for 30 min in 5% milk in PBS, 0.1% Tween. Each monoclonal scFv (2 μl) was dotted onto the membrane and incubated for 1 h at room temperature in a humidified chamber. The membrane was overlaid with 1:1000 protein A-HRP in PBS, 0.1% Tween with 5% milk for 1 h. Between each step, the membrane was washed extensively with PBS, 0.1% Tween, and antibody binding was detected using enhanced chemiluminescence. For amplification of phage and DNA extraction, KM13 helper phage (1 × 109) were added to 500 μl of TG1 bacterial cultures (2× TY, 100 μg/ml ampicillin, 1% glucose) infected with phage displaying anti-IRF-1 monoclonal antibodies and incubated without shaking at 37 °C for 30 min. The cultures were centrifuged at 3000 × g for 10 min, and the pellet was resuspended in 500 μl of 2× TY (100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.1% glucose) for overnight incubation with shaking at 30 °C. The culture was centrifuged (3300 × g for 30 min), and phage were precipitated from the supernatant by adding 200 μl of PEG/NaCl (20% polyethylene glycol 6000, 2.5 m NaCl) for 10–20 min at room temperature. Phage were pelleted (16,100 × g for 10 min at 4 °C) and resuspended in 100 μl of Iodide Buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 4 m NaI). The suspension was incubated with ethanol (250 μl) for 10–20 min at room temperature. Precipitated phage DNA was collected by centrifugation (16,100 × g for 10 min at 4 °C), washed with 0.5 ml of 70% (v/v) ethanol, recentrifuged, and dried briefly under vacuum. For the subsequent sequencing and PCR, the DNA was suspended in TE buffer and quantified using a spectrophotometer (Nanodrop ND-1000). The anti-IRF-1 scFv were cloned into pDEST 15, pDEST 14, and pDEST 53 for expression in bacterial and mammalian systems using Gateway® technology (Invitrogen). For in vitro studies, the scFv nanobodies were purified on Ni-NTA-agarose (Qiagen) or glutathione-Sepharose.
Immunoblots and Binding Assays
Peptide binding assays were carried out as described previously (11); scFv binding was detected using anti-His mAb and enhanced chemiluminescence. The protein binding assays were as described previously (8). For immunoblots, mammalian cells were lysed in 5× reporter lysis buffer (Promega) or 0.1% Triton lysis buffer and processed as described previously (8).
EMSA and Reporter Assays
EMSAs were carried out with a C1 probe using a protocol based on that of Fujita et al. (20). Briefly, 2 μl of 6× IRF-1 EMSA buffer (120 mm HEPES, pH 7.5, 300 mm KCl, 30% glycerol, 2.4 mm DTT, 0.6 mg/ml BSA, 3% Triton X-100), 1.5 μl of nonspecific DNA (1 μl of 1 μg/μl poly(dI-dC) and 0.5 μl of 1 μg/μl salmon sperm DNA), and GST-IRF-1 plus or minus various antibodies (as detailed in the figure legends) were preincubated for 30 min on ice prior to the addition of 32P-labeled C1 probe (1 μl). Following a further 30-min incubation at room temperature, the reactions were analyzed on a 5% polyacrylamide gel, and radiolabeled bands were detected using a phosphoimager. Luciferase reporter assays were carried out as described previously (5, 12) using 120 ng of either p125-luc IFNβ (which contains the human IFN-β promoter region −125 to +19) or a control plasmid p55-luc IFNβ (minus the ISRE; promoter region −55 to +19), which were the kind gifts from Dr. T. Fujita (Kyoto University), TLR3-Luc (hTLR3–588 or hTLR3IRF a mutant which is minus the ISRE) (19), -683Cdk2-Luc (5), TRAIL (pTRL3 or a mutant minus the ISRE/IRFE, pTRL3n6) (13), and IL-7 (−609-Luc or a mutant, −609-mtIRF-E-Luc which is missing the ISRE) (14). Reporter activity was determined 24 h post-transfection.
scFv Protein Pulldowns
Purified scFv (1 μg) in buffer A (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl) was incubated with Ni2+-NTA-agarose (15 μl) for 1 h at 4 °C and then washed two times for 5 min with buffer A plus 5 mm imidazole. The beads were subsequently incubated with HeLa cell lysate (500 μg) and mixed at 4 °C for 2 h. Unbound proteins were removed by washing three times with buffer A plus 25 mm imidazole, 0.5% Triton X-100, and 0.5% Tween 20, followed by three times with buffer A plus 25 mm imidazole. The beads were heated to 85 °C for 5 min in SDS sample buffer (100 μl). scFv bound protein were analyzed by immunoblot.
Subcellular Fractionation and Turnover
Fractionation was as described in the manufacturer's handbook (ThermoScientific subcellular fractionation kit). Fractions were analyzed by SDS-PAGE and immunoblotting. IRF-1 half-life was determined as described previously (6).
RESULTS
Screening for scFv Binding to a C-terminal Peptide from IRF-1
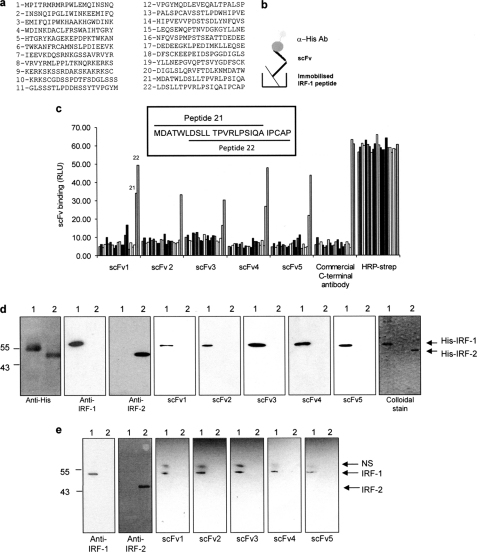
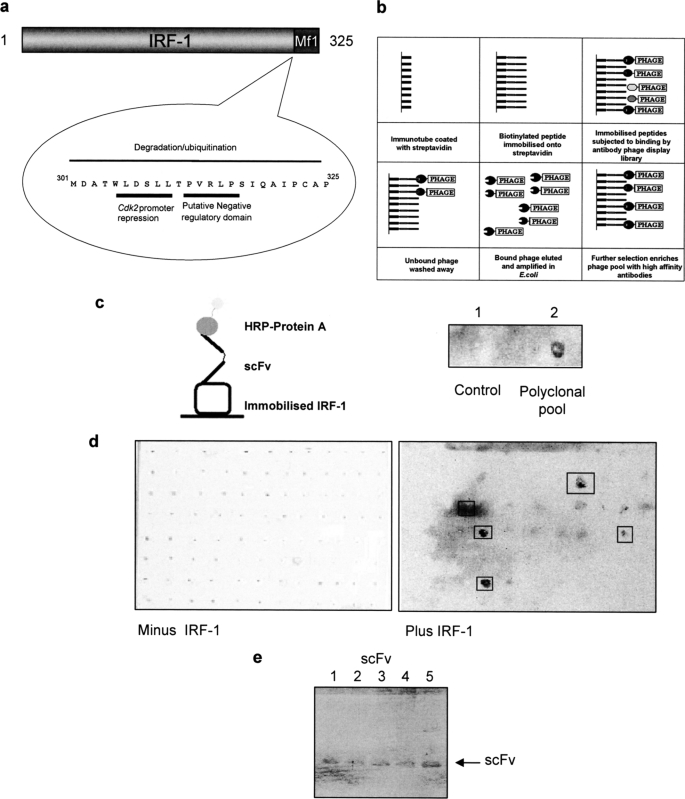
A biotinylated C-terminal IRF-1 peptide (LDSLLTPVRLPSIQAIPCAP, referred to as peptide 22 in Fig. 2) was immobilized on streptavidin-coated immunotubes and used to screen phage expressing single chain variable fragments (scFv) from the Tomlinson library (I or J) to generate Mf1 domain-specific reagents (Fig. 1a). Bound phage was eluted and amplified before being used for further rounds of selection as detailed under “Materials and Methods” (Fig. 1b). The pool of scFv (polyclonal pool) after the final round was infected into HB2151 bacteria, and 48 individual colonies were picked for each library (96 colonies in total). These were screened for monoclonal scFvs that bound to full-length IRF-1 following secretion from the phage-infected bacteria into culture medium upon induction with isopropyl 1-thio-β-d-galactopyranoside. First, the polyclonal pool of scFv was assayed to make sure it contained nanobodies that could bind peptide 22 when in the context of full-length IRF-1. A nitrocellulose filter coated with GST-tagged IRF-1 (0.3 mg/ml) was incubated with the polyclonal scFv and developed using HRP-linked to protein A. This gave a positive signal for the polyclonal scFv nanobodies when compared with a control (Fig. 1c). When soluble monoclonal scFvs, from the 96 individual bacterial colonies, were subsequently screened in this assay, five of the clones reproducibly gave a strong positive signal (Fig. 1d). These five monoclonal scFvs (Fig. 1e) were taken forward for characterization to determine both their specificity and ability to bind to IRF-1 under different conditions.
