Abstract
The mammalian target of rapamycin (mTOR) functions in cells at least as two complexes, mTORC1 and mTORC2. Intensive studies have focused on the roles of mTOR in the regulation of cell proliferation, growth, and survival. Recently we found that rapamycin inhibits type I insulin-like growth factor (IGF-1)-stimulated lamellipodia formation and cell motility, indicating involvement of mTOR in regulating cell motility. This study was set to further elucidate the underlying mechanism. Here we show that rapamycin inhibited protein synthesis and activities of small GTPases (RhoA, Cdc42, and Rac1), crucial regulatory proteins for cell migration. Disruption of mTORC1 or mTORC2 by down-regulation of raptor or rictor, respectively, inhibited the activities of these proteins. However, only disruption of mTORC1 mimicked the effect of rapamycin, inhibiting their protein expression. Ectopic expression of rapamycin-resistant and constitutively active S6K1 partially prevented rapamycin inhibition of RhoA, Rac1, and Cdc42 expression, whereas expression of constitutively hypophosphorylated 4E-BP1 (4EBP1-5A) or down-regulation of S6K1 by RNA interference suppressed expression of the GTPases, suggesting that both mTORC1-mediated S6K1 and 4E-BP1 pathways are involved in protein synthesis of the GTPases. Expression of constitutively active RhoA, but not Cdc42 and Rac1, conferred resistance to rapamycin inhibition of IGF-1-stimulated lamellipodia formation and cell migration. The results suggest that rapamycin inhibits cell motility at least in part by down-regulation of RhoA protein expression and activity through mTORC1-mediated S6K1 and 4E-BP1-signaling pathways.
Keywords: Cell Motility, Cytoskeleton, mTOR, Protein Synthesis, Rho
Introduction
The mammalian target of rapamycin (mTOR),3 a member of the phosphoinositide 3′-kinase-related kinase family, is a central controller of cell proliferation, growth, and survival (1). Rapamycin can form a complex with FK506 binding protein 12 and then bind mTOR, selectively inhibiting its kinase activity and function (1). Recently, two mTOR complexes (mTORC1 and mTORC2) have been identified in mammalian cells (1). mTORC1 is composed of mTOR, mLST8 (also termed G-protein β-subunit-like protein, GβL, a yeast homolog of LST8), PRAS40 (proline-rich Akt substrate 40 kDa), and raptor (regulatory-associated protein of mTOR) and is rapamycin-sensitive (2–8). In response to growth factors and nutrients, mTORC1 regulates cell proliferation and growth by modulating many processes, including protein synthesis and ribosome biogenesis through downstream effectors like 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) and S6K1 (ribosomal p70 S6 kinase 1) (1, 9). mTORC2 consists of mTOR, mLST8, mSin1 (mammalian stress-activated protein kinase-interacting protein 1), rictor (rapamycin-insensitive companion of mTOR), and protor (protein observed with rictor; also named PRR5 (proline-rich protein 5)), and is rapamycin-insensitive (10–16). mTORC2 phosphorylates Akt and PKC, signals to small GTPases (RhoA and Rac1), and controls cytoskeleton organization (11, 17–20). Most recently, mTORC2 has been reported to phosphorylate SGK1 (serum and glucocorticoid-inducible kinase 1) (21), although this remains controversial (22). Both mTORC1 and mTORC2 interact with a negative regulator DEPTOR (23).
Clinical trials have demonstrated that rapamycin and its analogs (CCI-779, RAD001, and AP23573) (termed rapalogs) are promising anticancer drugs. They share the same mechanism and specifically block the function of mTOR, inhibiting growth of numerous solid tumors (renal, breast, prostate, colon, and brain cancers) with only mild side effects (9). Intensive studies have focused on the crucial roles of mTOR in controlling cell proliferation, growth, and survival. Recently this laboratory and others have further revealed its pivotal role in regulation of tumor cell migration and cancer metastasis (18, 24–26). We found that rapamycin suppresses IGF-1-stimulated F-actin reorganization and migration in various tumor cell lines by inhibiting mTORC1-mediated 4E-BP1 and S6K1 pathways (24). This is in part associated with rapamycin inhibition of S6K1-mediated phosphorylation of focal adhesion proteins (FAK, paxillin, and p130Cas) (25). However, how mTOR regulates F-actin reorganization and cell motility, particularly how 4E-BP1 pathway regulates cell motility, remains to be elucidated.
RhoA, Rac1, and Cdc42 are Rho-family small GTPases that cycle between an active GTP-bound form and an inactive GDP-bound form and were identified as key regulators of actin cytoskeletal dynamics and cell motility (27). Specifically, RhoA induces formation of actin stress fibers and focal adhesions, Rac1 stimulates formation of lamellipodia and membrane ruffles, and Cdc42 promotes formation of filopodia and actin microspikes (27, 28). Recent studies further show that RhoA also spatiotemporally regulates tail detachment and lamellipodia formation (29, 30). In NIH 3T3 and HeLa cells, reduced Rac1 and RhoA activity was observed by disruption of mTORC2 using siRNAs to rictor, mLST8, or mTOR (11). Short time (30 min) rapamycin treatment does not affect Rac1 activity in HeLa cells (26), but prolonged rapamycin treatment (28 days) does inhibit RhoA protein expression and activity in an ex vivo organ culture model of human internal mammary arteries (31). However, the role of the small GTPases in mTOR-mediated cell motility and F-actin reorganization has not been studied.
Here we show that rapamycin inhibited protein expression and activities of RhoA, Cdc42, and Rac1 by suppressing their protein synthesis in a panel of tumor cells. Both mTORC1 and mTORC2 regulated the activities of these proteins. However, only disruption of mTORC1 mimicked the effect of rapamycin, inhibiting their protein expression. mTORC1-mediated 4E-BP1 and S6K1 pathways were essential for the expression of these small GTPases. Constitutively active RhoA, but not Cdc42 and Rac1, prevented rapamycin inhibition of IGF-1-stimulated F-actin reorganization and cell motility. The results suggest that rapamycin inhibits F-actin reorganization and cell motility at least in part by down-regulation of RhoA protein expression and activity through mTORC1-mediated S6K1- and 4E-BP1-signaling pathways.
