Abstract
Polycystin-1 (PC1) is a large membrane protein that is expressed along the renal tubule and exposed to a wide range of concentrations of urea. Urea is known as a common denaturing osmolyte that affects protein function by destabilizing their structure. However, it is known that the native conformation of proteins can be stabilized by protecting osmolytes that are found in the mammalian kidney. PC1 has an unusually long ectodomain with a multimodular structure including 16 Ig-like polycystic kidney disease (PKD) domains. Here, we used single-molecule force spectroscopy to study directly the effects of several naturally occurring osmolytes on the mechanical properties of PKD domains. This experimental approach more closely mimics the conditions found in vivo. We show that upon increasing the concentration of urea there is a remarkable decrease in the mechanical stability of human PKD domains. We found that protecting osmolytes such as sorbitol and trimethylamine N-oxide can counteract the denaturing effect of urea. Moreover, we found that the refolding rate of a structurally homologous archaeal PKD domain is significantly slowed down in urea, and this effect was counteracted by sorbitol. Our results demonstrate that naturally occurring osmolytes can have profound effects on the mechanical unfolding and refolding pathways of PKD domains. Based on these findings, we hypothesize that osmolytes such as urea or sorbitol may modulate PC1 mechanical properties and may lead to changes in the activation of the associated polycystin-2 channel or other intracellular events mediated by PC1.
Keywords: Protein Denaturation, Protein Folding, Protein Stability, Protein Structure, Single Molecule Biophysics, Urea, Atomic Force Microscopy, Force Spectroscopy, Mechanical Properties, Osmolytes
Introduction
Polycystin-1 (PC1)2 is a large transmembrane protein, which, when mutated, cause autosomal dominant polycystic kidney disease (PKD), one of the most common life-threatening genetic diseases, which is a leading cause of kidney failure (1). The available evidence suggests that PC1 acts as a mechanosensor, receiving signals from the primary (luminal) cilia, neighboring cells, and extracellular matrix and transduces them into cellular responses that regulate proliferation, adhesion, and differentiation that are essential for the control of renal tubules and kidney morphogenesis (1–4).
PC1 is expressed along the renal tubule, where it is exposed to a wide range of concentrations of urea, from 5 mm in the proximal tubule to up to ∼1 m in the collecting duct (5–7). Urea is known as a common denaturant that affects protein function by perturbing their structure (8). Several osmolytes have been found in the mammalian kidney that are known to counteract the effects of urea on proteins (9–15). Among epithelial cells of the kidney medulla, the principal organic osmolytes include the polyols sorbitol and inositol, the methylamines betaine and glycerophosphocholine, and the amino acid taurine. Protecting osmolytes are found in all taxa (11, 14). For example, cartilaginous fish concentrate trimethylamine N-oxide (TMAO), to offset the damaging effects of urea on protein function, and it is one of the best studied protecting osmolytes (e.g. 16–20).
The mechanisms by which protecting osmolytes promote protein folding, increase protein stability, and induce conformational changes have been the focus of intense investigation (15, 18, 19, 21–33). However, nothing is known about the effects of denaturing or protecting osmolytes on the mechanical properties of PKD domains. Here, we investigated the effects of several naturally occurring osmolytes on the mechanical properties of PKD domains using single-molecule force spectroscopy. This experimental approach more closely mimics the conditions found in vivo. In addition, single-molecule AFM experiments have provided novel and valuable insights into how osmolytes affect protein unfolding/folding dynamics (34, 35).
We found that upon increasing the concentration of urea there is a remarkable decrease in the mechanical stability of human PKD domains. Protecting osmolytes, such as sorbitol and TMAO, can effectively counteract the effect of urea. We also found that the mechanical refolding rate constant of a structurally homologous archaeal PKD domain is slowed by urea, an effect that can be reversed by adding sorbitol and TMAO. Our results suggest that the mechanical function of PC1 may be modulated by the interplay between denaturing and protecting osmolytes.