FIGURE 2.
scFv binding to denatured IRF-1. a, list of overlapping peptides covering the complete sequence of IRF-1 used in c. Peptide 22 was used for phage screening. b, schematic of scFv binding to biotinylated peptides in a peptide binding assay. c, immobilized overlapping peptides (peptides 1–22; given in a) were incubated with each scFv (1 μg/ml). Following extensive washing, scFv binding was detected with an anti-His monoclonal antibody and enhanced chemiluminescence. A commercial anti-IRF-1 C-terminal peptide antibody was used as a control, and peptide normalization was demonstrated using HRP-streptavidin to detect the biotin moiety. The sequences of two overlapping peptides (21 and 22) are shown in the inset. d, purified His-IRF-1 (lane 1) and His-IRF-2 (lane 2) (0.3 μg/lane) were analyzed on a 12% SDS-PAGE/immunoblot and probed with anti-His, anti-IRF-1, anti-IRF-2 and the scFv nanobodies (1 μg/ml). scFv binding was detected using anti-GST monoclonal antibody, and enhanced chemiluminescence. A colloidal stain shows the purity of His-IRF-1 and His-IRF-2. e, lysates from HeLa cells transfected with 1 μg of pcDNA3 IRF-1 (lane 1) or pcDNA3 IRF-2 (lane 2) were analyzed by 12% SDS-PAGE/immunoblot and probed with the antibodies listed in d with the exception of the anti-His antibody. NS, nonspecific band detected by the anti-GST antibody. The data are representative of at least two independent experiments.
FIGURE 1.
Screening for IRF-1 scFv nanobodies. a, Mf1 (multifunctional-1) domain of IRF-1 is located at the extreme C terminus and has been shown to play a role in transcriptional regulation and degradation. b, schematic of antibody phage display protocol. c, left panel, diagram of scFv binding to full-length IRF-1 in a dot blot format. Right panel, polyclonal scFvs secreted by phage-infected bacteria into culture media were spotted onto a nitrocellulose membrane coated with GST-IRF-1. After extensive washing, bound scFvs were detected using protein A-HRP and enhanced chemiluminescence. The control was scFv-free culture media. d, as in c except following the culture of individual monoclonal scFv colonies. e, SDS-polyacrylamide gel showing the five monoclonal scFv nanobodies identified in d, following expression by phage-infected HB2151 E. coli and secretion into 2× TY growth medium.
When the phagemid DNA corresponding to the five positive clones was sequenced, surprisingly all five clones consisted of only a light chain. The restriction sites SfiI/NcoI and XhoI, between which the heavy chain should have been cloned, were intact as was the intervening pIT2 vector sequence present in the unrestricted vector. However, numerous studies on functional single domain antibodies have shown that they can exhibit specific binding activity (15, 16) and that the light chain can prove to be a highly active paratope (17, 18).
Characterization of Recombinant scFv Nanobodies Binding to the C Terminus of IRF-1
To generate a reproducible supply of purified scFv, phagemid DNA was subcloned into a gateway donor vector. From there, the scFvs were transferred into pDEST14 for expression in Escherichia coli with the existing His/Myc tags or pDEST15, where an additional GST tag was added, as we found that this aided the expression of soluble scFv protein in E. coli. Following purification using glutathione-Sepharose, the GST-scFvs were used as primary antibodies in a series of assays aimed at establishing binding specificity for the target antigen. First, a biotin-tagged peptide library spanning the length of IRF-1 (Fig. 2a) was used in a binding assay (Fig. 2, b and c). The individual peptides were immobilized and incubated with the nanobodies (Fig. 2, b and c). Alternatively, the peptides were incubated with a commercial anti-peptide polyclonal sera to the C-terminal 20 amino acids of IRF-1 or with streptavidin-HRP to demonstrate peptide normalization. Fig. 2c shows that all the nanobodies bound to peptide 22, which was used in the original library screen, and that scFv1 and -3–5 also bound to the highly related peptide 21 (Fig. 2, a and c, inset).
The ability of the scFv nanobodies to bind IRF-1 under denaturing conditions was determined by immunoblot; this demonstrated that all the nanobodies were able to detect purified His-IRF-1 (Fig. 2d) and exogenous untagged-IRF-1 from crude cell lysate (Fig. 2e), but it did not bind to a closely related family member IRF-2 when either purified His-IRF-2 (Fig. 2d) or exogenous IRF-2 (Fig. 2e), present in cell lysates, was probed.
Capturing IRF-1 from Cells Using Nanobodies
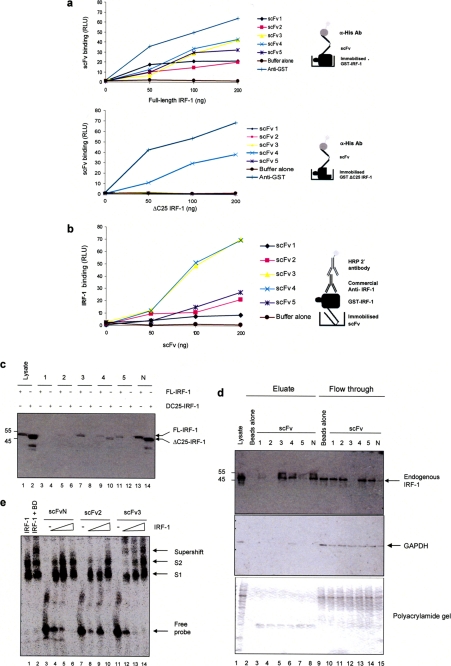
Having established that the five scFv nanobodies chosen were all able to recognize the peptide antigen (Fig. 2c) and the epitope present in denatured IRF-1 protein (Fig. 2, d and e), they were subsequently characterized for their ability to bind IRF-1 under nondenaturing conditions. First, a titration of GST-IRF-1 immobilized on a microtiter plate was probed with either the nanobodies or anti-GST (Fig. 3a, upper panel). (For all experiments involving GST-IRF-1, scFvs were expressed without the GST tag using pDEST14 and were purified on nickel-agarose.) This showed that all five of the scFvs were able to bind IRF-1, although there were reproducible differences in their ability to form stable complexes. Interestingly, when full-length IRF-1 was replaced by a truncation mutant (IRF-1ΔC25) from which the C-terminal 25 amino acids had been removed, as expected scFv1–3 and -5 were no longer able to detect the protein; however, scFv4 binding was unaffected, suggesting that this nanobody is not specific for its C-terminal epitope under the conditions of the assay (Fig. 3a, lower panel). When the assay was performed in the alternative orientation (Fig. 3b), a more pronounced difference in stable binding was detected. In this case, scFv3 bound significantly better than the remaining nanobodies, with the exception of the nonspecific nanobody scFv4.