EXPERIMENTAL PROCEDURES
Cell Lines and Cultures
Human rhabdomyosarcoma Rh30 and Ewing sarcoma Rh1 (gifts from Dr. Peter J. Houghton, Nationwide Children's Hospital, Columbus, OH) were grown in antibiotic-free RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) at 37 °C and 5% CO2. Human cervical adenocarcinoma (HeLa), prostate carcinoma (PC-3), and glioblastoma (U-373) (American Type Culture Collection, Manassas, VA) were grown in antibiotic-free Dulbecco's modified Eagle's medium (DMEM) (Mediatech) supplemented with 10% FBS at 37 °C and 5% CO2. Human embryonic kidney 293 (American Type Culture Collection), 293TD, and 293A cells (Invitrogen) were grown in antibiotic-free DMEM (Mediatech) supplemented with 10% heat-inactivated FBS and nonessential amino acid (Mediatech) at 37 °C and 5% CO2. For experiments where cells were deprived of serum, cell monolayers were washed with phosphate-buffered saline (PBS) and incubated in the serum-free DMEM.
Recombinant Adenoviral Constructs and Infection of Cells
The recombinant adenoviruses expressing HA-tagged 4E-BP1–5A (Ad-4EBP1-5A), HA-tagged constitutively active and rapamycin-resistant S6K1 (Ad-S6K1-ca), and the control virus expressing green fluorescence protein (GFP) alone (Ad-GFP) were described (24). To generate recombinant adenoviruses expressing mTOR mutants, FLAG-tagged mTOR (S2035T) and mTOR (S2035T/D2357E) were excised from expression vectors pIRESNeo-FLAG-mTOR-T and pIRESNeo-FLAG-mTOR-TE (32) (gifts from Dr. Jie Chen, University of Illinois, Urbana, IL) and subcloned to pShuttle-CMV vector. The recombinant adenoviruses were generated and amplified using “Ad-Easy system” (33). To construct recombinant adenoviruses expressing FLAG-tagged RhoA, Cdc42, and Rac1 mutants, DNA fragments encoding corresponding mutants were excised from retroviral vector mIEG3-RhoA-L63, mIEG3-RhoA-N19, mIEG3-Cdc42-L28, mIEG3-Cdc42-N17, mIEG3-Rac1-L61, and mIEG3-Rac1-N17 (34, 35) (gifts from Dr. Yi Zheng, University of Cincinnati, Cincinnati, OH) and then subcloned to FLAG-tagged pENTR11 shuttle vector. The recombinant adenoviruses including the control virus expressing LacZ were generated and amplified using ViraPowerTM Adenoviral GatewayTM Expression kit (Invitrogen) following the manufacture's instructions. All adenoviruses were amplified, titrated, and used as described (24).
Lentiviral shRNA Generation and Infection
Lentiviral shRNA constructs targeting raptor and rictor were gifts from Dr. David M. Sabatini (Massachusetts Institute of Technology, Cambridge, MA). Lentiviral shRNA to GFP and S6K1 were generated as described (24). Lentiviruses expressing the indicated shRNAs were produced, titrated, and used as described (25).
Semiquantitative RT-PCR
Total RNA from cells was isolated using TRIzol® reagent (Invitrogen) following the manufacturer's instructions. cDNA was constructed by using MML-VII reverse transcriptase (Invitrogen) and oligo(dT)12–18 primer (Invitrogen). PCR was performed using 10 ng of cDNA, Tag DNA polymerase (New England Biolabs, Ipswich, MA), and specific primer pairs for RhoA, Cdc42, and Rac1, respectively, including RhoA forward (5′-CTGGTGATTGTTGGTGATGG) and reverse, (5′-GCGATCATAATCTTCCTGCC), Cdc42 forward (5′-GTGCCTGAGATAACTCACCA) and reverse (5′-GTAGGTGCAGGGCATTTGTC), and Rac1 forward (5′-GCTGTAGGTAAAACTTGCCT) and reverse (5′-CTGTTTGCGGATAGGATAGG). β-Actin forward (5′-TGACGGGGTCACCCACACTGTGCCCAT) and reverse (5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG) was used as an internal control. The amplification was done for 30 cycles (94 °C 30 s, 60 °C 30 s, and 72 °C 40 s). PCR products were separated on 1% agarose gel, and the bands were recorded with a digital camera.
Western Blot Analysis
Western blotting was performed as described previously (24). The primary antibodies used included antibodies to FLAG, β-tubulin (Sigma), raptor, rictor (Bethyl Laboratories, Montgomery, TX), HA, p-S6K1 (Thr-389), S6K1, eIF4E, p-Akt (Thr-308), Akt, mTOR, RhoA, Cdc42, Rac1 (Santa Cruz Biotechnology, Santa Cruz, CA), 4E-BP1 (Zymed Laboratories Inc., South San Francisco, CA), p-Akt (Ser-473), p-S6 ribosomal protein (Ser-235/236), and S6 ribosomal protein (Cell Signaling, Beverly, MA).
Protein Synthesis and Degradation Assay
To determine the effect of rapamycin on protein synthesis of RhoA, Cdc42, and Rac1, in vivo [35S]Met/Cys labeling was used. Briefly, Rh30 or Rh1 cells grown in 100-mm dishes in serum-free DMEM medium were pretreated with or without rapamycin (100 ng/ml) in the presence or absence of IGF-1 (10 ng/ml) for 24 h. Subsequently, the cells were washed with PBS twice, switched to 3 ml of Met/Cys-free DMEM (Mediatech) containing 10 μm MG-132 (Calbiochem), a proteasome inhibitor (to prevent protein from degradation), and then pulsed with 0.3 mCi/ml [35S]Met/Cys (MP Biomedicals, Solon, OH) for up to 4 h in the presence or absence of rapamycin (100 ng/ml) and IGF-1 (10 ng/ml), as described previously (36). Subsequently, the labeled cells were lysed in 50 mm Tris, pH 7.2, containing 150 mm NaCl, 1% sodium deoxycholate, 0.1% SDS, 1%Triton X-100, 10 mm NaF, 1 mm Na3VO4, and protease inhibitor mixture (1:1000, Sigma). RhoA, Cdc42, and Rac1 and β-GAPDH (as control) were immunoprecipitated from cell lysates and separated on 12% SDS-PAGE gel followed by autoradiography. The rate of protein synthesis in cells was normalized by β-GAPDH values.
To determine the effect of rapamycin on protein degradation, cells were grown in complete medium and pretreated with/without rapamycin (100 ng/ml) for 24 h and then incubated with or without cycloheximide (50 μg/ml). After incubation for ∼12 h, cell lysates were collected followed by Western blot analysis using antibodies to RhoA, Cdc42, and Rac1 and β-tubulin (as control). The rate of protein degradation in cells was normalized by β-tubulin values.
