Abstract
We have isolated an enzyme classified as chlorogenate: glucarate caffeoyltransferase (CGT) from seedlings of tomato (Solanum lycopersicum) that catalyzes the formation of caffeoylglucarate and caffeoylgalactarate using chlorogenate (5-O-caffeoylquinate) as acyl donor. Peptide sequences obtained by trypsin digestion and spectrometric sequencing were used to isolate the SlCGT cDNA encoding a protein of 380 amino acids with a putative targeting signal of 24 amino acids indicating an entry of the SlCGT into the secretory pathway. Immunogold electron microscopy revealed the localization of the enzyme in the apoplastic space of tomato leaves. Southern blot analysis of genomic cDNA suggests that SlCGT is encoded by a single-copy gene. The SlCGT cDNA was functionally expressed in Nicotiana benthamiana leaves and proved to confer chlorogenate-dependent caffeoyltransferase activity in the presence of glucarate. Sequence comparison of the deduced amino acid sequence identified the protein unexpectedly as a GDSL lipase-like protein, representing a new member of the SGNH protein superfamily. Lipases of this family employ a catalytic triad of Ser-Asp-His with Ser as nucleophile of the GDSL motif. Site-directed mutagenesis of each residue of the assumed respective SlCGT catalytic triad, however, indicated that the catalytic triad of the GDSL lipase is not essential for SlCGT enzymatic activity. SlCGT is therefore the first example of a GDSL lipase-like protein that lost hydrolytic activity and has acquired a completely new function in plant metabolism, functioning in secondary metabolism as acyltransferase in synthesis of hydroxycinnamate esters by employing amino acid residues different from the lipase catalytic triad.
Keywords: Cell Wall, Electron Microscopy (EM), Enzymes, Lipase, Plant, Molecular Evolution, Protein Localization, Secondary Metabolism
Introduction
Plant metabolism is characterized by the formation of a vast number of secondary compounds, brought about by gene families coding for enzymes that modify various phenolic, terpenoid, alkaloid, or polyketide skeletons by oxidation and reduction as well as by methylation, glycosylation, prenylation, and acylation. Most of the phenolic structures in plants are synthesized via the shikimate/hydroxycinnamate pathway, which feeds into different types of hydroxycinnamate (HCA)4 side-chain reactions (1). Among them are extensions with formation of additional ring systems (e.g. flavonoids or stilbenes), degradation (e.g. hydroxybenzoates), reduction (e.g. hydroxycinnamyl alcohols feeding into lignin biosynthesis), oxidation and lactonization (e.g. coumarins), and conjugation with a wide range of different primary and secondary compounds to form esters or amides. Syntheses of HCA conjugates are catalyzed by hydroxycinnamoyltransferases that play a decisive role in catalyzing the formation of complex patterns of HCA esters (2). Such a pattern, for example, was recently identified from Brassica napus seeds and exhibited a mixture of sinapate esters containing choline, malate, mono- and disaccharides, as well as flavonoid glycosides and an unusual cyclic spermidine amide (3).
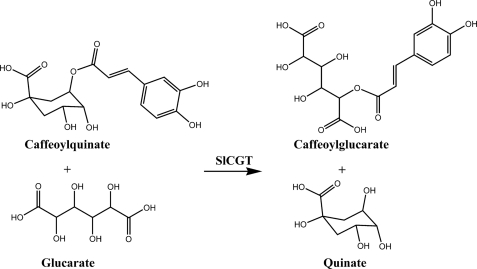
Tomato (Solanum lycopersicum) leaves accumulate caffeate esters with quinate (4), glucarate (5), and galactarate (6). Three enzymes are involved in the formation of these esters (7): (i) caffeate:CoA ligase, (ii) caffeoyl-CoA:quinate caffeoyltransferase, and (iii) chlorogenate:glucarate caffeoyltransferase (SlCGT). The reaction catalyzed by caffeate:CoA ligase leads to caffeoyl-CoA (8, 9) that is the substrate of caffeoyl-CoA:quinate caffeoyltransferase in the formation of chlorogenate (5-O-caffeoylquinate) (10), a reaction sequence that is widespread in plants (11). The third enzyme, SlCGT, is a unique acyltransferase accepting chlorogenate as the substrate to drive the transfer of the caffeoyl moiety of chlorogenate to the acceptor molecules glucarate (Fig. 1) or galactarate. Usually, most of the acyltransferases synthesizing HCA esters accept the common coenzyme A thioesters (e.g. caffeoyl-CoA) or β-acetal esters (e.g. 1-O-caffeoyl-β-glucose) (2, 12) as shown for the synthesis of caffeoylglucarate in Secale cereale (13) and in Cestrum elegans (14), respectively. Thus, the occurrence of chlorogenate is not a general precondition for the synthesis of hydroxycinnamoyl sugar carboxylic acids, as assumed by Maas et al. (15). At least the SlCGT has been unambiguously classified as a regiospecific chlorogenate-dependent caffeoyltransferase (EC 2.3.1.98) (7). The only other example known so far that chlorogenate is used as acyl donor in acyltransfer has been shown in a disproportionation reaction in the formation of isochlorogenate (3,5-di-O-caffeoylquinate) in Ipomoea batatas (16).
FIGURE 1.
Scheme of the SlCGT-catalyzed acyl transfer reaction.