EXPERIMENTAL PROCEDURES
Cloning and Expression of I27, Human PKD, and Archaea PKD Polyproteins for AFM Experiments
We cloned and expressed in bacteria a heteropolyprotein based on the first PKD domain from human PC1 (PKDd1, residues Val268–Glu354) and titin immunoglobulin domain 27 (I27). The I27 domain has been studied extensively by force spectroscopy and hence serves as an internal fingerprint (36). We assembled a polyI27-PKDd1 protein by using a multiple-step cloning technique (37–39). This protein has four copies of I27 and three copies of PKDd1 (I27-PKDd1)3-I27. A polyI27 polyprotein containing eight identical copies was constructed as described previously (36). The gene of the multimer containing seven copies of an Archaea PKD domain (ArPKD) was kindly supplied by Jane Clarke (University of Cambridge, UK). All three proteins, (I27)8, (I27-PKDd1)3-I27, and (ArPKD)7 were expressed in Escherichia coli C41 strain and purified by Ni-affinity chromatography as described previously (36, 38–40). The proteins were kept in PBS containing 5 mm DTT at 4 °C.
Single-molecule Force Spectroscopy
The mechanical properties of single proteins were studied using a home-built single molecule AFM as described previously (36, 41, 42). The spring constant of each individual cantilever (MLCT or Olympus OBL; Veeco Metrology Group, Santa Barbara, CA) was calculated using the equipartition theorem (43). In a typical experiment, a small aliquot of the purified proteins (∼1–10 μl, 10–100 μg/ml) was allowed to adsorb onto a nickel-nitrilotriacetic acid-coated glass coverslip (44, 45) and then rinsed with PBS alone or containing various concentrations of organic osmolytes. In a typical experiment, we first obtained several sawtooth patterns in PBS before switching to buffers containing osmolytes. We found that the effect of urea or urea plus protecting osmolytes was instantaneous. We collected force-extension curves for about 15 min before switching back to PBS. Proteins were picked up randomly by the cantilever tip and then stretched for several hundred nm. We found that the pick-up efficiency was typically much lower in the presence of osmolytes, making some of the experiments quite challenging. The pulling speed was in the range of 0.5–0.7 nm/ms.
RESULTS
Mechanical Stability of PKD Domains Is Remarkably Sensitive to the Urea Concentration
To study the effect of osmolytes on the mechanical stability of PKD domains, we used the first PKD domain from human PC1, PKDd1, because its structure is known (46) and its thermodynamic and mechanical stabilities have been characterized (38–40). We used a heteropolyprotein approach to study the mechanical properties of PKDd1 domains using AFM (38, 39). In these constructs we used the titin domain I27 as an internal mechanical fingerprint because it has been extensively studied with AFM techniques (36).
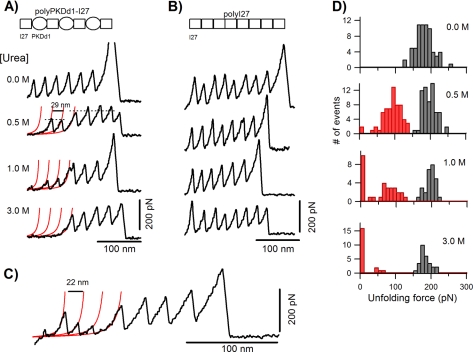
Fig. 1A shows the effects of increasing urea concentration on the mechanical stability of PKD and I27 domains in the polyPKDd1-I27 protein. At zero urea, both domains unfold at similar forces of about 200 pN (38–40). Increasing the urea concentration has a striking effect on the unfolding forces of PKDd1 but a relatively small effect on I27 unfolding forces. For example, the unfolding pattern under 0.5 m urea shows a total of six force peaks, four at ∼200 pN and two at ∼100 pN; given the construction of the polyprotein we attribute the high force peaks to the unfolding of I27 domains. This recording also shows that one of the PKDd1 domains in this recording is “missing.” The red lines correspond to fits to the worm-like chain equation (47, 48) using an increase in contour length of 29 nm. We interpret the spacer before the sawtooth pattern as a PKDd1 domain that is already unfolded before stretching or that unfolds at forces that are below our detection limit (∼20 pN). At 3 m urea all PKDd1 domains are unfolded as evidenced by the long spacer before the unfolding of the I27 domains. As a control, we studied the effect of urea on a polyprotein made of I27 domains (polyI27 protein, Fig. 1B). The data show that urea has a very small effect on the mechanical stability of I27 domains; at 3 m urea the unfolding forces decrease by only 10% (from ∼200 to ∼180 pN).
FIGURE 1.