Small GTPase Activity Assay
Activities of RhoA, Cdc42, and Rac1 were measured using Rho assay kit and Rac/Cdc42 assay kit (Millipore, Billerica, MA) following the manufacturer's instructions. Briefly, agarose beads conjugated with glutathione S-transferase (GST)-PAK1 (for Cdc42 and Rac1) and GST-rhotekin (for RhoA) were used to pull down endogenous Rac1-GTP, Cdc42-GTP, and RhoA-GTP from cell lysates. The active form of RhoA, Cdc42, or Rac1 was detected by immunoblotting with antibodies to individual GTPases, respectively.
Cell Motility Assay
The single cell motility assay and the wound healing assay were performed as described (24).
F-actin Staining
F-actin staining and visualization were performed as described (25).
Statistical Analysis
Results were expressed as the mean ± S.E.). Statistical analysis was performed by Student's t test (STATISTICA, Statsoft Inc, Tulsa, OK). A level of p < 0.05 was considered to be significant.
RESULTS
Rapamycin Inhibits Protein Expression and Activities of RhoA, Cdc42, and Rac1
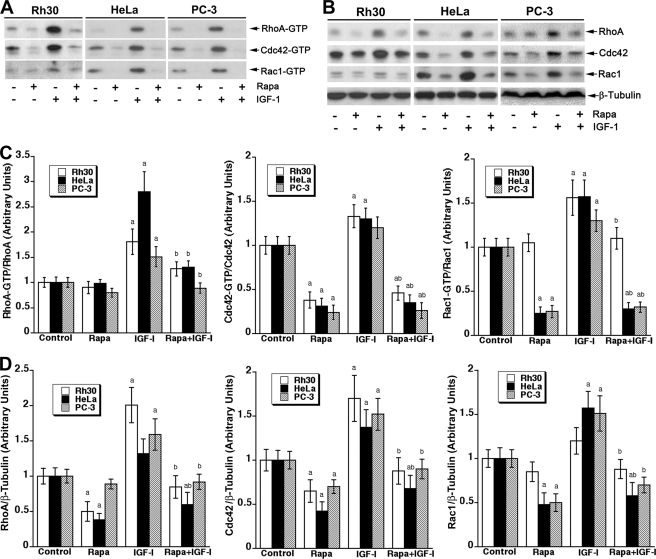
The small GTPases are critical for cell motility and F-actin reorganization (27). Because prolonged treatment with rapamycin inhibits RhoA protein expression and activity in an ex vivo organ culture model of human internal mammary arteries (31), we hypothesized that mTOR may regulate cell motility and F-actin reorganization in part by mediating the expression and activity of the small GTPases. To test this hypothesis, first we examined the expression and activities of RhoA, Cdc42, and Rac1 upon rapamycin and IGF-1 treatment. As shown in Fig. 1, IGF-1 (10 ng/ml, 22 h) stimulation markedly elevated both the active (GTP-bound) and total protein levels of RhoA, Cdc42, and Rac1 in Rh30 cells. Treatment with rapamycin (100 ng/ml, 24 h) reduced the basal or IGF-1-stimulated expression and activities of these proteins. This is not a cell-type context, as similar results were also observed in other tumor cell lines, including those derived from cervical cancer (HeLa), prostate cancer (PC-3) (Fig. 1), Ewing sarcoma (Rh1), and glioblastoma (U373) (data not shown).
FIGURE 1.
Rapamycin inhibits IGF-1-stimulated protein expression and activities of RhoA, Cdc42, and Rac1. Indicated cells were serum-starved for 24 h and then pretreated with or without rapamycin (100 ng/ml) for 2 h followed by stimulation with or without IGF-1 (10 ng/ml) for 22 h. Cells were harvested for the GTPase activity assay (A) and Western blot analysis (B) for RhoA, Cdc42, and Rac1 as described under “Experimental Procedures.” Densitometry for the bands in A and B was performed using NIH ImageJ, as shown in C and D, respectively. Note: for calculation of the ratio of GTP-bound/total protein of the small GTPases, shown in C, total protein values used were from D. Results are the means ± S.E. and are pooled from three independent experiments. a, p < 0.05, difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test).
Rapamycin Inhibits Protein Expression of RhoA, Cdc42, and Rac1 by Inhibiting Their Protein Synthesis Rather Than Altering Their mRNA Levels or Protein Degradation
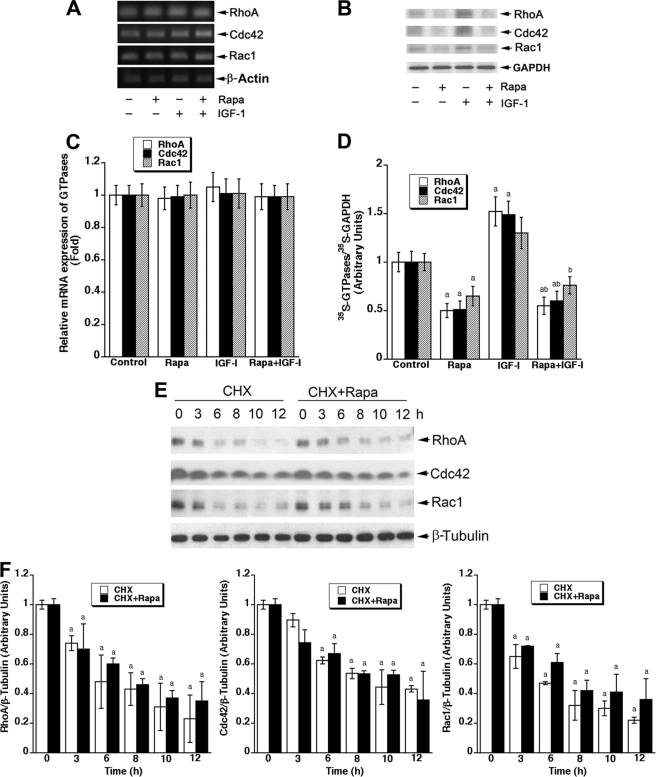
mTORC1 controls protein synthesis and coordinates transcription by nuclear RNA polymerases (37–39). It has been reported that rapamycin inhibits cyclin D1 protein expression at transcriptional, translational and post-translational levels (40). To elucidate how rapamycin inhibits expression of RhoA, Cdc42, and Rac1, we investigated whether rapamycin influences mRNA levels, protein synthesis, and degradation of these small GTPases. By semiquantitative RT-PCR, we found that rapamycin (100 ng/ml, 24 h) or IGF-1 (10 ng/ml, 22 h) did not alter their mRNA levels in Rh30 cells (Fig. 2, A and C), suggesting that rapamycin does not suppress expression of these small GTPases at transcriptional levels. However, when Rh30 cells, pretreated with or without rapamycin (100 ng/ml) in the presence or absence of IGF-1 (10 ng/ml) for 24 h, were labeled with [35S]Met/Cys for 4 h, IGF-1 enhanced the incorporation of [35S]Met/Cys into RhoA, Cdc42, and Rac1, which was remarkably suppressed by rapamycin treatment (Fig. 2, B and D). Furthermore, when Rh30 cells, grown in 10% FBS-RPMI medium and pretreated with or without rapamycin for 24 h, were exposed for ∼12 h to cycloheximide (50 μg/ml), an inhibitor of eukaryotic protein synthesis by preventing initiation and elongation on 80S ribosomes, we found that rapamycin treatment did not obviously alter their protein turnover rate (Fig. 2, E and F). The results suggest that rapamycin inhibits expression of RhoA, Cdc42, and Rac1 by suppression of their protein synthesis.