Although first purification of SlCGT and its enzymatic properties were described already in 1990 (7), the gene encoding this unique enzyme involved in the biosynthesis of caffeoylglucarate and its evolutionary recruitment were not known so far. Herein, we show the identification of the cDNA encoding SlCGT and present a combination of genetic, biochemical, and cellular evidence indicating that the enzyme encoded by this gene catalyzes the formation of caffeoylglucarate in the leaf apoplast. Moreover, the deduced amino acid sequence of the full-length clone unexpectedly identified the SlCGT as a GDSL lipase-like enzyme, thereby providing a new example on evolution and diversification of hydroxycinnamoyltransferases in plants.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Plants of S. lycopersicum Mill. cv. Moneymaker (Chrestensen, Germany) were grown in the greenhouse on standard soil at 24 °C and a 16-h light regiment. Two-week-old seedlings and organs from adult plants were frozen and stored at −80 °C.
Purification of SlCGT and Mass Spectrometric Sequencing
Enzyme purification was performed on an ÄKTAexplorer (Amersham Biosciences), essentially as described previously for a related enzyme (17). Briefly, frozen 14-day-old tomato seedlings (650 g) were homogenized in extraction buffer (100 mm Tris-Cl, 10% (v/v) glycerol, 0.5 mm EDTA, 1 mm mercaptoethanol, 50 mm ascorbate (pH 7.0)). After centrifugation, the supernatant was subjected to ammonium sulfate precipitation (80% saturation) and desalted on Sephadex G-25, and the resulting crude protein solution was fractionated on a Q-Sepharose FF 50/20 column using a gradient from 0 to 1 m NaCl in 0.02 m Tris-Cl buffer (pH 7.0). Pooled active fractions were precipitated with ammonium sulfate, redissolved in 0.02 m Tris buffer (pH 7.0) and 1 m ammonium sulfate, and further fractionated on a phenyl-Sepharose 16/10 column with a gradient of 0.02 m Tris buffer and 1 m ammonium sulfate to 0.02 m same buffer. Active fractions were pooled and concentrated by ultrafiltration, and the protein was further fractionated on a Superdex G-75 16/60 column eluted with 0.01 m citrate buffer (pH 5). Active fractions were finally subjected to a Mono Q column chromatography. Protein concentrations of all fractions were determined (18) using bovine serum albumin as standard. For mass spectrometric sequencing, the 40-kDa protein band on SDS-PAGE corresponding to SlCGT was excised, subjected to digestion with trypsin and processed as described previously (17).
Assay for SlCGT Activity
Enzyme solutions were incubated in 100 mm MES (Sigma-Aldrich) (pH 6.0), containing 20 mm chlorogenate and 5 mm glucarate in a total volume of 100 μl for 10 min at 30 °C. Enzyme reactions were stopped with 10 μl of trifluoracetate, and product formation was determined by HPLC with an Alliance High Throughput HPLC (Waters) equipped with an Alliance HPLC C18 column (5 μm; 200 mm × 4.6-mm internal diameter) using a 15-min linear gradient elution from 10% to 25% acetonitrile in 1.5% aqueous phosphoric acid. The eluates were monitored by max-plot UV spectroscopy. Identification and quantification were achieved by external standardization with chlorogenate as reference compound. For inhibition of enzyme activity, different concentrations of phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich) were added to the assays and incubated at 30 °C for 5 min prior to start of the reaction.
Lipase Activity Assay
Purified SlCGT enzyme solution was diluted to a final concentration of 0.5 mg of protein ml−1 with 50 mm sodium phosphate buffer (pH 7.0). As control, Candida rugosa lipase was diluted in 50 mm sodium phosphate buffer to the same concentrations as SlCGT enzyme. Lipase activity was estimated colorimetrically by determination of the liberation of p-nitrophenol from p-nitrophenyl laurate at 405 nm (19).
Isolation of SlCGT cDNA, Sequence Analysis, and Site-directed Mutagenesis
Extraction of total RNA from 5-day-old tomato seedlings was done as described (20). This RNA was used to enrich poly(A)+ RNA by selective binding to oligo(dT)-Oligotex beads (Qiagen). Degenerated primers deduced from two peptide sequences obtained from MS sequencing of purified SlCGT (supplemental Table S1) were used as forward primers in combination with a customized poly(dT) reverse primer for RT-PCR performed with the Omniscript RT kit (Qiagen) according to the supplied protocol. A 5′ incomplete cDNA fragment with sequence similarity to putative lipases was used to deduce gene-specific primers (CGT-R1 and CGT-R2; supplemental Table S1) and to amplify a full-length cDNA by 5′-rapid amplification of cDNA ends (RACE) with the BD Smart Race cDNA amplification Kit (BD Clontech). The resulting full-length cDNA was amplified by PCR using 5′-RACE ready cDNA from tomato seedlings as template and the primer pair SlCGTcom_F and SlCGTcom_R (supplemental Table S1). Sequence analysis and alignments of DNA and proteins were done using the software package Clone Manager (Sci-Ed, Cary, NC). Site-directed mutagenesis was performed with the QuikChange XL Site-directed Mutagenesis kit (Stratagene) according to the manufacturer's protocol. All constructs, wild type SlCGT, and the mutant variants were proven by sequence analysis.
Southern Blot Analysis
Genomic plant DNA was isolated from tomato seedlings using a Maxi kit for plant DNA purification (Qiagen), digested with EcoRI and NcoI, electrophoretically separated, and transferred to Hybond N+ membrane (Amersham Biosciences) according to standard protocols (21). Hybridization was performed in DIG Easy Hyb Buffer at 42 °C overnight with a full-length SlCGT cDNA labeled with dioxigenin-dUTP using the PCR DIG Probe Synthesis kit (Roche Diagnostics). The detection of DIG-dUTP-labeled hybrids was performed by enzyme immunoassay with the DIG DNA detection kit (Roche Diagnostics).