FIGURE 3.
Binding of the scFv to IRF-1 under nondenaturing conditions. a, titration (0–200 ng) of immobilized GST-IRF-1WT or GST-IRF-1ΔC25 was incubated with the scFv nanobodies (1 μg/ml). Binding was detected with anti-His/enhanced chemiluminescence and compared with anti-GST binding. The results are given as relative light units (RLU) for scFv binding plotted against IRF-1 amount and are representative of two separate experiments. b, as in a except that the scFv was immobilized and GST-IRF-1 was in the mobile phase; binding was detected using anti-IRF-1. c, scFv nanobodies immobilized on Ni-NTA-agarose beads were incubated with HeLa cell lysate (500 μg) expressing either wild-type or IRF-1ΔC25 protein, and bound proteins were analyzed by SDS-PAGE/immunoblots developed using an anti-IRF-1 monoclonal antibody and enhanced chemiluminescence. d, scFv nanobodies immobilized on Ni-NTA-agarose beads were incubated with untransfected HeLa cell lysate (500 μg). Following extensive washing, bound proteins were analyzed by SDS-PAGE/immunoblot developed with either anti-IRF-1 or anti-GAPDH; alternatively, the gel was stained with colloidal blue (bottom panel). The results are representative of two independent experiments. e, EMSA where purified GST-IRF-1 (200 ng) binds a 32P-labeled DNA probe (consisting of 4× ISRE; lane 1) to give a banding pattern consisting of two IRF-1-DNA complexes, labeled S1 and S2. DNA-bound IRF-1 can be supershifted by a commercial anti-IRF-1 antibody (lane 2). scFvN 2 and 3 were used at 200 ng per reaction with a titration of IRF-1 (0, 50, 100, and 200 ng; lanes 3–6, 7–10, and 11–14). The data are representative of at least three independent experiments.
To expand on the functionality of the scFv nanobodies, their binding to IRF-1 from cell lysates was investigated and compared with a control scFv (scFvN) selected from the ETH gold library for binding to a domain from an N-terminal region of IRF-1 (amino acids 60–124; data not shown). When the nanobodies were immobilized on nickel agarose-beads and incubated with HeLa cell lysate overexpressing full-length IRF-1, scFv3 and scFvN (Fig. 3c, lanes 7 and 13), and to a lesser extent scFv5 and scFv2, captured IRF-1 (lanes 11 and 5). scFvN could also capture IRF-1ΔC25 (Fig. 3c, lane 14), as it recognizes an N-terminal epitope, whereas scFv2, -3, and -5 were shown to be specific for the C terminus by their failure to pull down IRF-1ΔC25 (Fig. 3c, lanes 6, 8, and 12). Again the nonspecific nature of scFv4 was demonstrated as it bound weakly to both full-length and ΔC25 IRF-1 protein (Fig. 3c, lanes 9 and 10). We next determined if any of the nanobodies were able to deplete endogenous IRF-1 from cell lysates (Fig. 3d). scFv3 bound endogenous IRF-1 with sufficient affinity to deplete all the protein from the lysate (Fig. 3d, compare lanes 5 and 12). Consistent with the previous results, scFv5 showed weak binding (Fig. 3d, lanes 7 and 14) as did scFv4. The nonspecific nature of scFv4 was highlighted further by its ability to bind to GAPDH, whereas all the other scFv showed no GAPDH binding activity (Fig. 3d, middle panel).
As scFv3 could bind endogenous IRF-1, it appeared to be the best candidate for cell-based studies. However, before moving on to generate scFv3-based mammalian expression vectors, we first carried out an EMSA to establish whether scFv3 could bind to IRF-1 when it was in complex with DNA. Fig. 3e (lanes 12–14 compared with lane 1) shows that scFv3 binds to DNA-bound IRF-1 producing a “supershifted” IRF-1-DNA complex, whereas scFvN, although efficient at capture of endogenous IRF-1 from detergent-soluble lysates (Fig. 3d), showed no significant binding to the IRF-1- NA complex suggesting its epitope is masked in this situation (Fig. 3e, lanes 4–6). Consistent with its low affinity for IRF-1, scFv2 displayed a weak band shift activity.
Mapping the scFv Nanobody Epitopes
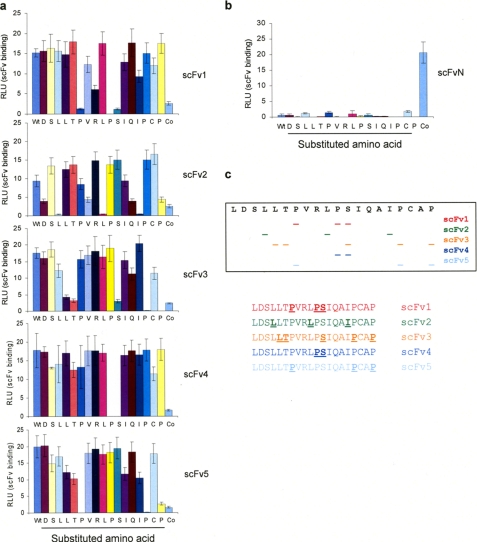
The data presented above demonstrate that although all five of the scFv nanobodies are able to bind to denatured IRF-1 with a similar affinity, they show considerable variation in their ability to bind both recombinant and cellular IRF-1 under nondenaturing conditions, suggesting that they have differing sequence specificities. To investigate this, we used a peptide library in which the amino acids of peptide 22 were sequentially mutated to Ala (Fig. 4a). The nanobodies showed distinct requirements for binding with no two scFvs having the same epitope specificity (Fig. 4c). Interestingly the two nanobodies that were able to specifically capture cell-expressed full-length IRF-1, scFv3, and scFv5, both require the Pro residues at positions 322 and 325. scFv3 also had a strong requirement for three other residues (Ser317, Leu310, and Thr311), whereas scFv5 required only one other residue (Pro312), and this is likely to explain why scFv3 binds with a higher apparent affinity than scFv5. The nonspecific nature of scFv4 is explained by its very limited epitope as it displayed a strong requirement for only two adjacent amino acids Pro316 and Ser317, a sequence that is also found in the main body of IRF-1 and in GAPDH. As expected, scFvN did not bind to any of the C-terminal peptides but was specific for its N-terminal epitope (Fig. 4b). Together, the characterization of the C-terminal scFv nanobodies suggested that scFv3 has properties that make it the best candidate for the development of an intracellular nanobody.
FIGURE 4.