FIGURE 2.
Rapamycin inhibits IGF-1-stimulated cellular protein expression of RhoA, Cdc42, and Rac1 by suppressing their protein synthesis. A, Rh30 cells were serum-starved for 24 h and then pretreated with or without rapamycin (100 ng/ml) for 2 h followed by stimulation with or without IGF-1 (10 ng/ml) for 22 h. Cells were harvested for semiquantitative RT-PCR. PCR products were separated on 1% agarose gel and stained with ethidium bromide. Bands were visualized under UV light and photographed with a digital camera. The effects of rapamycin on protein synthesis (B) and degradation (E) in Rh30 cells were determined as described under “Experimental Procedures.” Semiquantitative data for A, B and E by densitometry using NIH ImageJ are shown in C, D and F, respectively. Results are the means ± S.E. and are pooled from three independent experiments. a, p < 0.05, difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test). CHX, cycloheximide.
Inhibition of mTOR Kinase Activity Is Necessary for Rapamycin Inhibition of Expression of Small GTPases
During skeletal myogenesis, mTOR regulates the production of IGF-II mRNA, which is independent of its kinase activity (41). To determine whether rapamycin inhibition of expression of RhoA, Cdc42, and Rac1 is through inhibition of mTOR activity, we generated recombinant adenoviruses encoding FLAG-tagged rapamycin-resistant but kinase active mTOR (S2035T, designated mTOR-T) or kinase-dead mTOR-T (S2035T/D2357E, designated mTOR-TE) (32). The function of adenoviral constructions was confirmed by determining the expression of FLAG-tagged mTOR mutants and phosphorylation status of S6K1 (Thr-389) and 4E-BP1. Consistent with previous observation (42), ectopic expression of FLAG-tagged mTOR-T, but not FLAG-tagged mTOR-TE or GFP (control), conferred high resistance to rapamycin inhibition of phosphorylation of 4E-BP1 and rendered mild resistance to rapamycin inhibition of phosphorylation of S6K1 in Rh30 cells (Fig. 3A). Of interest, expression of FLAG-mTOR-T, but not mTOR-TE or GFP, rendered high resistance to rapamycin inhibition of expression of RhoA, Cdc42, and Rac1 in the cells, indicating that rapamycin inhibits expression of RhoA, Cdc42, and Rac1 in an mTOR kinase activity-dependent manner (Fig. 3, A and B).
FIGURE 3.
Inhibition of mTOR kinase activity is necessary for rapamycin inhibition of GTPase expression. Rh30 cells infected with recombinant adenovirus expressing GFP or FLAG-tagged mTOR-T and mTOR-TE mutants were serum-starved for 24 h and then pretreated with or without rapamycin (100 ng/ml) for 2 h followed by stimulation with IGF-1 (10 ng/ml) for 22 h. Cells were harvested for Western blot analysis using indicated antibodies (A). Semiquantitative data for A by densitometry using NIH ImageJ are shown in B. Results are the means ± S.E. and are pooled from three independent experiments. a,p < 0.05, difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test).
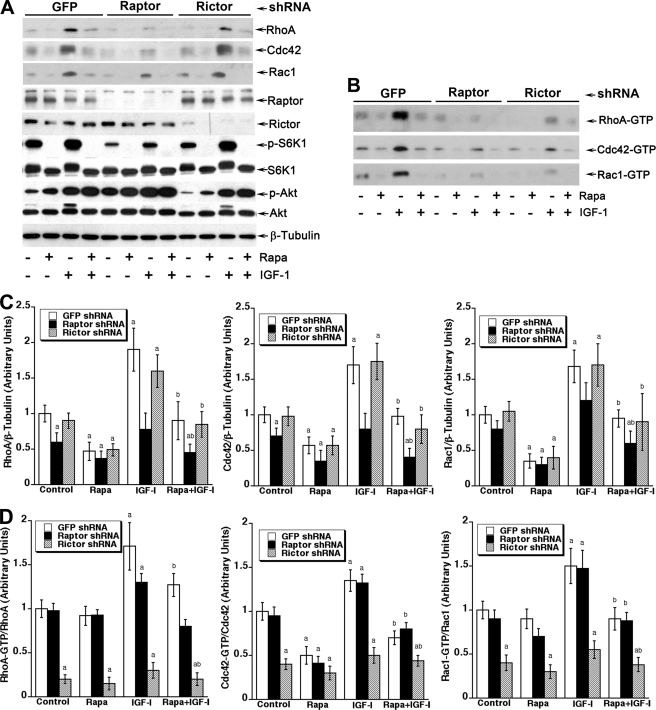
Both mTORC1 and mTORC2 Regulate the Small GTPases Activities, but Only mTORC1 Controls Their Cellular Protein Expression
Because mTORC2, which was originally thought to be rapamycin-insensitive, has recently been found to be sensitive to prolonged (>24 h) rapamycin treatment (17), we next determined which of the mTOR complexes is responsible for rapamycin inhibition of small GTPase expression. To this end, mTORC1 and mTORC2 were disrupted, respectively, by knocking down raptor or rictor in cultured cells using corresponding lentiviral shRNAs. As expected, down-regulation of raptor and rictor inhibited phosphorylation of mTORC1-mediated S6K1 (Thr-389) and mTORC2-mediated Akt (Ser-473), respectively (Fig. 4A). However, only down-regulation of raptor, but rictor, inhibited the basal or IGF-1-stimulated protein expression of RhoA, Cdc42, and Rac1 (Fig. 4, A and C). Next we assessed the activity of these small GTPases in Rh30 cells with disrupted mTORC1 or mTORC2. Knocking down raptor reduced the total activity of RhoA, Cdc42, or Rac1 in the cells (Fig. 4B), which was consistent with inhibition of protein expression of the small GTPases (Fig. 4A). However, down-regulation of raptor did not significantly alter the GTP-bound form (ratio of GTP-bound/total protein) of RhoA, Cdc42, or Rac1 in the cells (Fig. 4D). In contrast, knocking down rictor did not affect protein expression of RhoA, Cdc42, or Rac1 (Fig. 4, A and C) but decreased GTP-bound forms of the small GTPases sharply (Fig. 4, B and D). The results indicate that the two mTOR complexes regulate the small GTPases through different mechanisms, i.e. mTORC1 controls protein expression of RhoA, Cdc42, and Rac1, and mTORC2 mediates the activities of these small GTPases.