Real-time RT-PCR
Total RNA for analysis via Real-time RT-PCR was prepared with the RNeasy Plant Mini kit and the RNase-free DNase set (Qiagen). cDNA synthesis was performed using oligo(dT)16 primer and the Omniscript Reverse Transcription kit (Qiagen). Real-time PCR was performed with the MxPro-Mx3005P real-time PCR system (Stratagene) and SYBR® Green PCR Master Mix (Applied Biosystems) as described in the manufacturer's protocol with primer pairs specific for SlCGT and elongation factor EF1α (supplemental Table S1). PCR efficiency of the primer pairs was calculated with the program LineRegPCR (22). Transcript levels of SlCGT were normalized to EF1α levels using the formula ΔCt = Ct(SlCGT) − Ct(EF1α). Relative expression levels were calculated using the highest ΔCt value as standard and the formula ER = 2−(ΔCtSample−ΔCtStandard).
Heterologous expression of SlCGT in Nicotiana benthamiana
For heterologous expression, SlCGT cDNA (wild type and mutations) was cloned into the vector pImpact1.1 (Plant Research International, Wageningen, The Netherlands). The expression cassette, consisting of rbcs promoter from Asteraceous chrysanthemum (23), coding sequence (CDS) and rbcs transcription terminator, was then cloned into the binary vector pBINPLUS (Plant Research International). The resulting recombinant vectors were transformed into Agrobacterium tumefaciens strain GV2260 (24). Recombinant agrobacteria were infiltrated into fully developed leaves of N. benthamiana. Agrobacteria harboring the empty vector pBINPLUS served as a negative control. As a control for the transformation efficiency, agrobacteria harboring a construct carrying the GUS gene (25) under control of the 35S promoter were co-infiltrated. Plants were cultivated for 5 days under greenhouse conditions. Soluble proteins extracted from leaves, desalted with a PD-10 column (Sephadex G-25; GE Healthcare) and concentrated, were used for activity assays for SlCGT and GUS (25). Calculation of CGT activity was done using GUS activity as an interna1 standard for each transformation assay as basically described by (26).
Production and Characterization of SlCGT-specific Antibody
SlCGT purified from 7-day-old tomato seedlings was used to immunize rabbits (27, 28). The serum was taken 2 weeks after the last immunization. Rabbit IgG was enriched by affinity chromatography using protein A-Sepharose CL-4B (Sigma-Aldrich) followed by elution at pH 3.0. Protein extracts from tomato seedlings and purified SlCGT were separated on SDS gels (29) and transferred to nitrocellulose. Immunological detection of SlCGT using the SlCGT-specific antibody in a dilution of 1:500 followed by a goat anti-rabbit IgG antibody conjugated with alkaline phosphatase (diluted 1:1000, BIOMOL) was performed as described (30).
Immunogold Detection of SlCGT
Small pieces of cotyledons of 7-day-old plantlets were fixed with 4% (v/w) paraformaldehyde and 0.5% (v/v) glutaraldehyde in 0.1 m sodium phosphate buffer (pH 7.4) for 2 h. After washing with buffer and dehydration by a graded series of ethanol, specimen were infiltrated with LR-White (Polysciences). After addition of accelerator, polymerization of LR-White was done at 4 °C for 2 h according to the manufacturer's instructions. Ultrathin sections mounted on cupper grids were blocked with 3% (w/v) BSA in phosphate-buffered saline (PBS) and incubated with the anti-SlCGT antibody diluted 1:1000 in 1% (v/w) BSA in PBS for 3 h. Control experiments were performed by use of preimmune serum at the same dilution. After washing with BSA/PBS, sections were incubated with protein A conjugated with 10 nm colloidal gold (Sigma-Aldrich). Sections were poststained with uranyl acetate and lead citrate and visualized with a Philips CM 10 electron microscope.
RESULTS
SlCGT Purification, Sequence Analysis, and Heterologous Expression
Protein extracts from 14-day-old tomato seedlings exhibiting high SlCGT activities (7) were used to purify the enzyme. Ammonium sulfate-precipitated protein extract from these seedlings was applied to a combination of chromatographic separation steps including adsorption, ion-exchange, and size-exclusion techniques (Table 1), essentially as described earlier for purification of a related protein (17). The final protein fraction with the highest enzymatic activity (24 nanokatals (mg of protein)−1) resulted in a 585-fold enrichment of enzyme activity. SDS-PAGE of this fraction showed a major protein band at the expected molecular mass of approximately 40 kDa (7) (supplemental Fig. S1). To gain SlCGT sequence information, this protein was excised and subjected to trypsin digestion and mass spectrometric sequencing. Sequences of four peptides were used to derive degenerated primers for reverse transcription PCR with RNA from tomato seedlings and resulted after 5′-RACE in the identification of full-length cDNA encoding SlCGT. The cDNA consists of 1143 bp, and translation of the ORF results in a protein of 380 amino acids (Fig. 2) with a calculated molecular mass of 42.58 kDa and pI of pH 5.25. Analysis of the ORF for the presence of a targeting sequence by using the SignalP and TargetP programs (31) revealed a predicted 24-amino acid N-terminal signal putatively directing SlCGT into the secretory pathway. Cleaving off this signal sequence, a mature polypeptide of 39.95 kDa and pI of pH 5.01 would result corresponding to the size determined after purification of the enzyme from seedlings. Therefore, the protein does not seem to be highly glycosylated, although there are six potential N-glycosylation sites. At least two of them (N82AT and N146SS), however, were predominantly occupied by the plant-typical N-glycan structure Man3HexNAc2FucXyl as deduced from tandem mass spectrometry of the corresponding glycopeptides, whereas N326YT was found not to be modified. No data are available from the other potential N-glycosylation sites.
TABLE 1.