Identification of scFv epitopes. a, immobilized biotinylated peptides, including an alanine scan of the last 20 amino acids of IRF-1 LDSLLTPVRLPSIQAIPCAP, were incubated with 1 μg/ml scFv1–5. scFv binding was detected with anti-His. A peptide containing the epitope for scFvN is labeled as Co, and the data are representative of two independent experiments. b, as in a except using scFvN. c, diagrammatic representation of the amino acid motifs targeted by scFv1–5.
scFv Nanobodies Activate Exogenous IRF-1 in Cells
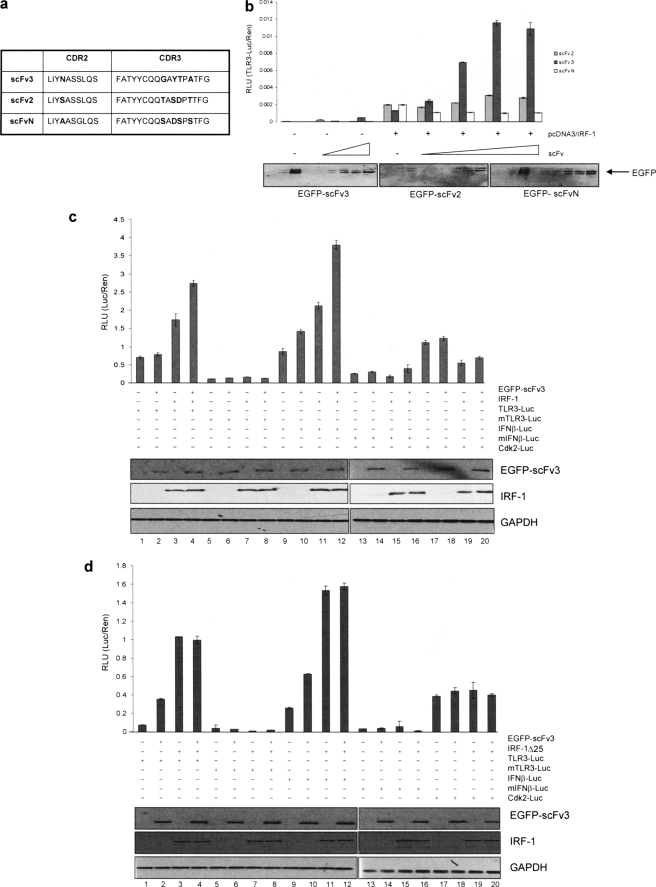
To determine whether the specific interaction of a nanobody with the Mf1 domain of IRF-1 in a cellular environment had consequences for IRF-1 transcriptional activity, scFv3, together with control nanobodies (scFv2 and scFvN), was cloned into a mammalian GFP expression vector. scFv2 was chosen to act as a control for scFv3 as these two nanobodies are highly similar in sequence (Fig. 5a), with only five variant amino acids through the CDR2 and CDR3 regions, but they have differing affinities and amino acid specificities. scFvN was used as a second control antibody, in preference to a nonspecific nanobody, as it again has good sequence conservation with scFv3 (Fig. 5a) but binds out with the IRF-1 C-terminal domain.
FIGURE 5.
scFv3 nanobody activates exogenous IRF-1-dependent transcription. a, table showing sequence variation between CDRs (complementarity determining regions) 2 and 3 of scFv2, -3, and -N. b, titration (0–250 ng) of EGFP-scFv2, -scFv3, and -scFvN were expressed in HeLa cells together with a TLR3(+ISRE)-luc reporter (120 ng), Renilla (60 ng), and IRF-1 (75 ng) as shown. DNA levels were normalized using an empty vector. Reporter gene activity was measured in relative light units (RLU) and is expressed as the ratio of Luc/Ren. The assays were carried out in duplicate; results are given as mean ± half the range and are representative of at least three independent sets of experiments. The immunoblot shows the levels of EGFP-scFv detected using anti-EGFP. c, HeLa cells were transfected with TLR3-Luc, TLR3(-ISRE)-Luc, IFNβ-p125luc, IFNβ(-ISRE)-p55luc, or Cdk2-luciferase reporter constructs (120 ng), Renilla (60 ng), and IRF-1 (100 ng) plus EGFP-scFv3 (100 ng) as indicated and reporter activity determined as in a. Expressed proteins were detected by immunoblot using anti-EGFP and anti-IRF-1, and GAPDH was detected as a loading control. d, as in c except that IRF-1 was replaced with IRF-1ΔC25. Reporter assays are representative of at least two independent experiments.
scFv3 effects on transcriptional activity of IRF-1 were assessed using a TLR3 reporter (19). Fig. 5b shows the effect on reporter activity when a fixed amount of IRF-1 was expressed together with a titration of either the C-terminal binding nanobodies, scFv3 and scFv2, or the N-terminal nanobody scFvN. scFv3 significantly increased gene expression from the TLR3 promoter, under conditions where, although expressed at comparable levels, scFvN if anything slightly suppressed IRF-1 activity. Consistent with its weak C-terminal binding activity, scFv2 showed very weak activity as an activator of IRF-1.
scFv3 was further tested for the ability to modulate IRF-1 activity on other known target promoters, specifically IFNβ (20) and Cdk2 (5, 21). Fig. 5c shows that as well as activating IRF-1 at the TLR3 promoter (lane 4 versus lane 3), scFv3 also stimulated expression from the IFNβ promoter (lane 12 versus lane 11). The scFv3 effect is specific for the IRF-1 response element as mutant forms of the TLR3 (Fig. 5c, lanes 5–8) and IFNβ (lanes 13–16) promoter, in which the ISRE is absent, were not stimulated by the nanobody. scFv3 appears to specifically target the transcription activating function of IRF-1 as no enhancement of its ability to repress expression from the Cdk2 promoter was detected (Fig. 5c, lanes 17–20).
To control for off-target effects of scFv3 on IRF-1 activity, the above experiment was repeated using IRF-1ΔC25 instead of wild-type protein (Fig. 5d). As expected from our previous studies (5), IRF-1ΔC25 had a higher intrinsic transcriptional activity against the IFNβ (Fig. 5d, lanes 3) and TLR3 (lane 11) promoters and was unable to repress Cdk2 promoter activity (lane 19) when compared with the wild-type protein (Fig. 5c). However, no additional enhancement of IRF-1ΔC25 activity was observed in the presence of scFv3 (Fig. 5d, compare lanes 3 and 4, 11 and 12), suggesting the scFv3 nanobody affects IRF-1-mediated transcription through a direct interaction with the C-terminal regulatory domain rather than through another domain of IRF-1 or through binding to other proteins in the initiator complex.
scFv3 Activates Endogenous IRF-1 Activity
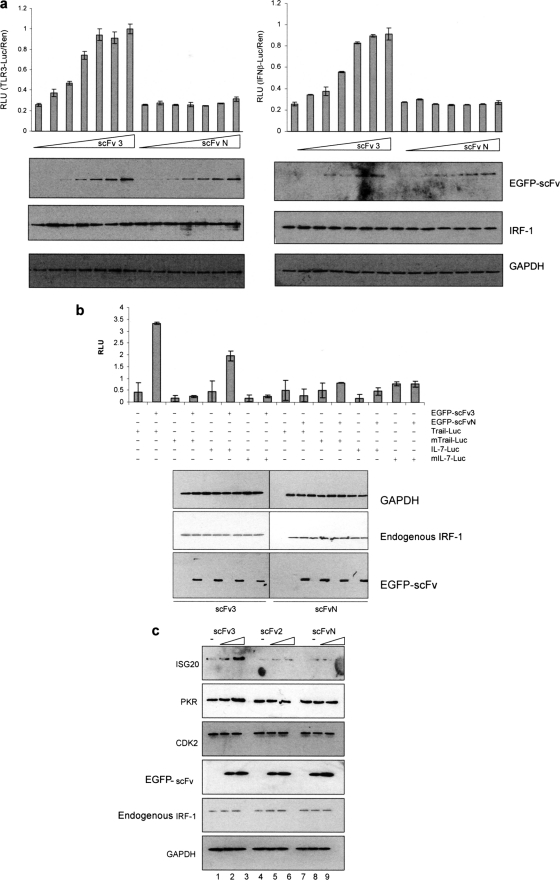
As scFv3 could activate exogenous IRF-1 in a cellular environment, we extended the study to look at the potential effect of scFv3 on endogenous IRF-1 using both reporter constructs (Fig. 6, a and b) and endogenous readouts (Fig. 6c). When a titration of scFv3 and scFvN was introduced into HeLa cells (Fig. 6a), it showed scFv3 could increase luciferase production from both the TLR3 and IFNβ promoters by 4–5-fold at the peak. In contrast, scFvN had no significant effect on IRF-1 activity with either of the promoters used. Furthermore, when the result was confirmed using additional physiologically relevant reporters (Fig. 6b) based on the IRF-1-responsive promoter regions from TRAIL (13) and IL-7 (14) scFv3 could activate the WT reporters but not reporters previously shown to be defective in IRF-1 binding (13, 14). As expected from Fig. 6a, scFvN had no significant effect on either TRAIL or IL-7 expression (Fig. 6b).