Increasing the urea concentration has a striking effect on the unfolding forces of PKDd1 but a relatively small effect on I27 unfolding forces. A, force-extension curves of polyPKDd1-I27 proteins under increasing concentrations of urea (from 0 to 3 m) showing the unfolding patterns of PKD and I27 domains. At 0 m urea both domains unfold at similar forces of about 190 pN (188 ± 25 pN, n = 67), whereas at 3 m PKD domains are all denatured with little effect on I27 domains. The red lines correspond to fits to the worm-like chain equation. As shown some PKD domains are missing from the unfolding pattern obtained under urea. B, force-extension curves of polyI27 proteins under the same range of urea concentrations. C, example of a force-extension curve of a polyPKDd1-I27 protein in 1 m urea showing partially folded PKD domains. The increase in contour length upon PKD domain unfolding is 22 nm instead of 29 nm. D, unfolding force histograms for the polyPKDd1-I27 protein at 0.0, 0.5, 1.0, and 3.0 m. The missing unfolding events are counted as points below the noise level (∼20 pN) in the histograms. The unfolding events for I27 and PKD domains in the polyPKDd1-I27 protein are shown as gray and red bars, respectively. The average unfolding forces for I27 and PKDd1 domains are: 195 ± 19 pN (n = 64) and 103 ± 28 pN (n = 63) in 0.5 m, 187 ± 22 pN (n = 27) and 78 ± 42 pN (n = 31) in 1 m, 178 ± 18 pN (n = 24) and <20 pN (n = 19) in 3 m urea.
We also observed other types of unfolding patterns after stretching the polyPKDd1-I27 protein in urea. Fig. 1C shows an example obtained in 1 m urea where the spacing between the PKD unfolding events is 22 nm instead of 29 nm. We observed this type of event in about 15% of the recordings. An increase in contour length of 22 nm corresponds to the unfolding of about 70 amino acids, suggesting that these domains were partially unfolded under these conditions. This shows that urea has a strong effect on the unfolding pathways of PKDd1 domains.
Figs. 1D and 2 show an analysis of the effect of urea on the unfolding forces of the polyPKDd1-I27 protein. Unfolding force histograms obtained at 0, 0.5, 1, and 3 m urea are shown in Fig. 1D. At 0.5 m there is a significant shift of the unfolding forces for PKD domains to about 100 pN (red bars, 103 ± 28 pN, n = 63) but not for I27 domains (gray bars, 195 ± 19 pN, n = 64). We found that the fraction of missing PKD domains increases as a function of the urea concentration (8% in 0.5 m, 32% in 1 m, and 76% in 3 m). Fig. 2 shows a plot that compares the effects of urea on the unfolding forces of PKDd1 domains (red circles) and I27 domains (black and open squares). There is a linear relationship between the mechanical stability and urea concentration for both domains. However, urea has a much stronger effect on the mechanical stability of PKDd1 than I27 domains. The slopes in Fig. 2 are 75 pN/m for PKDd1 and 7 pN/m for I27 domains. These data demonstrate that the mechanical stability of PKDd1 domains is very sensitive to urea. Our results suggest that physiological levels of urea may have a deleterious effect on the mechanical strength of PKD domains.
FIGURE 2.
Comparison of the effects of urea on the mechanical stability of I27 and PKD domains. A plot of the unfolding forces for PKD (red circles) and I27 (black squares) domains in the polyPKDd1-I27 protein as a function of the urea concentration is shown. The open squares correspond to the unfolding forces of I27 in the polyI27 protein. The lines are linear fits to the experimental data. The slopes are 7 pN/m for I27 and 75 pN/m for PKD domains.
Effects of Protecting Osmolytes on the Mechanical Stability of Urea-weakened PKDd1 Domains
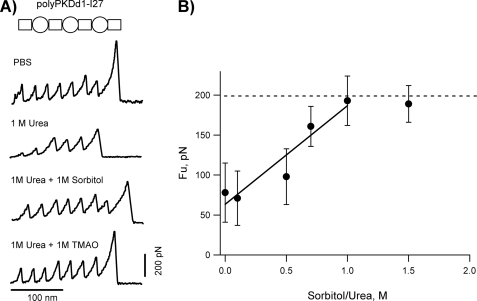
As discussed above, several naturally occurring osmolytes have been shown to counteract the perturbing effects of urea on proteins. Based on these observations we hypothesized that osmolytes such as sorbitol may offset urea effects on PKDd1 domains. Fig. 3A shows representative force-extension curves obtained in different combination of denaturing and protecting osmolytes. The trace obtained in 1 m urea shows two PKDd1 domains that unfold at ∼35 and 80 pN and three I27 domains that unfold all about ∼200 pN. In the presence of a 1:1 mixture of urea plus sorbitol, all PKDd1 domains unfold at forces identical to I27 domains (∼200 pN). A similar effect was observed when sorbitol was substituted by TMAO (Fig. 3A, bottom recording). In the absence of urea these osmolytes did not affect the unfolding forces of PKDd1 or I27 domains at concentrations as high as 2 m.