Purification scheme of SlCGT from seedlings
| Purification step | Total protein | Total activity | Specific activity | Enrichment | Yield |
|---|---|---|---|---|---|
| mg | nanokatals | nanokatals (mg protein)−1 | -fold | % | |
| Sephadex G-25 | 3,500 | 145 | 0.041 | 1 | 100 |
| Q-Sepharose | 351 | 100 | 0.28 | 7 | 68 |
| Phenyl-Sepharose | 17.7 | 30 | 1.7 | 41 | 21 |
| Superdex G-75 | 2.7 | 21 | 7.8 | 188 | 14 |
| Mono Q | 0.53 | 12.8 | 24.2 | 585 | 9 |
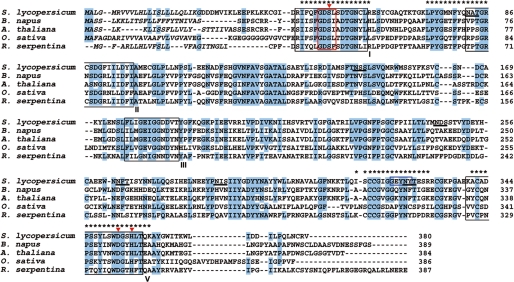
FIGURE 2.
Alignment of the deduced amino acid sequence of SlCGT with characterized related GDSL lipase-like proteins. SlCGT amino acid sequences were deduced from cDNA and aligned with a carboxylic ester hydrolase (A. thaliana; U38916) (33), sinapine esterase (B. napus; AAX59709) (17), lanatoside 15′-O-acetylesterase-like enzyme (Oryza sativa; AP002866) (60), and acetylajmalan acetylesterase (R. serpentina; AY762990) (36). The predicted N-terminal leader sequences of all enzymes are shown in italics. Highly conserved residues are shaded in blue. Asterisks, peptides identified by sequencing and used to identify SlCGT; blue box, amino acid motif used to deduce degenerated primers for PCR, red box, GDSL motif; red triangles, catalytic triad (Ser27, Asp328, His331); black box, conserved blocks in the SGNH hydrolase family (I, II, III, V) (35, 42).
To estimate the number of gene copies of SlCGT, genomic tomato DNA was digested with the restriction enzymes EcoRI and NcoI and hybridized with the cDNA resulting in a single band and two bands, respectively (supplemental Fig. S2). Because the cDNA has no cleavage site for EcoRI, but one for NcoI, we suggest that SlCGT is a single-copy gene in the tomato genome.
The SlCGT cDNA was cloned in vectors for gene expression in Escherichia coli or Saccharomyces cereviseae, but all attempts to obtain bacterial or fungal expression under various conditions failed (data not shown). Therefore, we transiently transformed leaves of Nicotiana benthamiana (32) that were subjected to protein extraction. Activity of SlCGT was proven by incubating the leaf protein extract with chlorogenate and glucarate as substrates. High specific enzyme activity was detected (2.9 nanokatals (mg of protein)−1) that was 69 times higher than that extracted from tomato leaves (0.042 nanokatals (mg of protein)−1) (supplemental Fig. S3). Controls, the nontransformed leaves, and leaves transformed with the empty vector, did not show any SlCGT activity.
Localization of the SlCGT Protein
Because the N-terminal region of SlCGT predicted entry into the secretory pathway, we investigated subcellular localization of the protein in cotyledons of 7-day-old seedlings by immunogold electron microscopy. The enzyme used to immunize the rabbits was isolated by an optimized purification protocol as described (7). Specificity of the anti-SlCGT antibodies was tested by Western blot analysis (supplemental Fig. S4). All lanes on SDS-PAGE, from crude extracts to purified enzyme, gave a single distinct signal at the expected molecular mass of 40 kDa.
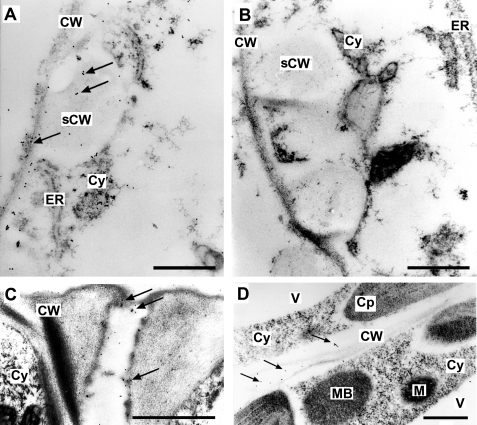
As visualized by the gold particles, SlCGT was detected in the apoplastic space in all cell types examined, such as developing xylem elements characterized by the typical secondary cell wall depositions of young vessels in the bundle sheets, the typically thickened and elaborated inner wall of guard cells, and cell walls between mesophyll cells (Fig. 3). The enzyme was also detected in some cytosolic areas and appeared to be secreted from vesicles to the apoplast (data not shown). In control experiments, in which preimmune serum was used, we did not observe any labeling, neither in the cellular cytoplasm nor in the apoplastic space of the leaf.
FIGURE 3.
SlCGT is located in the apoplastic space of cotyledons of seven-day-old seedlings. Ultrathin sections of cotyledons were immunostained with the anti-SlCGT antibody (A, C, and D) or the respective preimmune serum (B). The presence of SlCGT is visualized by colloidal gold (arrows). A, detail of a developing xylem element characterized by the typical secondary cell wall (sCW) depositions. SlCGT is located mainly in the area of cell wall including secondary cell wall depositions. B, concomitant section to A treated with preimmune serum. Note the absence of gold particles. C, detail of a guard cell pair showing the thickened and elaborated inner wall labeled by the anti-SlCGT antibody. D, cell walls between leaf mesophyll cells exhibiting label. CW, cell wall; Cy, cytoplasm; ER, endoplasmic reticulum; M, mitochondrion; MB, microbody; V, vacuole. Scale bars, 0.5 μm.