FIGURE 4.
mTORC1 controls GTPase protein expression, whereas mTORC2 mediates GTPase activity. A, disruption of mTORC1, but not mTORC2, impaired RhoA, Cdc42, and Rac1 expression. Rh30 cells infected with lentiviral shRNA to raptor, rector, or GFP were serum-starved for 24 h and then pretreated with or without rapamycin (100 ng/ml) for 2 h followed by stimulation with or without IGF-1 (10 ng/ml) for 22 h. Cells were harvested for Western blot analysis using indicated antibodies. B, down-regulation of rictor or raptor impaired the activities of RhoA, Cdc42, and Rac1. Rh30 cells infected with lentiviral shRNA to raptor, rictor or GFP were serum-starved for 24 h and then stimulated with IGF-1 for 24 h. Cells were harvested for RhoA GTPase activity assay and Rac1/Cdc42 GTPase activity assay, as described under “Experimental Procedures.” Active small GTPases are shown as GTP-bound forms (RhoA-GTP, Cdc42-GTP, and Rac1-GTP). Densitometry for the bands in A and B was performed using NIH ImageJ, as shown in C and D, respectively. Note: for calculation of the ratio of GTP-bound/total protein of the small GTPases, shown in D, total protein values used were from C. Results are the means ± S.E. and are pooled from three independent experiments. a, p < 0.05, difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test).
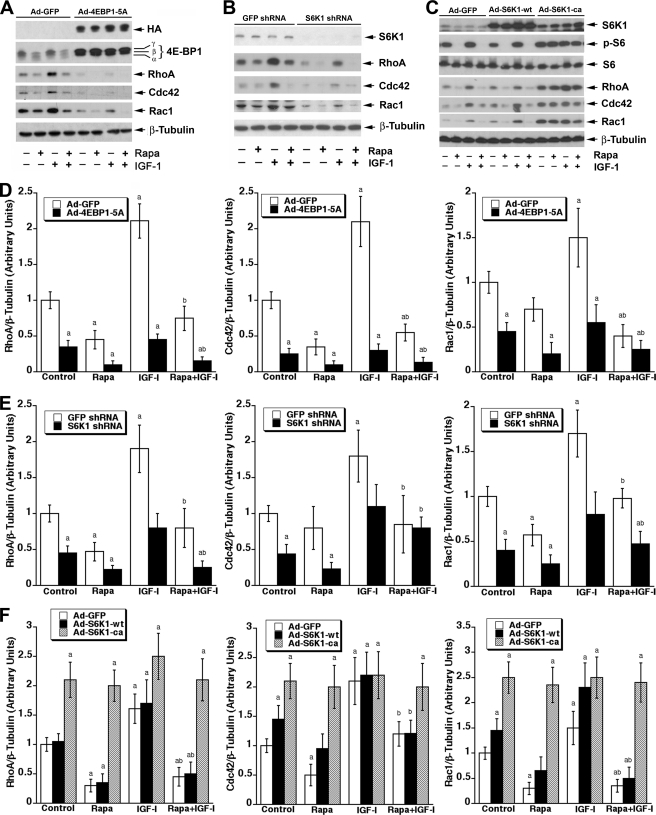
Both mTORC1-mediated 4E-BP1 and S6K1 Pathways Are Involved in Rapamycin Inhibition of RhoA, Cdc42, and Rac1 Expression
Our recent data indicate that rapamycin suppresses IGF-1-stimulated F-actin reorganization and migration, in part through inhibition of S6K1-mediated phosphorylation of focal adhesion proteins (FAK, paxillin, and p130Cas) (25). It is not clear how the 4E-BP1 pathway regulates cell motility and F-actin reorganization. Because 4E-BP1/eIF4E pathway is crucial for protein synthesis (1), we hypothesized that this pathway may regulate cell motility by controlling protein synthesis of the small GTPases. To test this hypothesis, Rh30 cells were infected with recombinant adenoviruses expressing HA-tagged constitutively hypophosphorylated 4E-BP1 (4EBP1-5A) and Ad-GFP (as control). 4EBP1-5A expressed in Rh30 cells, as evidenced by Western blot analysis of tagged HA and elevated protein levels of 4E-BP1 (Fig. 5A). As expected, forced expression of 4EBP1-5A, which tightly binds eIF4E and suppresses its activity (24), substantially down-regulated the basal or IGF-1-stimulated protein levels of RhoA, Cdc42, and Rac1 in the cells (Fig. 5, A and D).
FIGURE 5.
Both 4E-BP1 and S6K1 pathways are involved in rapamycin inhibition of RhoA, Cdc42, and Rac1 expression. Rh30 cells infected with recombinant adenovirus expressing GFP or constitutively hypophosphorylated 4EBP1-5A (A) with lentiviral shRNA to GFP or S6K1 (B) or with recombinant adenovirus expressing GFP, wild type S6K1 (S6K1-wt), or constitutively active S6K1 (S6K1-ca) (C) were serum-starved for 24 h and then pretreated with or without rapamycin (100 ng/ml) for 2 h followed by stimulation with or without IGF-1 (10 ng/ml) for 22 h. Cells were harvested for Western blot analysis using indicated antibodies. Semiquantitative data for A–C by densitometry using NIH ImageJ are shown in D–F, respectively. Results are the means ± S.E. and are pooled from three independent experiments. a, p < 0.05, difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test).
As the S6K1 pathway is also important for protein synthesis (1), next we investigated whether the S6K pathway controls protein synthesis of the small GTPases as well. As shown in Fig. 5, B and E, lentiviral shRNA to S6K1, but not to GFP (control), down-regulated protein expression of S6K1 by ∼90%. Similar to forced expression of 4EBP1-5A, down-regulation of S6K1 also suppressed expression of RhoA, Cdc42, and Rac1. In contrast, cells expressing constitutively active and rapamycin-resistant S6K1 (S6K1-ca), but not GFP (control), showed partial resistance to rapamycin inhibition of expression of the small GTPases (Fig. 5, C and F). Collectively, our results suggest that both 4E-BP1 and S6K1 pathways are essential for mTORC1-regulated RhoA, Cdc42, and Rac1 expression, mediating cell motility.