FIGURE 6.
scFv activates endogenous IRF-1. a, HeLa cells were transfected with a titration of EGFP-scFv3 or EGFP-scFvN (0–250 ng) plus Renilla (60 ng) and either TLR3-Luc (120 ng) (left-hand graph) or IFNβ-p125luc (right-hand graph) (120 ng). DNA levels were normalized using an empty vector. Reporter gene activity was measured in relative light units (RLU) and is expressed as the ratio of Luc/Ren. The assays were carried out in duplicate; results are given as mean ± half the range and are representative of at least three independent sets of experiments. Expressed proteins were detected by immunoblot using anti-EGFP and anti-IRF-1, and GAPDH was detected as a loading control. b, HeLa cells were transfected with either TRAIL-Luc (WT or mutant-ISRE) or IL-7-Luc (WT or mutant -ISRE) (120 ng), in each case with or without EGFP-scFv3 or EGFP-scFvN (120 ng). Reporter assays and immunoblots were carried out as above. c, lysate from HeLa cells transfected with 0, 0.5, or 1 μg of EGFP-scFv3 (lanes 1–3), EGFP-scFv2 (lanes 4–6), or EGFP-scFvN (lanes 7–9) were analyzed by SDS-PAGE/immunoblot and probed with anti-ISG20, anti-PKR, anti-CDK2, anti-EGFP, anti-IRF-1, or anti-GAPDH antibody.
We next sought to use changes in the level of endogenous proteins for IRF-1 target genes as a readout of its activity as a transcriptional activator for PKR (22, 23) and ISG20 (24) gene expression. Fig. 6c shows that scFv3 expression induced the levels of the ISG20 and PKR proteins by ∼8- and 2.5-fold, respectively (lane 3 versus lane 1, band intensity quantified using SynGene Imaging Systems; data not shown). In agreement with the data in Fig. 5c, scFv3 had little effect on the levels of Cdk2. Moreover, neither scFv2 nor scFvN had any measurable effect on ISG20, PKR, or Cdk2 levels. The data showing an increase in the levels of both ISG20 and PKR suggest that scFv3-activated IRF-1 is able to induce transcription from endogenous promoters as well as reporter constructs.
One interpretation of the data presented here, showing that the scFv3 nanobody can significantly increase IRF-1 transcriptional activity, is that the C terminus of the endogenous protein is rate-limiting for IRF-1-mediated gene activation and that the nanobody functions by relieving Mf1-imposed negative regulation. Alternatively, as the Mf1 domain is involved in protein interactions, which impact on IRF-1 localization (8) and degradation (6), scFv3 may affect IRF-1-mediated transcription through an indirect mechanism.
scFv3 Nanobody Decreases the Half-life of IRF-1
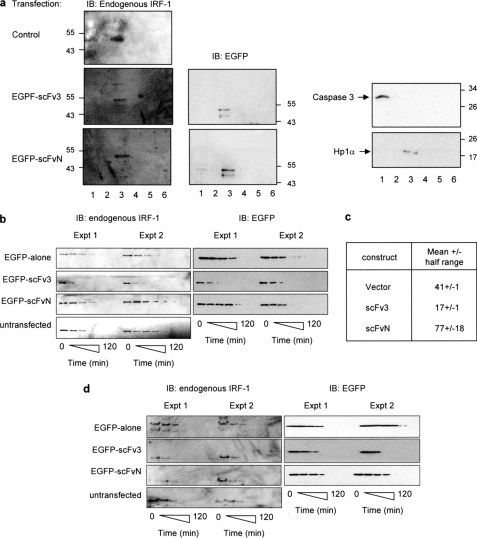
Possible changes in the localization of IRF-1 were investigated using subcellular fractionation. Fig. 7a shows that in untransfected HeLa cells IRF-1 is almost exclusively detected in the soluble nuclear compartment (fraction 3), and this is not affected by the presence of either scFv3 or scFvN, providing evidence that the nanobodies co-localize with IRF-1 (Fig. 7a, left panel) in the nucleus but do not affect its localization. The C terminus of IRF-1 is known to be required for its efficient turnover (6). The half-life of soluble endogenous IRF-1 protein was therefore measured in cycloheximide-treated cells that had been transfected with scFv3 or scFvN, and this was compared with the half-life in untransfected or EGFP alone expressing cells. Two independent experiments are shown that are representative of a total of five such experiments (Fig. 7, b and c; GAPDH control given in supplemental figure). In each case, scFv3 caused a decrease in the half-life of detergent-extractable IRF-1 to around 17 min (Fig. 7, b and c; scFv3), from a value of 41 min in the EGFP alone expressing cells and 77 min in cells expressing scFvN (Fig. 7, b and c; vector or EGFP and scFvN). Interestingly, the rate of degradation of scFv3 itself was similar to that of IRF-1 when the nanobody was present (Fig. 7b) suggesting that scFv3-IRF-1 may be co-degraded. It is possible that binding of scFv3 to IRF-1 changes detergent solubility rather than the half-life per se; IRF-1 remaining in the pellet following re-extraction under denaturing conditions was therefore measured. As with the soluble fraction, the half-life of insoluble IRF-1 appeared to be reduced in the presence of scFv3, compared with the controls (Fig. 7d).
FIGURE 7.
scFv3 decreases the half-life of IRF-1. a, HeLa cells were untransfected (control) or transfected with EGFP-scFv3 or EGFP-scFvN (1.2 μg) and fractionated after 24 h. Fraction 1, cytosol; fraction 2, membrane/organelle; fraction 3, soluble nuclear proteins; fraction 4, detergent-insoluble nuclear fraction; fraction 5, cytoskeletal proteins. Fraction 6 was generated by resuspending the residual pellet in SDS sample buffer. The fractions were analyzed by SDS-PAGE/immunoblot (IB) and probed with anti-IRF-1, anti-EGFP, anti-HP1α (a nuclear localization marker), and anti-caspase 3 (a cytosolic localization marker). The data are representative of two independent experiments. b, HeLa cells were untransfected or transfected with pDEST53 (EGFP alone), EGFP-scFvN, or scFv3 (0.6 μg). Post-transfection (24 h), the cells were treated with cycloheximide (30 μg/ml) and harvested at 0, 15, 30, 45, 60, 90, and 120 min. Detergent-soluble proteins were analyzed by SDS-PAGE immunoblot developed using anti-EGFP or anti-IRF-1 monoclonal antibody. Two independent experiments are shown, and they are representative of a total of five such experiments. c, natural logarithm (ln) of (% protein remaining) was calculated for the data in b, and plotted against time (in minutes) to obtain a linear graph of the form y = mx + c (m = slope, c = y-intercept). A mean graph was drawn, and the time at y = ln(50) was calculated from the equation of the mean line. This value represents the calculated half-life (t0.5) ± half the range. d, detergent-insoluble pellet from b was solubilized in sample buffer and analyzed by SDS-PAGE/immunoblot developed using an anti-IRF-1 monoclonal antibody.