Structure-Function Analysis
The deduced SlCGT amino acid sequence matches the conservation of the serine catalytic motif GXSXXDXG within the first consensus block near the N terminus (33, 34). Multiple sequence alignment to four characterized GDSL lipase-like enzymes shows high sequence identities and indicates that SlCGT is a novel member of the GDSL lipase family (Fig. 2). Therefore, it represents another rare example of a defined catalytic function of such an enzyme. Due to this sequence identity with GDSL lipases, the mature enzyme could possibly employ a catalytic triad of Ser-Asp-His with the seryl side chain as nucleophile. The conserved amino acid residues of the proposed catalytic triad of SlCGT have been identified at sequence position 27 of the mature protein within the GDSL motif (Ser), at positions 328 (Asp) and 331 (His), which forms part of the conserved sequence motif DXXH. As part of the GDSL motif near the N terminus, the consensus sequence GDSXXD found in all members of the respective enzyme family (35, 36) is also present in the SlCGT. To prove the involvement of a catalytic seryl residue in the caffeoyl transfer reaction, the enzyme was subjected to treatment with PMSF, a potent inhibitor that phosphorylates seryl residues of proteins, which has also been successfully applied to hydrolytic GDSL proteins (34, 37). Treatment of purified SlCGT with 1, 10, and 50 mm PMSF led to strong decreases in caffeoyl transfer activities to 60%, 40%, and 0% activity, respectively (supplemental Table S2). Thus, a serine moiety of the GDSL protein may be part of the catalytic center in SlCGT.
To confirm Ser27 as a member of the active catalytic triad of the SlCGT and to gain insight into other functional elements of the caffeoyltransferase, site-directed mutagenesis was employed to replace eligible amino acid residues by alanine. From the assumed catalytic triad of SlCGT, Ser27 as well as Asp328 and His331 were changed. As an additional proof, we also mutated Asp161 and Asp162 to exclude their potential involvement in the catalytic triad instead of Asp328, as has been proposed for acetylajmalan esterase from Rauvolfia serpentina (36). The mutated cDNAs were expressed transiently in N. benthamiana. As a control for the transformation efficiency, N. benthamiana leaves were co-transformed with the GUS gene expressed from the CaMV 35S promoter. The enzyme activities of the mutant proteins are illustrated in Table 2. Results revealed for the mutant variants S27A and D328A decreased SlCGT activities by 13 and 40%, respectively. The enzymatic activity of the H331A mutant did not differ from that of the WT enzyme. The enzymes mutated at Asp161 and Asp162 lost 30 and 39% activity, respectively. The unexpectedly low reduction in enzyme activities caused by site-directed mutation indicates that the potential catalytic triad deduced from known GDSL lipases is not involved in the acyltransferase reaction of SlCGT. Therefore, the SlCGT might employ an as yet unknown catalytic motif that is involved in the specific chlorogenate-dependent caffeoyl-transferase activity.
TABLE 2.
Relative activities of recombinant SlCGT wild type (WT) and mutant variants designed by site-directed mutagenesis
Activity of wild type SlCGT (10 nanokatals (mg of protein)−1) was set to 100%. Mean values ± S.D. (n = 3) are shown.
| Variant | Activity |
|---|---|
| % | |
| WT | 100 ± 1.3 |
| S27A | 87 ± 12 |
| D161A | 70 ± 10 |
| D162A | 61 ± 2.0 |
| D328A | 60 ± 15 |
| H331A | 99 ± 12 |
To test whether the enzyme retained its lipolytic activity, possible liberation of p-nitrophenol from p-nitrophenyl laurate was measured (19). A defined lipase from C. rugosa was used as positive control. As demonstrated in supplemental Fig. S5, the SlCGT is unable to hydrolyze the laurate ester, whereas the control assay exhibits the expected activity, indicated by the appearance of yellow colored p-nitrophenol. This result agrees with those obtained from the acyltransferase assays with the SlCGT mutant variants, excluding a decisive role of the conserved GDSL lipase-specific catalytic triad.
Patterns of SlCGT Transcript Accumulation, Enzyme Activity, and Metabolite Accumulation
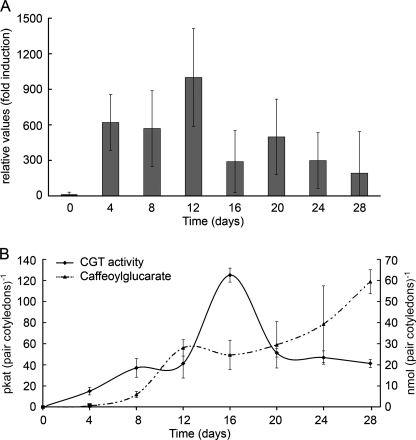
Previously published data showed a continuous increase in SlCGT activity during the development of tomato cotyledons (7). To get insights into expression of the SlCGT gene in these leaves, quantitative RT-PCR was performed to analyze the transcript accumulation of SlCGT during seedling development up to 28 days (Fig. 4A). Whereas in seeds SlCGT transcripts were barely detectable, a significant increase was measurable from 4 days after germination onward showing a maximum at 12 days. This maximum preceded the maximum in SlCGT activity, which reached highest enzyme activity at day 16 with 127 picokatals (pair of cotyledons)−1 (Fig. 4B). The metabolic product caffeoylglucarate continuously accumulates until day 28 reaching 59 nmol (pair of cotyledons)−1. A similar pattern was observed in developing primary leaves (data not shown) reaching 358 picokatals of enzyme activity and approximately 1 nmol of caffeoylglucarate (mg of leaf tissue)−1.