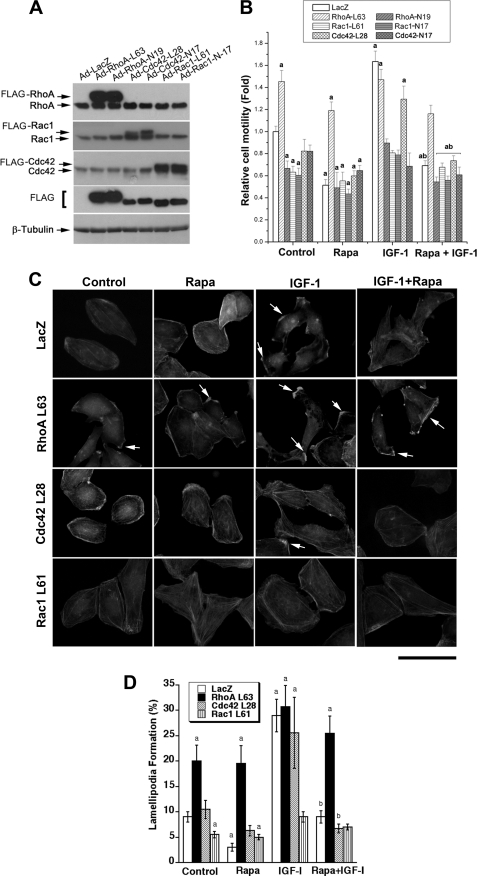
Overexpression of Constitutively Active RhoA, but Not Rac1 and Cdc42, Prevents Rapamycin Inhibition of Lamellipodia Formation and Cell Migration
RhoA, Cdc42, and Rac1 regulate cytoskeleton organization and cell migration (27). To demonstrate that these small GTPases are really involved in mTOR-mediated cell motility and F-actin reorganization, we constructed recombinant adenoviruses encoding constitutively active or dominant negative forms of RhoA, Cdc42, and Rac1. Rh30 cells infected with these adenoviruses were treated with or without rapamycin with and without IGF-1 followed by cell motility assays and F-actin staining. As shown in Fig. 6A, considerable levels of constitutively active RhoA (RhoA-L63), Cdc42 (Cdc42-L28), and Rac1 (Rac1-L61) and dominant negative RhoA (RhoA-N19), Cdc42 (Cdc42-N17), and Rac1 (Rac1-N17) were, respectively, expressed in Rh30 cells, as detected by Western blotting using antibodies to FLAG and individual GTPases. Expression of RhoA-L63 mimicked the effect of IGF-1 stimulation and increased the basal cell motility by ∼150% and conferred resistance to rapamycin inhibition of the basal and IGF-1-stimulated cell motility (Fig. 6B). To our surprise, expression of Rac1-L61 and Cdc42-L28 inhibited the basal cell motility by ∼35 and 20%, respectively, and did not render significant resistance to rapamycin inhibition of the basal or IGF-1-stimulated cell motility. Furthermore, IGF-1 was able to stimulate the motility of cells expressing RhoA-L63 and Cdc42-L28 but not Rac1-L61 (Fig. 6B). In addition, expression of RhoA-N19, Cdc42-N17, or Rac1-N17 not only inhibited the basal cell motility, but also suppressed the IGF-1-stimulated cell motility.
FIGURE 6.
Overexpression of constitutively active RhoA, but not Cdc42 or Rac1, prevents rapamycin inhibition of IGF-1-stimulated lamellipodia formation and cell migration. Rh30 cells were infected with recombinant adenovirus expressing FLAG-tagged constitutively active or dominant negative RhoA (RhoA-L63 or RhoA-N19), Cdc42 (Cdc42-L28 or Cdc42-N17), and Rac1 (Rac1-L61 or Rac1-N17) as well as control adenovirus expressing LacZ for 24 h. Infected cells were used for Western blot analysis (A), wound healing assay (B) and F-actin staining (C). A, Western blots show expression of corresponding GTPase mutants. B, wound healing assay results are the means ± S.E. and are pooled from three independent experiments. a, p < 0.05 (unpaired Student's t test). C, F-actin staining was visualized and photographed with a Nikon TE300 digital inverted microscope. Representative images are shown. Note: arrows indicate lamellipodia formation. Bar = 40 μm. Quantitative results for lamellipodia formation are shown in D as the means ± S.E. (n = 3). a, p < 0.05 difference versus control group; b, p < 0.05, difference versus IGF-1 group (unpaired Student's t test).
Consistent with the above findings, expression of RhoA-L63 not only increased the lamellipodia formation even in the absence of IGF-1 but also conferred resistance to rapamycin inhibition of IGF-1 stimulated lamellipodia formation (Fig. 6, C and D). Expression of RhoA-N19 mimicked the effect of rapamycin, blocking IGF-1 stimulated lamellipodia formation (data not shown). Expression of Cdc42-L28 had no apparent effect on either IGF-1-stimulated or rapamycin-inhibited lamellipodia formation (Fig. 6, C and D). Expression of Rac1-L61 inhibited IGF-1-stimulated lamellipodia formation and noticeably increased the cellular stress fibers (Fig. 6, C and D), suggesting a distinct role of Rac1 in regulating cytoskeleton organization in the cells. The data indicate that rapamycin may target RhoA, inhibiting IGF-1-stimulated F-actin reorganization and cell motility.
DISCUSSION
Recently we demonstrated that rapamycin inhibits F-action reorganization and cell motility by inhibition of mTOR kinase activity (24, 25). mTOR controls synthesis of a variety of proteins, such as cyclin D1, c-Myc, ornithine decarboxylase, vascular endothelial growth factor, etc. (43). Small GTPases (RhoA, Rac1, and Cdc42) are crucial for cytoskeleton organization and cell migration (27). Therefore, we hypothesized that rapamycin may inhibit mTOR-mediated protein synthesis or activities of the small GTPases, leading to inhibition of F-action reorganization and cell motility. To test this hypothesis, we first studied the effect of rapamycin on cellular protein levels and activities of RhoA, Rac1, and Cdc42. By Western blotting and small GTPase activity assay, we found that rapamycin did inhibit the basal and IGF-1-stimulated activities and protein expression of RhoA, Rac1, and Cdc42 in Rh30 cells. The inhibitory effect of rapamycin on expression of the small GTPases was also observed in other tumor cell lines, including those derived from cervical cancer (HeLa), prostate cancer (PC-3) (Fig. 1), Ewing sarcoma (Rh1), and glioblastoma (U-373) (data not shown), suggesting that this is not cell type-dependent.