Together the above results suggest that the observed increase in IRF-1 activity caused by binding of scFv3 to the Mf1 domain (Figs. 5 and 6) was not due to a change in localization of IRF-1 or to a decrease in the rate of its degradation. It also suggests that, in addition to its previously documented role as a requirement for efficient degradation of IRF-1, the Mf1 domain may also be involved in preventing IRF-1 from being degraded too rapidly.
Introduction of a Single Point Mutation at the C Terminus of IRF-1 Mimics the Effect of scFv3 Nanobody Binding
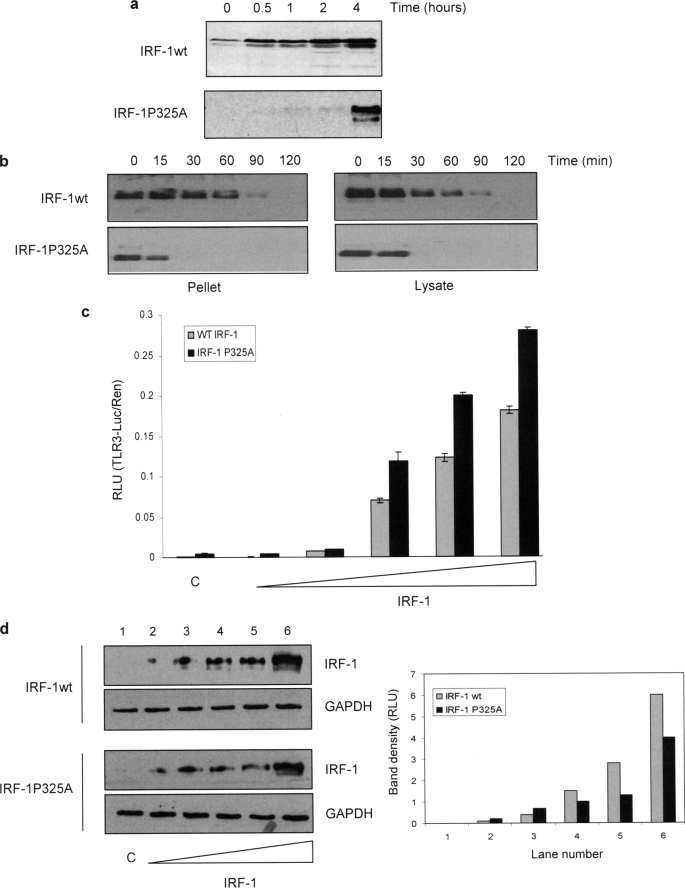
We found it of interest that the increase in IRF-1 transcriptional activity seen in the presence of scFv3 was accompanied by a significant decrease in the half-life of the protein, especially as deletion of the Mf1 domain has previously been associated with an increase in IRF-1 t0.5 (6). We therefore sought independent evidence that the Mf1 domain, as well as being required for efficient degradation of IRF-1 (6, 7), might also act as a “brake” on IRF-1 turnover. During characterization of C-terminal truncation mutants, we have previously noted that a construct from which the C-terminal four residues had been deleted was expressed in cells at much lower levels than wild-type IRF-1 (5). We therefore investigated whether mutations in the extreme C terminus of IRF-1 increased its rate of degradation and whether this was reflected in a gain of transcriptional activity. The minimal mutation that affected the level of IRF-1 expression was found to be the substitution of Pro325 at the extreme C terminus with Ala (IRF-1P325A). Fig. 8a shows that this mutant is expressed at low levels, but it accumulates in the presence of the proteasome inhibitor MG132. To investigate the difference in expression of wild-type IRF-1 and the P325A mutant, their half-lives were determined in cycloheximide-treated cells. The t0.5 for both the detergent-soluble and -insoluble fraction of P325A-IRF-1 was significantly less than that of the wild-type protein (Fig. 8b). Thus, mutation of Pro325 is sufficient to signal an increase in the rate of IRF-1 degradation.
FIGURE 8.
Mutation of Pro325 mimics scFv3 binding to IRF-1. a, A375 cells were transfected with IRF-1WT and IRF-1P325A (0.5 μg); 24 h later they were treated with MG132 (50 μm) and harvested at the times shown. IRF-1 was detected following analysis of the lysates by 10% SDS-PAGE/immunoblot. The data are representative of two separate experiments. b, HeLa cells were transfected with IRF-1WT or IRF-1P325A and 24 h later treated with cycloheximide (30 μg/ml) and then harvested at the times shown. Detergent-soluble lysate (right panel) and the residual pellet from the detergent extraction solubilized using SDS-PAGE sample buffer (left panel) were analyzed by SDS-PAGE immunoblot developed using anti-IRF-1, and the data are representative of at least three experiments. c and d, HeLa cells were transfected with TLR3-luc (120 ng), Renilla (60 ng), and a titration of either IRF-1WT (0–0.25 μg) or IRF-1P325A (0–0.5 μg) DNA to allow for normalized protein expression (varying expression levels of WT and mutant IRF-1 previously determined, data not shown). Reporter gene activity was measured in relative light units (RLU) and is expressed as the ratio of Luc:Ren (c). The assays were carried out in duplicate; results are given as mean ± half the range and are representative of at least two independent sets of experiments. Wild-type and IRF-1P325A protein normalization was shown by SDS-PAGE/immunoblot developed using anti-IRF-1 monoclonal antibody (d, left panel),and the data from the gel was quantified using a SynGene imaging system and are given as relative light units against gel lane number (d, right panel).
Having identified a residue within the C-terminal 25 amino acids of IRF-1, which when mutated had a similar effect on IRF-1 degradation to scFv3 nanobody binding, we asked whether this mutant had a higher intrinsic transcriptional activity (Fig. 8c). When the activity of IRF-1P325A was determined on the TLR3 promoter, the mutant consistently displayed significantly higher activity (Fig. 8c) than the wild-type protein. The difference in activity is accentuated when expression levels are taken into account (Fig. 8d). Thus, although at each point 2-fold more IRF-1P325A vector was transfected than wild-type vector, to compensate for the shorter half-life of the mutant protein, quantitation demonstrates that the wild-type protein was still expressed at higher levels (Fig. 8d, right panel). Thus the P325A mutant is more active that the wild-type protein as a transcriptional activator, providing independent evidence that the C terminus of IRF-1 can limit the rate at which the protein is degraded and that changes in the rate of degradation correlate with increases in the activity of IRF-1 as an activator of gene expression.
DISCUSSION
Data are presented suggesting that the Mf1 domain of IRF-1 limits its ability to activate transcription and that this negative regulation can be relieved using nanobodies that bind to the Mf1 domain or by mutating the C-terminal residue of the protein (Pro325). In addition, the data highlight a possible role for the Mf1 domain in coordinating a link between the rate of IRF-1 degradation and its transcriptional activity. Previous studies have suggested that IRF-1 activity is regulated primarily at the level of transcription as IRF-1 steady state levels increase in response to agents such as interferon and DNA damage, which induce its transcriptional activity (3, 25). However, we demonstrate here that IRF-1 is also subject to post-translational regulation; it is held in a latent or partially active state, and activity can be induced in the absence of an increase in its steady state levels.