FIGURE 4.
Relative transcript accumulation of SlCGT in developing cotelydons is followed by increasing enzyme activity and product accumulation. A, developing cotelydons of tomato seedlings were harvested at the time points indicated, and total RNA was analyzed by real-time RT-PCR. SlCGT transcript levels were normalized to SlEF1α levels, the lowest value (seeds) was set to 1. B, time course of changes in enzyme activity of SlCGT and accumulation of 2-O-caffeoylglucaric acid as one of the two isomeric structures (2-O- or 5-O-isomer). Data represent the mean ± S.D. (N ≥ 3) and are calculated on the basis of one pair of cotyledons.
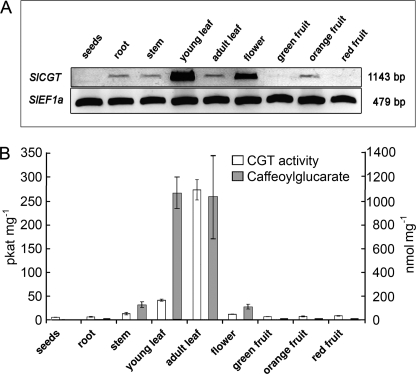
Inspection of various organs showed that young leaves exhibited highest transcript levels, followed by flowers. Transcript could also be detected in the root, stem, and young orange fruits, but only at trace levels (Fig. 5A). The SlCGT activity occurred predominantly in leaves reaching highest values in adult leaves accompanied by accumulation of caffeoylglucarate (Fig. 5B). The discrepancy between high transcript levels and low enzyme activities in the flowers may be due to distinct tissue-specific localization that awaits further studies.
FIGURE 5.
Transcripts of SlCGT, enzyme activity, and caffeoylglucarate accumulate preferentially in tomato leaves and flowers. A, Total RNA was isolated from tomato organs indicated and used for RT-PCR analysis of SlCGT transcript accumulation. Transcripts of SlEF1α served as loading control. PCR products were separated by agarose gel electrophoresis and stained by ethidium bromide. B, SlCGT enzyme activities and amount of caffeoylglucaric acid in tomato organs. Data represent the mean ± S.D. (N ≥ 3) and are calculated on the basis of mg, fresh weight.
DISCUSSION
In the course of our studies on hydroxycinnamoyltransferases in plants (38–41), we investigated the SlCGT, a tomato-specific acyltransferase that catalyzes the transfer of the caffeoyl moiety from chlorogenate (5-O-caffeoylquinate) to glucarate and galactarate, forming caffeoylglucarate and caffeoylgalactarate, respectively. SlCGT has been classified as a regiospecific chlorogenate-dependent caffeoyltransferase involved in a rather unusual mechanism found in plants leading to the formation of HCA esters (7).
After purification from tomato seedlings, fractions with highest SlCGT activities revealed enzyme enrichment to near homogeneity and were used to identify specific peptide sequences followed by PCR-based cDNA cloning. The isolated full-length SlCGT cDNA sequence was unexpectedly found to encode a GDSL lipase-like protein classified as member of the SGNH hydrolase superfamily (42). A broad range of substrates has been found to be accepted by enzymes of this superfamily (43), which hydrolyze ester bonds, e.g. of complex polysaccharides (44), phospholipids (45), or fatty acyl esters (46). Other GDSL proteins were described as cell wall-secreted lipases, such as an enzyme in the secretome of Arabidopsis thaliana (47) that may play a role in plant resistance. The SlCGT, however, obviously exhibits a completely new catalytic function, resulting in regiospecificity toward chlorogenate (5-O-caffeoylquinate) (7). The enzyme does not accept the 3-O- or 4-O-isomers or the related 5-O- and 3-O-(4-coumaroyl)-quinates as donor molecules nor other sugar acids besides glucarate and galactarate.
Expression of the gene, enzyme activity, and accumulation of caffeoylglucarate as the main product were highest in vegetative green tissues and depended on the developmental stage of leaves. Young leaves exhibited high transcript levels, resulting in accumulating enzyme activities toward mature ones. With regard to the localization of the enzyme in these leaves, the N-terminal region of SlCGT predicted entry into the secretory pathway. This assumption was supported by immunogold electron microscopy that indicated that the enzyme is present in the apoplastic space. This observation agrees with some enzyme characteristics (7). The enzyme is relatively stable and does not show the typical transient activity increase, which is characteristic for many secondary enzymes such as the caffeate:CoA ligase and caffeoyl-CoA:quinate caffeoyltransferase, which are most likely under cytoplasmic control, such as the BAHD acyltransferases (48). In addition, it exhibits some functional properties similar to typical cell wall enzymes, such as operation with stable substrates without requirement of labile cofactors or a pH optimum at 5.7 with steep decline of enzyme activity caused by higher values (49).
It is tempting to speculate on the as yet unknown physiological role of SlCGT and its main product caffeoylglucarate in the apoplast. This subcellular compartment provides not only a physical barrier against pathogen attack but also plays an important role in defense against plant pathogens through the presence of extracellular pathogenesis-related proteins (50, 51) and cell wall-located phenolics (52). Moreover, GDSL proteins were described as cell wall-secreted lipases, like a secreted enzyme from A. thaliana (47, 53), shown to disrupt fungal spore integrity and to inhibit spore germination. Given a continuous supply with chlorogenate and glucarate, the presence of the SlCGT in the apoplastic space of leaves should lead to the extracellular accumulation of caffeoylglucarate. Whether caffeoylglucarate alone or in combination with chlorogenate acts as the bioactive agent in defense reactions against herbivores (5, 54, 55), bacteria and fungi (56–58), or against viruses (59) remains elusive. Biological activity of these caffeate esters has been related to their assumed prooxidant effects through quinone formation.