FIGURE 3.
Effect of protecting osmolytes on the mechanical stability of urea-weakened PKDd1 domains. A, typical force-extension curves for the polyPKDd1-I27 protein obtained in PBS, 1 m urea, 1 m urea + 1 m sorbitol, and 1 m urea + 1 m TMAO. B, plot of the unfolding forces for PKD domains as a function of the sorbitol:urea ratio (at a fixed 1 m urea). The average unfolding forces of PKDd1 are: 78 ± 37 pN (n = 31) at 0 m, 71 ± 42 pN (n = 16) at 0.1 m, 98 ± 27 pN (n = 19) at 0.5 m, 161 ± 25 pN (n = 11) at 0.7 m, 193 ± 22 pN (n = 18) at 1 m, 189 ± 19 pN (n = 6) at 1.5 m. The line is a fit to the experimental data obtained below 1 m sorbitol. The slope is 123 pN/m sorbitol.
To quantify the effects of osmolytes, we carried out unfolding experiments of the polyPKDd1-I27 protein at different molar ratios of sorbitol and urea. Fig. 3B shows a plot of the unfolding forces for PKDd1 domains as a function of the ratio of sorbitol:urea, at a fixed urea concentration (1 m). Upon increasing sorbitol concentration there is a clear increase in the mechanical stability of PKDd1, from ∼80 pN at 0 m to ∼190 pN at 1 m sorbitol. Hence, these experiments provide direct evidence a polyol (sorbitol) and a methylamine (TMAO) can effectively counteract the urea weakening effect on the mechanical stability of PKDd1 domains.
Effects of Osmolytes on the Mechanical Refolding Rates Constant of PKD Domains
We have previously shown that PKD domains of the PC1 ectodomain refold very slowly with a mechanical folding rate of about 0.09 s−1 (39) (>10× slower than I27 domains). Similar experiments with the polyPKDd1-I27 protein revealed that the refolding of unfolded PKDd1 domains is not complete even after a waiting time of 20 s (supplemental Fig. S1), making it very difficult to study the effect of osmolytes on PKDd1 folding rate (this is mainly because of mechanical drift of the AFM). For this reason, a different PKD domain was used. The PKD domain from Methanosarcina archaeobacteria (Protein Data Bank code 1L0Q, termed ArPKD) has a structure very similar to human PKDd1 (the two structures superimpose with an root mean square deviation of 2.2 Å) (49). Interestingly, as recently demonstrated by Forman et al., both ArPKD and PKDd1 domains have a similar mechanical stability (they unfold at ∼200 pN; supplemental Fig. S2, A and B) (50). In addition, we found that naturally occurring missense mutations alter the thermodynamic stability of ArPKD and the mechanical stability of PKDd1 to a similar extent (38). ArPKD therefore presents itself as a good model system to study the effects of denaturing and protecting osmolytes on the folding kinetics of PKD domains. In these experiments we used a polyprotein that has seven identical repeats of the ArPKD domain. As shown in supplemental Fig. S2C, urea also has a clear weakening effect on ArPKD domains.
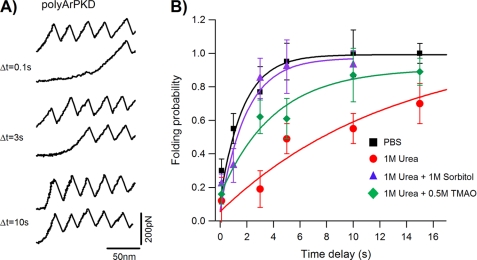
We used a standard double-pulse protocol (41, 51) to measure the folding kinetics of ArPKD domains. We found that ArPKD domains readily refold after complete unfolding in PBS (Fig. 4A). It was observed that the fraction of refolded ArPKD domains depended exponentially on the amount of time that the protein remained relaxed (Fig. 4B, black squares). For example, the data show that the folding probability is about 25% after waiting 100 ms and 100% after waiting 10 s (Fig. 4B, black squares). In 1 m urea, the folding probability measured after waiting 10s is only about 50% (Fig. 4B, red circles).