The Mf1 domain is a 25-amino acid region in the extreme C terminus of IRF-1, which appears to be a “hot spot” for the regulation of both its function and turnover. Thus, the Mf1 domain has been shown to house an LXXLL coregulator signature motif (amino acids 306–310), which is essential for IRF-1-mediated repression of Cdk2 (5) and a binding site for members of the Hsp70 family of molecular chaperones that play a role in maintaining the normal cellular function of IRF-1 (8). The Mf1 domain lies within a region originally defined as an enhancer region (amino acids 257–325) required for maximal IRF-1 transcriptional activity (26) that most likely functions through the recruitment of coactivators such as p300 (27). More detailed analysis of the IRF-1 C terminus has revealed that removal of the last 25 residues, rather than the entire enhancer domain, does not compromise its ability to activate gene expression. Rather a ΔC-terminal 25-residue IRF-1 truncation mutant displayed an increase in intrinsic transactivation potential when compared with the full-length protein (Fig. 5) (5). This suggests that the extreme C terminus, later named multifunctional-1 (Mf1) domain (8), might be a negative regulator of IRF-1 activity. As these studies relied on the use of transiently expressed IRF-1 mutant proteins, it has not been possible to determine the effect of the Mf1 domain on IRF-1 as a transcriptional activator under normal cellular conditions. By developing a nanobody that can bind to both endogenous and exogenous IRF-1, we have been able to demonstrate that the Mf1 domain is normally rate-limiting for IRF-1-mediated gene expression. The data presented in this study are in good agreement with our previous observations using Ala scanning and a series of truncation mutants to narrow down the possible negative regulatory domain (5) to amino acids 311–317. As the scFv3 interaction with IRF-1 requires both Tyr311 and Ser317 (Fig. 4), its activation of IRF-1-mediated gene expression lends strong support to the idea that these residues include a negative regulatory motif (5).
Previous studies have shown that single chain nanobodies can be used to modulate the function of their target antigens in a cellular environment by, for example, altering the intracellular localization of the target proteins (18, 28–30), neutralizing enzyme activity (31), disrupting normal protein-protein interactions (32–35), or affecting DNA-protein interactions (32). In this study, the scFv3 nanobody was able to activate IRF-1-mediated transcription between 4- and 8-fold making it a powerful tool to study the functions of the Mf1 domain in a cell system. Conventional monoclonal antibodies and nanobodies have been used previously to study a negative regulatory domain in p53. Monoclonals and scFv, which bind to the 421 epitope, have been used to explore the role of the p53 C-terminal domain in negative regulation of its DNA binding and transcriptional activity (36–38). In addition, 421 antibody-based scFvs have been suggested as a way to partially reactivate mutant forms of the p53 protein in cells (39). The mechanism of p53 activation by 421 antibodies is through an increase in the sequence-specific DNA binding activity of the protein. In this study, we found no evidence for an increased DNA binding activity of IRF-1 in the presence of scFv3 (Fig. 3e) nor was there any obvious difference in the subcellular localization of IRF-1 when bound to the activating nanobody (Fig. 7a). Nanobody binding may impact on the conformation/dynamics of the IRF-1 structure or attenuate inhibition mediated via an intramolecular interaction. The IRF-1-related protein IRF-3 is subject to this type of regulation. In the case of IRF-3, autoinhibition occurs due to an interaction between the C terminus and a region from the N-terminal domain (40), and this process is regulated by C-terminal phosphorylation (41). However, structural information is only available for the DNA binding domain of IRF-1 (42), and how the Mf1 domain relates to the overall tertiary structure of the protein remains to be determined. It is also possible that the Mf1 domain can act as an allosteric modulatory site in response to C-terminal binding proteins. To date, few IRF-1 interactions have been documented, and the number of proteins known to interact with the C terminus is even more limited. However, Hsp70 family members have recently been identified as physiologically relevant Mf1 interacting factors that impact on IRF-1 localization, transcriptional activity, and its rate of degradation (8) suggesting that the activity of IRF-1 is likely to be modulated by agents that mimic or disrupt Mf1 protein-protein interactions.
Increased IRF-1 degradation upon scFv3 binding (Fig. 7, b–d) seems contrary to our previous observation that the Mf1 domain is required for efficient IRF-1 turnover (6). However, the Pro325 IRF-1 mutant protein displays a similar property as it has a reduced t0.5 compared with the wild-type protein (Fig. 8b). As nanobody epitope mapping (Fig. 4) shows that scFv3 also has a strong requirement for Pro325, its effect on degradation seems unlikely to be an artifact. The Mf1 domain therefore appears to contain elements that are both required for efficient degradation of IRF-1 (6) and that prevent the rate of degradation from being too rapid (Figs. 7 and 8), i.e. the Mf1 can act like a rheostat “fine-tuning” IRF-1 turnover dependent on cellular conditions.
Both scFv3-bound IRF-1 and P325A IRF-1 have increased transactivation activity, as well as an increased rate of degradation, making it interesting to speculate that these two Mf1 domain functions may be linked. It is increasingly clear that both the proteolytic and nonproteolytic functions of the proteasome are required for correct regulation of the transcriptional machinery, with evidence of both the 19 S and 26 S proteasomes being associated with chromatin, components of the basal transcription machinery, and/or various transcription factors (43–48). The precise mechanism(s) linking transcriptional activation with protein degradation remain unclear. However, evidence suggests roles for proteasome-mediated degradation in establishing limits for transcription, promoting the exchange of transcription factors on chromatin, and stimulating multiple rounds of transcription initiation (49–54). It will therefore be of interest to determine exactly how changes in the half-life of IRF-1 relate to its function as a transcription factor.
Supplementary Material
Acknowledgments
We thank Prof. Ted Hupp for help with setting up the antibody phage display screening and for critical reading of the manuscript. We also thank Dr. Mirjam Eckert for generation of mutant IRF-1 constructs.
This work was supported by Programme Grant 377/A6355 (to K. L. B.) from Cancer Research UK.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
- scFv
- single chain variable fragment
- Ni-NTA
- nickel-nitrilotriacetic acid
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- ISRE
- interferon-stimulated response element
- EGFP
- enhanced GFP
- PKR
- protein kinase RNA-activated.