It is well known that rapamycin selectively inhibits mTOR kinase activity and function (1). However, studies have also shown that rapamycin inhibits differentiation of C2C12 cells in mTOR kinase activity-independent manner (44), although this remains controversial (45). During skeletal myogenesis, mTOR regulates the production of IGF-II mRNA, which is also independent of the kinase activity of mTOR (41). This prompted us to study whether rapamycin inhibition of small GTPase expression is through inhibition of mTOR kinase activity. Here we found that rapamycin failed to inhibit RhoA, Cdc42, and Rac1 expression in cells expressing rapamycin-resistant mTOR (mTOR-T) but not in control cells expressing GFP or rapamycin-resistant but kinase dead mTOR (mTOR-TE), suggesting that the kinase activity of mTOR is essential for expression of RhoA, Cdc42, and Rac1. This is consistent with our previous finding that the kinase activity of mTOR is necessary for IGF-1-stimulated F-actin reorganization and cell motility (24, 25).
mTOR may control protein expression at transcriptional, translational, or post-translational levels (40). In this study we found that rapamycin did not alter mRNA levels or protein turnover of RhoA, Cdc42, and Rac1 but inhibited protein synthesis of these small GTPases, suggesting that mTOR controls protein expression of RhoA, Cdc42, and Rac1 at translational level.
Previous studies only showed that mTORC2 regulates the activities of RhoA and Rac1 in NIH 3T3, HeLa cells, and human umbilical vein endothelial cells (11, 18, 26). Here for the first time we demonstrate that mTOR not only controls the cellular activities of RhoA, Rac1, and Cdc42 but also regulates the cellular protein expression of these small GTPases. Our data support the notion that mTORC1 mediates protein synthesis of RhoA, Cdc42, and Rac1; mTORC2 regulates the activities of these proteins. This is supported by the findings that disruption of mTORC1 by down-regulation of raptor inhibited the expression and activities of the small GTPases, whereas disruption of mTORC2 by down-regulation of rictor only inhibited the activities of RhoA, Cdc42, and Rac1. This is consistent with the previous findings that a 20–30% decrease in GTP-bound Rac1 was observed in mTOR-, mLST8-, or mAVO3-siRNA-transfected NIH 3T3 cells and little-to-no decrease in GTP-bound Rac1 in raptor siRNA-transfected cells (11). Recent studies have revealed that mTORC2 regulates Rac1 activation through P-Rex1 (26). How mTORC2 regulates RhoA and Cdc42 activity remains to be determined.
4E-BP1 and S6K1 are the best characterized mTORC1 effectors, which were found to be involved in the regulation of F-actin reorganization and cell motility (24, 25). Recently we observed that only S6K1 pathway regulated phosphorylation of the focal adhesion proteins (FAK, paxillin, and p130Cas) (25), which is related to F-actin reorganization and cell migration (46). It was not clear how 4E-BP1/eIF4E pathway regulates F-actin reorganization and cell motility. To address this question, we used 4EBP1-5A, which is a constitutively hypophosphorylated mutant 4E-BP1 (T36A, T45A, S64A, T69A, and S82A) (47) and can tightly binds and sequester eIF4E, inhibiting Cap-dependent translation (47). We found that expression of 4EBP1-5A mimicked the effect of rapamycin, remarkably inhibiting expression of RhoA, Cdc42, and Rac1 in Rh30 cells stimulated with or without IGF-1. We also investigated the role of S6K1 pathway in the regulation of the GTPase expression. Interestingly, down-regulation of S6K1 also impaired the expression of RhoA, Cdc42, and Rac1. In contrast, expression of a rapamycin-resistant and constitutively active of S6K1 mutant (S6K1-F5A-E389-R3A, S6K1-ca) (24, 48) was able to confer partial resistance to rapamycin inhibition of the GTPase expression. Collectively, we conclude that both 4E-BP1 and S6K1 pathways participate in the regulation of expression of RhoA, Cdc42, and Rac1.
RhoA, Cdc42, and Rac1 are all critical molecules for cytoskeleton organization and cell migration, but they play different roles during cytoskeletal dynamics (27, 28). Our data indicate that although the activities and expression of RhoA, Cdc42, and Rac1 were inhibited by rapamycin, only overexpression of constitutively active RhoA (RhoA-L63), but not Cdc42 (Cdc42-L28) and Rac1 (Rac1-L61), prevented rapamycin inhibition of IGF-1 stimulated lamellipodia formation and cell motility, implying a crucial role of RhoA in mTOR-mediated cell motility. This is strongly supported by our observations that expression of constitutively active RhoA enhanced the basal level of lamellipodia formation and cell motility to a level stimulated by IGF-1, whereas expression of dominant negative RhoA (RhoA-N19) blocked the effect of IGF-1 stimulation. Interestingly, although expression of constitutively active Cdc42 or Rac1 failed to rescue rapamycin inhibition of IGF-1 stimulated F-actin reorganization and cell migration, expression of dominant negative Cdc42 (Cdc42-N17) or Rac1 (Rac1-N17), like dominant negative RhoA (RhoA N19), abolished IGF-1-stimulated cell motility. These results suggest that a certain level of RhoA, Cdc42, or Rac1 activity may be required for either cell polarization/protrusion or adhesion/de-adhesion, leading to cell migration. In this study we also noticed that expression of constitutively active Rac1 actually increased the cellular stress fibers and inhibited IGF-1-stimulated lamellipodia formation and cell motility in Rh30 cells (Fig. 6). This is in contrast to the finding in NIH 3T3 cells in which expression of constitutively active Rac1 induces formation of membrane ruffles and lamellipodia, whereas expression of constitutively active RhoA results in formation of stress fibers (11). The discrepancy may likely be due to different cell types used. It has been described that Rac1 is essential for actin stress fiber formation in primary mouse embryonic fibroblasts (49). In colon carcinoma cells (50), hepatocarcinoma cells (51), and human microvascular endothelial cells (52), activation of RhoA promotes lamellipodia formation and cell migration. Therefore, the roles of the small GTPases in the regulation of F-actin reorganization and cell motility depend on cell types studied.
In summary, here we found that rapamycin inhibited RhoA, Cdc42, and Rac1 expression and activity in an mTOR kinase activity-dependent manner. mTORC1 controls the protein synthesis, whereas mTORC2 regulates the activities of the small GTPases. Both mTORC1-mediated S6K1 and 4E-BP1/eIF4E pathways are essential for RhoA, Cdc42, and Rac1 expression. However, inhibition of RhoA activity is primarily responsible for rapamycin inhibition of IGF-1-stimulated lamellipodia formation and cell motility.