FIGURE 4.
Effects of different combinations of osmolytes on the refolding rate of ArPKD domains. A, measuring the refolding rate for ArPKD domains using a two-pulse stretching/relaxation protocol. Three examples obtained with time delays of 0.1, 3, and 10 s are shown. B, plot of the folding probability of as a function of the time delay between stretching pulses obtained in PBS (black squares), 1 m urea (red circles), 1 m urea + 1 m sorbitol (purple triangles), and 1 m urea + 0.5 m TMAO (green diamonds). The lines correspond to fits of the data to the function Pf(t) = 1 − exp (−βot), using the following folding rate constants: βo, 0.58 ± 0.16 s −1 in PBS, 0.07 ± 0.04 s−1 in 1 m urea, 0.52 ± 0.19 s−1 in 1 m urea + 1 m sorbitol, and 0.24 ± 0.07 s−1 in 1 m urea + 0.5 m TMAO.
For a simple two-state model, the folding probability is given by Pf(t) = 1 − exp (−βot), where βo is the folding rate constant of ArPKD at zero force. Fits of the data to this function measure the refolding rate constants of ArPKD in PBS and in osmolytes. The refolding rate constant in PBS is 0.58 ± 0.16 s−1. By contrast, this value decreases by about 8-fold in 1 m urea (0.07 ± 0.04 s−1). In a 1 m urea/1 m sorbitol mixture (Fig. 4B, purple triangles) the folding rate is very similar to that in PBS (0.52 ± 0.19 s−1). A similar folding rate is obtained when sorbitol is replaced with 1 m TMAO (0.54 ± 0.23 s−1; data not shown), but it was slower in a 2:1 molar ratio of urea and TMAO (0.24 ± 0.07 s−1; Fig. 4B, green diamonds). Our results clearly show that 1 m urea slows down the refolding rate of ArPKD domains, and this effect is effectively counteracted by the protecting osmolytes sorbitol and TMAO.
DISCUSSION
Osmolytes are a class of small organic molecules found in all taxa (11, 14) that can profoundly affect the stability and function of proteins (15). Here, we discovered that naturally occurring denaturing and protecting osmolytes such as urea and sorbitol have remarkable effects on the mechanical stability and folding kinetics of PKD domains.
How can we explain the effects of osmolytes on the mechanical properties of PKD domains? The denaturing properties of urea have been studied extensively (52–55). The evidence suggests that urea denaturation likely occurs through two possible pathways (56). Urea can act “indirectly” by altering the water structure and orienting the distribution of water molecules and perturbing hydrophobic interactions (57–59). It can also interact “directly” through hydrogen bonding with both the peptide backbone and exposed side chains. This results in a negative free-energy contribution that forces protein to unfold (22, 60, 61). Recent data suggest that urea-induced denaturation may occur through a combination of both indirect and direct actions (19). Molecular dynamics simulations suggest that the mechanical resistance of human and archaea PKD domains mainly comes from a few hydrogen bonds between the A–A′ loop and the G-strand (50). We propose a mechanism where urea has a direct action on the mechanical stability of PKD domains by weakening these force-bearing hydrogen bonds. A similar mechanism was proposed for the effect of guanidinium chloride on the mechanical stability of the B1 immunoglobulin-binding domain of protein G (35).
Our results show that protecting osmolytes such as sorbitol and TMAO are very efficient in counteracting the effect of urea on the mechanical stability of PKD domains. Indeed, it has been shown that the stabilizing effects of methylamines and denaturing effects of urea on protein stability and function are additive (14, 16, 17, 27, 62–64). As shown by Bolen's group, the protein backbone is the key determinant for the stabilization or denaturation by osmolytes (17, 22, 25). According to this model, the favorable interactions of urea with the backbone provide the main driving force for protein unfolding, and the unfavorable interaction of TMAO with backbone is the main force opposing urea denaturation. This model is consistent with our findings. For example, we found that 1 m urea slows the refolding rate of ArPKD domains and this effect is counteracted by 1 m TMAO or 1 m sorbitol. After the domains are completely unfolded by force, urea accumulates preferentially with the protein backbone stabilizing the unfolded state and hence slowing down the refolding rate. In 1 m TMAO, unfolding is opposed by a large unfavorable interaction with the backbone, and this is the dominant force opposing the effect of urea on the refolding rate.