It is worth paying special attention to the fact that the SlCGT has obviously been recruited from the GDSL lipase family to function as a highly specific acyltransferase in the secondary phenylpropanoid metabolism of tomato. According to recent findings, members of the highly diverse GDSL lipase enzyme family might be involved not only in lipid metabolism but in various other functions in primary and secondary metabolism of plants (42). Most of these enzymes are usually characterized by hydrolytic activities toward synthetic substrates, whereas the functions in planta remain elusive. Because GDSL lipases have a flexible substrate binding pocket that enables these enzymes to bind different substrates in conformations that are optimal for their catalytic activities (42), it is difficult to arrive at a conclusion regarding their functions in planta from in vitro assays. So far there are only few examples of GDSL lipases for which the endogenous substrates in plant secondary metabolism are known. Among these are a specific acetylajmalan esterase from R. serpentina (36), a sinapine esterase from Brassicaceae (17) and the regiospecific CGT in tomato leaves, described herein, as the most prominent examples. Whereas the first two enzymes retained hydrolytic activities, SlCGT turned out to be the first known GDSL lipase-like protein that was evolutionary driven to lose lipolytic activity and to take over the role of an acyltransferase in plant phenylpropanoid metabolism. It will be a formidable task to elucidate the structure-function relationship of this novel enzyme and determine the kinetic mechanism of the acyl transfer reaction. It will also be of utmost interest to approach the function of the apoplastic-located caffeoylglucarate synthesis in biotic interactions.
Supplementary Material
This work was supported by the Deutsche Forschungsgemeinschaft (D. S. and B. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) FR667689.
- HCA
- hydroxycinnamate
- CGT
- chlorogenate:glucarate caffeoyltransferase
- RACE
- rapid amplification of cDNA ends
- SlCGT
- Solanum lycopersicum CGT.
REFERENCES
- 1.Barz W., Köster J., Weltring K.-M., Strack D. (1985) in The Biochemistry of Plant Phenolics (Van Sumere C., Lea P. eds) pp. 307–347, Clarendon Press, Oxford [Google Scholar]
- 2.Strack D., Mock H. (1993) in Enzymes in Secondary Metabolism (Dey P., Harborne J. eds) pp. 45–97, Academic Press, London [Google Scholar]
- 3.Baumert A., Milkowski C., Schmidt J., Nimtz M., Wray V., Strack D. (2005) Phytochemistry 66, 1334–1345 [DOI] [PubMed] [Google Scholar]
- 4.Aronoff S., Perkins H. J. (1956) Arch. Biochem. Biophys. 64, 506–507 [DOI] [PubMed] [Google Scholar]
- 5.Elliger C., Lundin R., Haddon W. (1981) Phytochemistry 20, 1133–1134 [Google Scholar]
- 6.Strack D., Gross W., Wray V., Grotjahn L. (1987) Plant Physiol. 83, 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strack D., Gross W. (1990) Plant Physiol. 92, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross G. G., Zenk M. H. (1974) Eur. J. Biochem. 42, 453–459 [DOI] [PubMed] [Google Scholar]
- 9.Rhodes M., Wooltorton L. (1973) Phytochemistry 12, 2381–2387 [Google Scholar]
- 10.Stöckigt J., Zenk M. H. (1974) FEBS Lett. 42, 131–134 [DOI] [PubMed] [Google Scholar]
- 11.Ulbrich B., Zenk M. (1979) Phytochemistry 18, 929–933 [Google Scholar]
- 12.Steffens J. (2000) Plant Cell Environ. 12, 1253–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strack D., Keller H., Weissenböck G. (1987) J. Plant Physiol. 131, 61–73 [Google Scholar]
- 14.Strack D., Gross W., Heilemann J., Keller H., Ohm S. (1988) Z. Naturforsch. 43, 32–36 [Google Scholar]
- 15.Maas M., Petereit F., Hensel A. (2009) Molecules 14, 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villegas R., Shimokawa T., Okuyama H., Kojima M. (1987) Phytochemistry 26, 1577–1581 [Google Scholar]
- 17.Clauss K., Baumert A., Nimtz M., Milkowski C., Strack D. (2008) Plant J. 53, 802–813 [DOI] [PubMed] [Google Scholar]
- 18.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19.Ruiz C., Falcocchio S., Xoxi E., Pastor F. I., Diaz P., Saso L. (2004) Biochim. Biophys. Acta 1672, 184–191 [DOI] [PubMed] [Google Scholar]
- 20.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003) Neurosci. Lett. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- 23.Outchkourov N. S., Peters J., de Jong J., Rademakers W., Jongsma M. A. (2003) Planta 216, 1003–1012 [DOI] [PubMed] [Google Scholar]
- 24.McBride K. E., Summerfelt K. R. (1990) Plant Mol. Biol. 14, 269–276 [DOI] [PubMed] [Google Scholar]
- 25.Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z. B., Ulmasov T., Shi X., Hagen G., Guilfoyle T. J. (1994) Plant Cell 6, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper T. (1981) Biochemische Arbeitsmethoden, Walter de Gruyter Verlag, Berlin [Google Scholar]