REFERENCES
- 1.Boultwood J., Fidler C., Lewis S., MacCarthy A., Sheridan H., Kelly S., Oscier D., Buckle V. J., Wainscoat J. S. (1993) Blood 82, 2611–2616 [PubMed] [Google Scholar]
- 2.Green W. B., Slovak M. L., Chen I. M., Pallavicini M., Hecht J. L., Willman C. L. (1999) Leukemia 13, 1960–1971 [DOI] [PubMed] [Google Scholar]
- 3.Willman C. L., Sever C. E., Pallavicini M. G., Harada H., Tanaka N., Slovak M. L., Yamamoto H., Harada K., Meeker T. C., List A. F., et al. (1993) Science 259, 968–971 [DOI] [PubMed] [Google Scholar]
- 4.Lee E. J., Jo M., Park J., Zhang W., Lee J. H. (2006) Biochem. Biophys. Res. Commun. 347, 882–888 [DOI] [PubMed] [Google Scholar]
- 5.Eckert M., Meek S. E., Ball K. L. (2006) J. Biol. Chem. 281, 23092–23102 [DOI] [PubMed] [Google Scholar]
- 6.Pion E., Narayan V., Eckert M., Ball K. L. (2009) Cell. Signal. 21, 1479–1487 [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa K., Yokosawa H. (2000) Eur. J. Biochem. 267, 1680–1686 [DOI] [PubMed] [Google Scholar]
- 8.Narayan V., Eckert M., Zylicz A., Zylicz M., Ball K. L. (2009) J. Biol. Chem. 284, 25889–25899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G. P. (1985) Science 228, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 10.McCafferty J., Griffiths A. D., Winter G., Chiswell D. J. (1990) Nature 348, 552–554 [DOI] [PubMed] [Google Scholar]
- 11.Dornan D., Hupp T. R. (2001) EMBO Rep. 2, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wawrzynow B., Pettersson S., Zylicz A., Bramham J., Worrall E., Hupp T. R., Ball K. L. (2009) J. Biol. Chem. 284, 11517–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke N., Jimenez-Lara A. M., Voltz E., Gronemeyer H. (2004) EMBO J. 23, 3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshima S., Nakamura T., Namiki S., Okada E., Tsuchiya K., Okamoto R., Yamazaki M., Yokota T., Aida M., Yamaguchi Y., Kanai T., Handa H., Watanabe M. (2004) Mol. Cell. Biol. 24, 6298–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wesolowski J., Alzogaray V., Reyelt J., Unger M., Juarez K., Urrutia M., Cauerhff A., Danquah W., Rissiek B., Scheuplein F., Schwarz N., Adriouch S., Boyer O., Seman M., Licea A., Serreze D. V., Goldbaum F. A., Haag F., Koch-Nolte F. (2009) Med. Microbiol. Immunol. 198, 157–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saerens D., Ghassabeh G. H., Muyldermans S. (2008) Curr. Opin. Pharmacol. 8, 600–608 [DOI] [PubMed] [Google Scholar]
- 17.Colby D. W., Garg P., Holden T., Chao G., Webster J. M., Messer A., Ingram V. M., Wittrup K. D. (2004) J. Mol. Biol. 342, 901–912 [DOI] [PubMed] [Google Scholar]
- 18.Cossins A. J., Harrison S., Popplewell A. G., Gore M. G. (2007) Protein Expr. Purif. 51, 253–259 [DOI] [PubMed] [Google Scholar]
- 19.Heinz S., Haehnel V., Karaghiosoff M., Schwarzfischer L., Müller M., Krause S. W., Rehli M. (2003) J. Biol. Chem. 278, 21502–21509 [DOI] [PubMed] [Google Scholar]
- 20.Fujita T., Kimura Y., Miyamoto M., Barsoumian E. L., Taniguchi T. (1989) Nature 337, 270–272 [DOI] [PubMed] [Google Scholar]
- 21.Xie R. L., Gupta S., Miele A., Shiffman D., Stein J. L., Stein G. S., van Wijnen A. J. (2003) J. Biol. Chem. 278, 26589–26596 [DOI] [PubMed] [Google Scholar]
- 22.Beretta L., Gabbay M., Berger R., Hanash S. M., Sonenberg N. (1996) Oncogene 12, 1593–1596 [PubMed] [Google Scholar]
- 23.Kirchhoff S., Koromilas A. E., Schaper F., Grashoff M., Sonenberg N., Hauser H. (1995) Oncogene 11, 439–445 [PubMed] [Google Scholar]
- 24.Gongora C., Degols G., Espert L., Hua T. D., Mechti N. (2000) Nucleic Acids Res. 28, 2333–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romeo G., Fiorucci G., Chiantore M. V., Percario Z. A., Vannucchi S., Affabris E. (2002) J. Interferon Cytokine Res. 22, 39–47 [DOI] [PubMed] [Google Scholar]
- 26.Kirchhoff S., Oumard A., Nourbakhsh M., Levi B. Z., Hauser H. (2000) Eur. J. Biochem. 267, 6753–6761 [DOI] [PubMed] [Google Scholar]
- 27.Dornan D., Eckert M., Wallace M., Shimizu H., Ramsay E., Hupp T. R., Ball K. L. (2004) Mol. Cell. Biol. 24, 10083–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beerli R. R., Wels W., Hynes N. E. (1994) J. Biol. Chem. 269, 23931–23936 [PubMed] [Google Scholar]
- 29.Zhou P., Goldstein S., Devadas K., Tewari D., Notkins A. L. (1998) J. Immunol. 160, 1489–1496 [PubMed] [Google Scholar]
- 30.Tellez C., Jean D., Bar-Eli M. (2004) Methods 34, 233–239 [DOI] [PubMed] [Google Scholar]
- 31.Paz K., Brennan L. A., Iacolina M., Doody J., Hadari Y. R., Zhu Z. (2005) Mol. Cancer Ther. 4, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 32.Visintin M., Melchionna T., Cannistraci I., Cattaneo A. (2008) J. Biotechnol. 135, 1–15 [DOI] [PubMed] [Google Scholar]
- 33.Riley C. J., Engelhardt K. P., Saldanha J. W., Qi W., Cooke L. S., Zhu Y., Narayan S. T., Shakalya K., Croce K. D., Georgiev I. G., Nagle R. B., Garewal H., Von Hoff D. D., Mahadevan D. (2009) Cancer Res. 69, 1933–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen P. A., Mani J. C., Lane D. P. (1998) Oncogene 17, 2445–2456 [DOI] [PubMed] [Google Scholar]
- 35.Bai J., Sui J., Zhu R. Y., Tallarico A. S., Gennari F., Zhang D., Marasco W. A. (2003) J. Biol. Chem. 278, 1433–1442 [DOI] [PubMed] [Google Scholar]
- 36.Hupp T. R., Lane D. P. (1994) Curr. Biol. 4, 865–875 [DOI] [PubMed] [Google Scholar]
- 37.Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. (1992) Cell 71, 875–886 [DOI] [PubMed] [Google Scholar]
- 38.Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. (1993) Nucleic Acids Res. 21, 3167–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caron de Fromentel C., Gruel N., Venot C., Debussche L., Conseiller E., Dureuil C., Teillaud J. L., Tocque B., Bracco L. (1999) Oncogene 18, 551–557 [DOI] [PubMed] [Google Scholar]
- 40.Lin R., Mamane Y., Hiscott J. (1999) Mol. Cell. Biol. 19, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Lin C. H., Ma G., Orr M., Baffi M. O., Wathelet M. G. (2002) Eur. J. Biochem. 269, 6142–6151 [DOI] [PubMed] [Google Scholar]
- 42.Escalante C. R., Yie J., Thanos D., Aggarwal A. K. (1998) Nature 391, 103–106 [DOI] [PubMed] [Google Scholar]
- 43.Muratani M., Tansey W. P. (2003) Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 44.Lipford J. R., Deshaies R. J. (2003) Nat. Cell Biol. 5, 845–850 [DOI] [PubMed] [Google Scholar]
- 45.Reid J., Svejstrup J. Q. (2004) J. Biol. Chem. 279, 29875–29878 [DOI] [PubMed] [Google Scholar]
- 46.Somesh B. P., Reid J., Liu W. F., Søgaard T. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2005) Cell 121, 913–923 [DOI] [PubMed] [Google Scholar]
- 47.Dhananjayan S. C., Ismail A., Nawaz Z. (2005) Essays Biochem. 41, 69–80 [DOI] [PubMed] [Google Scholar]
- 48.Ransom M., Williams S. K., Dechassa M. L., Das C., Linger J., Adkins M., Liu C., Bartholomew B., Tyler J. K. (2009) J. Biol. Chem. 284, 23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillette T. G., Gonzalez F., Delahodde A., Johnston S. A., Kodadek T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5904–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins G. A., Tansey W. P. (2006) Curr. Opin. Genet. Dev. 16, 197–202 [DOI] [PubMed] [Google Scholar]
- 51.Kang Z., Pirskanen A., Jänne O. A., Palvimo J. J. (2002) J. Biol. Chem. 277, 48366–48371 [DOI] [PubMed] [Google Scholar]
- 52.Saccani S., Marazzi I., Beg A. A., Natoli G. (2004) J. Exp. Med. 200, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka T., Grusby M. J., Kaisho T. (2007) Nat. Immunol. 8, 584–591 [DOI] [PubMed] [Google Scholar]
- 54.Ostendorff H. P., Peirano R. I., Peters M. A., Schlüter A., Bossenz M., Scheffner M., Bach I. (2002) Nature 416, 99–103 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.