PC1 is a membrane protein found in kidney tubule epithelial cells, and it localizes to the cell-cell contacts such as adhesions junctions and focal densities at the basal membrane in contact with the extracellular matrix and also in the primary cilium (2–4). It is likely that PC1 must be under mechanical tension and that PKD domains are likely to deform or partially unfold during these mechanical interactions (39). Because urea is permeable to cell membranes, it will affect the stability of PC1 expressed in all of these subcellular locations. Here, we made the unexpected finding that the mechanical stability of the PKDd1 domain is strongly dependent on urea in the physiological range (<1 m). The unfolding forces rapidly decrease as a function of urea (about 75 pN/m increase in urea concentration). PKD domains comprise about 50% of the structure of the PC1 ectodomain (65, 66). PKD domains display a wide range of mechanical stabilities requiring ∼50 pN of force to unfold the weakest domains and ∼200 pN for the most stable domains (39). We speculate that urea may alter the mechanical properties of the PC1 ectodomain and hence may affect its mechanical sensing and cell adhesion properties. Based on our findings, we hypothesize that naturally occurring osmolytes such as urea or sorbitol may modulate PC1 function and may lead to changes in the activation of the associated polycystin-2 channel or other intracellular events mediated by PC1 (e.g. JAK/STAT pathway) (67).
Protecting osmolytes such as sorbitol, inositol, and glycerophosphorylcholine are found in the mammalian kidney at concentrations as high as 400 mm (12, 68–72). It has been reported that the distribution of sorbitol increases from the cortex to the papillary tip in mammalian kidneys (68, 69). This is consistent with an increasing pattern of urea from the proximal tubule to the collecting duct. Moreover, a similar distribution pattern was observed for other osmolytes such as betaine and glycerophosphorylcholine (12, 68). Hence, PC1 proteins are likely to be exposed to many different types of osmolytes. We are currently studying the effects of mixtures of protecting osmolytes on the mechanical stability of PKD domains weakened by urea and determining whether their actions on PKD domain stability are independent or synergistic.
Supplementary Material
Acknowledgments
We thank Wayne Bolen for valuable discussions on osmolytes and Drs. Bryan Sutton and Matthew Auton for comments on this work.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK073394. This work was also supported by the John Sealy Memorial Endowment Fund for Biomedical Research and by the Polycystic Kidney Foundation Grant 116a2r.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PC1
- polycystin-1
- AFM
- atomic force microscopy
- ArPKD
- Archea PKD
- I27
- titin immunoglobulin domain 27
- PKD
- polycystic kidney disease
- pN
- piconewtons
- TMAO
- trimethylamine N-oxide.
REFERENCES
- 1.Harris P. C., Torres V. E. (2009) Annu. Rev. Med. 60, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibraghimov-Beskrovnaya O., Bukanov N. (2008) Cell. Mol. Life Sci. 65, 605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., Zhou J. (2003) Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 4.Streets A. J., Newby L. J., O'Hare M. J., Bukanov N. O., Ibraghimov-Beskrovnaya O., Ong A. C. (2003) J. Am. Soc. Nephrol. 14, 1804–1815 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt-Nielsen B. (1979) Yale J. Biol. Med. 52, 545–561 [PMC free article] [PubMed] [Google Scholar]
- 6.Goyton A. C., Hall J. E. (1997) Human Physiology and Mechanisms of Disease, W.B. Saunders Co., Philadelphia, PA [Google Scholar]
- 7.MacMillen R. E., Lee A. K. (1967) Science 158, 383–385 [DOI] [PubMed] [Google Scholar]
- 8.Pace C. N. (1975) CRC Crit. Rev. Biochem. 3, 1–43 [DOI] [PubMed] [Google Scholar]
- 9.Sizeland P. C., Chambers S. T., Lever M., Bason L. M., Robson R. A. (1993) Kidney Int. 43, 448–453 [DOI] [PubMed] [Google Scholar]