- 28.Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Hause B., Stenzel I., Miersch O., Maucher H., Kramell R., Ziegler J., Wasternack C. (2000) Plant J. 24, 113–126 [DOI] [PubMed] [Google Scholar]
- 31.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 32.Kapila J., De Rycke R., Van Montagu M., Angenon G. (1997) Plant Sci. 122, 101–108 [Google Scholar]
- 33.Brick D. J., Brumlik M. J., Buckley J. T., Cao J. X., Davies P. C., Misra S., Tranbarger T. J., Upton C. (1995) FEBS Lett. 377, 475–480 [DOI] [PubMed] [Google Scholar]
- 34.Cummins I., Edwards R. (2004) Plant J. 39, 894–904 [DOI] [PubMed] [Google Scholar]
- 35.Ling H., Zhao J., Zuo K., Qiu C., Yao H., Qin J., Sun X., Tang K. (2006) J. Biochem. Mol. Biol. 39, 297–303 [DOI] [PubMed] [Google Scholar]
- 36.Ruppert M., Woll J., Giritch A., Genady E., Ma X., Stöckigt J. (2005) Planta 222, 888–898 [DOI] [PubMed] [Google Scholar]
- 37.Teissère M., Borel M., Caillol B., Nari J., Gardies A. M., Noat G. (1995) Biochim. Biophys. Acta 1255, 105–112 [DOI] [PubMed] [Google Scholar]
- 38.Milkowski C., Strack D. (2004) Phytochemistry 65, 517–524 [DOI] [PubMed] [Google Scholar]
- 39.Milkowski C., Strack D. (2010) Planta 232, 19–35 [DOI] [PubMed] [Google Scholar]
- 40.Stehle F., Brandt W., Milkowski C., Strack D. (2006) FEBS Lett. 580, 6366–6374 [DOI] [PubMed] [Google Scholar]
- 41.Stehle F., Brandt W., Schmidt J., Milkowski C., Strack D. (2008) Phytochemistry 69, 1826–1831 [DOI] [PubMed] [Google Scholar]
- 42.Akoh C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. (2004) Progr. Lipid Res. 43, 534–552 [DOI] [PubMed] [Google Scholar]
- 43.Reina J. J., Guerrero C., Heredia A. (2007) J. Exp. Bot. 58, 2717–2731 [DOI] [PubMed] [Google Scholar]
- 44.Dalrymple B. P., Cybinski D. H., Layton I., McSweeney C. S., Xue G. P., Swadling Y. J., Lowry J. B. (1997) Microbiology 143, 2605–2614 [DOI] [PubMed] [Google Scholar]
- 45.Lo M., Taylor C., Wang L., Nowack L., Wang T. W., Thompson J. (2004) Plant Physiol. 135, 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beisson F., Gardies A., Teissere M., Ferte N., Noat G. (1997) Plant Physiol. Biochem. 35, 761–765 [Google Scholar]
- 47.Oh I. S., Park A. R., Bae M. S., Kwon S. J., Kim Y. S., Lee J. E., Kang N. Y., Lee S., Cheong H., Park O. K. (2005) Plant Cell 17, 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujiwara H., Tanaka Y., Yonekura-Sakakibara K., Fukuchi-Mizutani M., Nakao M., Fukui Y., Yamaguchi M., Ashikari T., Kusumi T. (1998) Plant J. 16, 421–431 [DOI] [PubMed] [Google Scholar]
- 49.Fry F. (1995) Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 497–520 [Google Scholar]
- 50.Hückelhoven R. (2007) Annu. Rev. Phytopathol. 45, 101–127 [DOI] [PubMed] [Google Scholar]
- 51.Floerl S., Druebert C., Majcherczyk A., Karlovsky P., Kües U., Polle A. (2008) BMC Plant Biol. 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franke R., Fry F., Kauss H. (1998) Plant Cell Rep. 17, 379–383 [DOI] [PubMed] [Google Scholar]
- 53.Lee L. C., Lee Y. L., Leu R. J., Shaw J. F. (2006) Biochem. J. 397, 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leiss K. A., Maltese F., Choi Y. H., Verpoorte R., Klinkhamer P. G. (2009) Plant Physiol. 150, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennett R., Wallsgrove R. (1994) New Phytol. 127, 617–633 [DOI] [PubMed] [Google Scholar]
- 56.Lizzi Y., Roggero J., Coulomb P. (1995) J. Phytopathol. 143, 619–627 [Google Scholar]
- 57.Lyons P., Wood K., Nicholson R. (1990) Phytochemistry 29, 97–101 [Google Scholar]
- 58.Ravn H., Brimer L. (1988) Phytochemistry 27, 3433–3437 [Google Scholar]
- 59.Cheminat A., Zawatzky R., Becker H., Brouillard R. (1988) Phytochemistry 27, 2787–2794 [Google Scholar]
- 60.Sasaki T., Matsumoto T., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Antonio B. A., Kanamori H., Hosokawa S., Masukawa M., Arikawa K., Chiden Y., Hayashi M., Okamoto M., Ando T., Aoki H., Arita K., Hamada M., Harada C., Hijishita S., Honda M., Ichikawa Y., Idonuma A., Iijima M., Ikeda M., Ikeno M., Ito S., Ito T., Ito Y., Ito Y., Iwabuchi A., Kamiya K., Karasawa W., Katagiri S., Kikuta A., Kobayashi N., Kono I., Machita K., Maehara T., Mizuno H., Mizubayashi T., Mukai Y., Nagasaki H., Nakashima M., Nakama Y., Nakamichi Y., Nakamura M., Namiki N., Negishi M., Ohta I., Ono N., Saji S., Sakai K., Shibata M., Shimokawa T., Shomura A., Song J., Takazaki Y., Terasawa K., Tsuji K., Waki K., Yamagata H., Yamane H., Yoshiki S., Yoshihara R., Yukawa K., Zhong H., Iwama H., Endo T., Ito H., Hahn J. H., Kim H. I., Eun M. Y., Yano M., Jiang J., Gojobori T. (2002) Nature 420, 312–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.