Acknowledgments
We thank Drs. John Blenis, Jie Chen, Peter J. Houghton, John Lawrence, Jr., David M. Sabatini, and Yi Zheng for providing cell lines or constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA115414 (to S. H.). This work was also supported by American Cancer Society Grant RSG-08-135-01-CNE (to S. H.).
- mTOR
- mammalian target of rapamycin
- 4E-BP1
- eukaryotic initiation factor 4E binding protein 1
- IGF-1
- type I insulin-like growth factor
- mTORC1/2
- mTOR complex 1/2
- raptor
- regulatory-associated protein of mTOR
- rictor
- rapamycin (Rapa)-insensitive companion of mTOR
- S6K1
- ribosomal p70 S6 kinase 1.
REFERENCES
- 1.Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 2.Fonseca B. D., Smith E. M., Lee V. H., MacKintosh C., Proud C. G. (2007) J. Biol. Chem. 282, 24514–24524 [DOI] [PubMed] [Google Scholar]
- 3.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 4.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 5.Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 6.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 7.Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 8.Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 9.Fasolo A., Sessa C. (2008) Expert Opin. Investig. Drugs 17, 1717–1734 [DOI] [PubMed] [Google Scholar]
- 10.Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. (2006) Curr. Biol. 16, 1865–1870 [DOI] [PubMed] [Google Scholar]
- 11.Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 12.Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 13.Pearce L. R., Huang X., Boudeau J., Pawłowski R., Wullschleger S., Deak M., Ibrahim A. F., Gourlay R., Magnuson M. A., Alessi D. R. (2007) Biochem. J. 405, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 15.Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo S. Y., Kim D. H., Jun C. B., Kim Y. M., Haar E. V., Lee S. I., Hegg J. W., Bandhakavi S., Griffin T. J., Kim D. H. (2007) J. Biol. Chem. 282, 25604–25612 [DOI] [PubMed] [Google Scholar]
- 17.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 18.Dada S., Demartines N., Dormond O. (2008) Biochem. Biophys. Res. Commun. 372, 875–879 [DOI] [PubMed] [Google Scholar]
- 19.Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Martínez J. M., Alessi D. R. (2008) Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 22.Hong F., Larrea M. D., Doughty C., Kwiatkowski D. J., Squillace R., Slingerland J. M. (2008) Mol. Cell 30, 701–711 [DOI] [PubMed] [Google Scholar]
- 23.Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Chen L., Chung J., Huang S. (2008) Oncogene 27, 4998–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., Li F., Cardelli J. A., Martin K. A., Blenis J., Huang S. (2006) Oncogene 25, 7029–7040 [DOI] [PubMed] [Google Scholar]
- 26.Hernández-Negrete I., Carretero-Ortega J., Rosenfeldt H., Hernández-García R., Calderón-Salinas J. V., Reyes-Cruz G., Gutkind J. S., Vázquez-Prado J. (2007) J. Biol. Chem. 282, 23708–23715 [DOI] [PubMed] [Google Scholar]
- 27.Heasman S. J., Ridley A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 28.Tzima E. (2006) Circ. Res. 98, 176–185 [DOI] [PubMed] [Google Scholar]
- 29.Kurokawa K., Matsuda M. (2005) Mol. Biol. Cell 16, 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 31.Guérin P., Sauzeau V., Rolli-Derkinderen M., Al Habbash O., Scalbert E., Crochet D., Pacaud P., Loirand G. (2005) J. Vasc. Res. 42, 21–28 [DOI] [PubMed] [Google Scholar]
- 32.Vilella-Bach M., Nuzzi P., Fang Y., Chen J. (1999) J. Biol. Chem. 274, 4266–4272 [DOI] [PubMed] [Google Scholar]
- 33.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F., Gao Y., Wang L., Zheng Y. (2003) J. Biol. Chem. 278, 14414–14419 [DOI] [PubMed] [Google Scholar]
- 35.Guo F., Zheng Y. (2004) Mol. Cell. Biol. 24, 1426–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunavala-Dossabhoy G., Fowler M., De Benedetti A. (2004) BMC Mol. Biol. 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer C., Grummt I. (2006) Oncogene 25, 6384–6391 [DOI] [PubMed] [Google Scholar]
- 38.Peng T., Golub T. R., Sabatini D. M. (2002) Mol. Cell. Biol. 22, 5575–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang C. K., Zheng X. F. (2007) Cell Cycle 6, 25–29 [DOI] [PubMed] [Google Scholar]
- 40.Hashemolhosseini S., Nagamine Y., Morley S. J., Desrivières S., Mercep L., Ferrari S. (1998) J. Biol. Chem. 273, 14424–14429 [DOI] [PubMed] [Google Scholar]
- 41.Erbay E., Park I. H., Nuzzi P. D., Schoenherr C. J., Chen J. (2003) J. Cell Biol. 163, 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahon L. P., Choi K. M., Lin T. A., Abraham R. T., Lawrence J. C., Jr. (2002) Mol. Cell. Biol. 22, 7428–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S., Houghton P. J. (2003) Curr. Opin. Pharmacol. 3, 371–377 [DOI] [PubMed] [Google Scholar]
- 44.Erbay E., Chen J. (2001) J. Biol. Chem. 276, 36079–36082 [DOI] [PubMed] [Google Scholar]
- 45.Shu L., Zhang X., Houghton P. J. (2002) J. Biol. Chem. 277, 16726–16732 [DOI] [PubMed] [Google Scholar]
- 46.Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 47.Mothe-Satney I., Brunn G. J., McMahon L. P., Capaldo C. T., Abraham R. T., Lawrence J. C., Jr. (2000) J. Biol. Chem. 275, 33836–33843 [DOI] [PubMed] [Google Scholar]
- 48.Schalm S. S., Tee A. R., Blenis J. (2005) J. Biol. Chem. 280, 11101–11106 [DOI] [PubMed] [Google Scholar]
- 49.Guo F., Debidda M., Yang L., Williams D. A., Zheng Y. (2006) J. Biol. Chem. 281, 18652–18659 [DOI] [PubMed] [Google Scholar]
- 50.O'Connor K. L., Nguyen B. K., Mercurio A. M. (2000) J. Cell Biol. 148, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genda T., Sakamoto M., Ichida T., Asakura H., Kojiro M., Narumiya S., Hirohashi S. (1999) Hepatology 30, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 52.Abécassis I., Olofsson B., Schmid M., Zalcman G., Karniguian A. (2003) Exp. Cell Res. 291, 363–376 [DOI] [PubMed] [Google Scholar]