- 10.Bagnasco S., Balaban R., Fales H. M., Yang Y. M., Burg M. (1986) J. Biol. Chem. 261, 5872–5877 [PubMed] [Google Scholar]
- 11.Burg M. B., Ferraris J. D. (2008) J. Biol. Chem. 283, 7309–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Perez A., Burg M. B. (1991) Physiol. Rev. 71, 1081–1115 [DOI] [PubMed] [Google Scholar]
- 13.Yancey P. H. (2005) J. Exp. Biol. 208, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 14.Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982) Science 217, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 15.Bolen D. W. (2001) Methods Mol. Biol. 168, 17–36 [DOI] [PubMed] [Google Scholar]
- 16.Lin T. Y., Timasheff S. N. (1994) Biochemistry 33, 12695–12701 [DOI] [PubMed] [Google Scholar]
- 17.Wang A., Bolen D. W. (1997) Biochemistry 36, 9101–9108 [DOI] [PubMed] [Google Scholar]
- 18.Zou Q., Bennion B. J., Daggett V., Murphy K. P. (2002) J. Am. Chem. Soc. 124, 1192–1202 [DOI] [PubMed] [Google Scholar]
- 19.Beck D. A. C., Bennion B. J., Alonso D. O. V., Daggett V., Dieter H., Helmut S. (2007) in Methods in Enzymology, Vol. 428, pp. 373–396, Academic Press, Orlando, FL: [DOI] [PubMed] [Google Scholar]
- 20.Baskakov I., Wang A., Bolen D. W. (1998) Biophys. J. 74, 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo T. W., Clarke D. M. (2007) Expert Rev. Mol. Med. 9, 1–18 [DOI] [PubMed] [Google Scholar]
- 22.Auton M., Bolen D. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15065–15068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baskakov I., Bolen D. W. (1998) J. Biol. Chem. 273, 4831–4834 [DOI] [PubMed] [Google Scholar]
- 24.Kumar R., Baskakov I. V., Srinivasan G., Bolen D. W., Lee J. C., Thompson E. B. (1999) J. Biol. Chem. 274, 24737–24741 [DOI] [PubMed] [Google Scholar]
- 25.Street T. O., Bolen D. W., Rose G. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13997–14002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Street T. O., Krukenberg K. A., Rosgen J., Bolen D. W., Agard D. A. (2010) Protein Sci. 19, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesu P., Lee M. J., Lin H. M. (2009) J. Phys. Chem. B 113, 5327–5338 [DOI] [PubMed] [Google Scholar]
- 28.Ignatova Z., Gierasch L. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13357–13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratnaparkhi G. S., Varadarajan R. (2001) J. Biol. Chem. 276, 28789–28798 [DOI] [PubMed] [Google Scholar]
- 30.Fischer H., Fukuda N., Barbry P., Illek B., Sartori C., Matthay M. A. (2001) Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L52–L57 [DOI] [PubMed] [Google Scholar]
- 31.Howard M., Welch W. J. (2002) Methods Mol. Med. 70, 267–275 [DOI] [PubMed] [Google Scholar]
- 32.Holthauzen L. M., Rösgen J., Bolen D. W. (2010) Biochemistry 49, 1310–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns J. N., Orwig S. D., Harris J. L., Watkins J. D., Vollrath D., Lieberman R. L. (2010) ACS Chem. Biol. 5, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Manyes S., Dougan L., Fernández J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10540–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y., Li H. (2008) J. Mol. Biol. 375, 316–324 [DOI] [PubMed] [Google Scholar]
- 36.Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steward A., Toca-Herrera J. L., Clarke J. (2002) Protein Sci. 11, 2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L., Xu M., Forman J. R., Clarke J., Oberhauser A. F. (2009) J. Biol. Chem. 284, 32942–32949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian F., Wei W., Germino G., Oberhauser A. (2005) J. Biol. Chem. 280, 40723–40730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forman J. R., Qamar S., Paci E., Sandford R. N., Clarke J. (2005) J. Mol. Biol. 349, 861–871 [DOI] [PubMed] [Google Scholar]
- 41.Oberhauser A. F., Marszalek P. E., Erickson H. P., Fernandez J. M. (1998) Nature 393, 181–185 [DOI] [PubMed] [Google Scholar]
- 42.Carrion-Vazquez M., Oberhauser A. F., Fisher T. E., Marszalek P. E., Li H., Fernandez J. M. (2000) Prog. Biophys. Mol. Biol. 74, 63–91 [DOI] [PubMed] [Google Scholar]
- 43.Florin E. L., Rief M., Lehmann T., Ludvig M., Dornmair C., Moy V. T., Gaub H. E. (1995) Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 44.Sakaki N., Shimo-Kon R., Adachi K., Itoh H., Furuike S., Muneyuki E., Yoshida M., Kinosita K., Jr. (2005) Biophys. J. 88, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh H., Takahashi A., Adachi K., Noji H., Yasuda R., Yoshida M., Kinosita K. (2004) Nature 427, 465–468 [DOI] [PubMed] [Google Scholar]
- 46.Bycroft M., Bateman A., Clarke J., Hamill S. J., Sandford R., Thomas R. L., Chothia C. (1999) EMBO J. 18, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bustamante C., Marko J. F., Siggia E. D., Smith S. (1994) Science 265, 1599–1600 [DOI] [PubMed] [Google Scholar]
- 48.Marko J. F., Siggia E. D. (1995) Phys. Rev. E 52, 2912–2938 [DOI] [PubMed] [Google Scholar]
- 49.Jing H., Takagi J., Liu J. H., Lindgren S., Zhang R. G., Joachimiak A., Wang J. H., Springer T. A. (2002) Structure 10, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 50.Forman J. R., Yew Z. T., Qamar S., Sandford R. N., Paci E., Clarke J. (2009) Structure 17, 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrion-Vazquez M., Marszalek P. E., Oberhauser A. F., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11288–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schellman J. A. (2002) Biophys. Chem. 96, 91–101 [DOI] [PubMed] [Google Scholar]
- 53.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 54.Tanford C. (1968) Adv. Protein Chem. 23, 121–282 [DOI] [PubMed] [Google Scholar]
- 55.Greene R. F., Jr., Pace C. N. (1974) J. Biol. Chem. 249, 5388–5393 [PubMed] [Google Scholar]
- 56.Caballero-Herrera A., Nordstrand K., Berndt K. D., Nilsson L. (2005) Biophys. J. 89, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennion B. J., Daggett V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5142–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Idrissi A. (2005) Spectrochim. Acta A Mol. Biomol. Spectrosc. 61, 1–17 [DOI] [PubMed] [Google Scholar]
- 59.Daggett V. (2006) Chem. Rev. 106, 1898–1916 [DOI] [PubMed] [Google Scholar]
- 60.Wu J. W., Wang Z. X. (1999) Protein Sci. 8, 2090–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mountain R. D., Thirumalai D. (2003) J. Am. Chem. Soc. 125, 1950–1957 [DOI] [PubMed] [Google Scholar]
- 62.Yancey P. H., Somero G. N. (1979) Biochem. J. 183, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yancey P. H. (2001) Am. Zool. 41, 699–709 [Google Scholar]
- 64.Holthauzen L. M., Bolen D. W. (2007) Protein Sci. 16, 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandford R., Sgotto B., Aparicio S., Brenner S., Vaudin M., Wilson R. K., Chissoe S., Pepin K., Bateman A., Chothia C., Hughes J., Harris P. (1997) Hum. Mol. Genet. 6, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 66.Hughes J., Ward C. J., Peral B., Aspinwall R., Clark K., San Millán J. L., Gamble V., Harris P. C. (1995) Nat. Genet. 10, 151–160 [DOI] [PubMed] [Google Scholar]
- 67.Bhunia A. K., Piontek K., Boletta A., Liu L., Qian F., Xu P. N., Germino F. J., Germino G. G. (2002) Cell 109, 157–168 [DOI] [PubMed] [Google Scholar]
- 68.Schmolke M., Schilling A., Keiditsch E., Guder W. G. (1996) Eur. J. Clin. Chem. Clin. Biochem. 34, 499–501 [PubMed] [Google Scholar]
- 69.Schmolke M., Bornemann A., Guder W. G. (1996) Am. J. Physiol. Renal Physiol. 40, F645–F652 [DOI] [PubMed] [Google Scholar]
- 70.Guder W. G., Beck F. X., Schmolke M. (1990) Klin. Wochenschr. 68, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 71.Schmolke M., Guder W. G. (1989) Renal Physiol. Biochem. 12, 347–358 [DOI] [PubMed] [Google Scholar]
- 72.Wirthensohn G., Lefrank S., Schmolke M., Guder W. G. (1989) Am. J. Physiol. Renal Physiol. 256, F128–F135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.