Abstract
We have previously demonstrated that histone deacetylase 7 (HDAC7) expression and splicing play an important role in smooth muscle cell (SMC) differentiation from embryonic stem (ES) cells, but the molecular mechanisms of increased HDAC7 expression during SMC differentiation are currently unknown. In this study, we found that platelet-derived growth factor-BB (PDGF-BB) induced a 3-fold increase in the transcripts of HDAC7 in differentiating ES cells. Importantly, our data also revealed that PDGF-BB regulated HDAC7 expression not through phosphorylation of HDAC7 but through transcriptional activation. By dissecting its promoters with progressive deletion analysis, we identified the sequence between −343 and −292 bp in the 5′-flanking region of the Hdac7 gene promoter as the minimal PDGF-BB-responsive element, which contains one binding site for the transcription factor, specificity protein 1 (Sp1). Mutation of the Sp1 site within this PDGF-BB-responsive element abolished PDGF-BB-induced HDAC7 activity. PDGF-BB treatment enhanced Sp1 binding to the Hdac7 promoter in differentiated SMCs in vivo as demonstrated by the chromatin immunoprecipitation assay. Moreover, we also demonstrated that knockdown of Sp1 abrogated PDGF-BB-induced HDAC7 up-regulation and SMC differentiation gene expression in differentiating ES cells, although enforced expression of Sp1 alone was sufficient to increase the activity of the Hdac7 promoter and expression levels of SMC differentiation genes. Importantly, we further demonstrated that HDAC7 was required for Sp1-induced SMC differentiation of gene expression. Our data suggest that Sp1 plays an important role in the regulation of Hdac7 gene expression in SMC differentiation from ES cells. These findings provide novel molecular insights into the regulation of HDAC7 and enhance our knowledge in SMC differentiation and vessel formation during embryonic development.
Keywords: Cell Differentiation, Gene Regulation, Histone Deacetylase, Sp1, Stem Cell, Embryonic Stem Cells, Histone Deacetylase 7, Platelet-derived Growth Factor-BB, Smooth Muscle Cell Differentiation
Introduction
Embryonic stem (ES)4 cells are the most promising pluripotent stem cell sources and are derived from the inner cell mass of blastocysts. They have unlimited cell growth and the capacity to self-renew and differentiate into all types of mature tissue cells. For instance, ES cells may serve as a source of vascular smooth muscle cells (SMCs) that is essential for vascular tissue engineering. In addition, stem/progenitor cells were reported to differentiate into vascular cells to repair damaged vascular tissues and form neointima in vivo (1–4). Our previous studies demonstrated that ES cells can differentiate into SMCs (5–8) and endothelial cells in response to growth factor stimulation (3) and mechanical force (9). We also demonstrated that extracellular matrix protein collagen IV plays an important role in SMC differentiation from ES cells and that the collagen type IV-integrin signaling pathways are involved in SMC differentiation (6). However, the detailed molecular mechanisms of SMC differentiation from stem cells have not been fully clarified.
Platelet-derived growth factor-BB (PDGF-BB) has been identified as one of the key modulators for SMC differentiation from stem/progenitor cells (1, 10, 11) and mature vascular SMC phenotype modulation (12, 13). Our previous experiments showed that SMC differentiation from ES cells was enhanced through short stimulations with PDGF-BB (within 24 h) in the absence of serum, indicating that the duration of serum deprivation and the proliferation stage of cells are important parameters in SMC differentiation mediated by PDGF-BB (7). This highlights the fact that PDGF-BB has different functions in mature SMCs and ES cells. In proteomic analysis, we also demonstrated that ES-derived SMCs were different from mature SMCs and that PDGF-BB induced SMC differentiation gene expression during stem cell differentiation (14). Therefore, it seems that PDGF-BB can initiate and enhance stem/progenitor cell differentiation toward SMCs through transcriptional regulation.
In addition, we recently found that PDGF-BB induced ES cell differentiation toward SMCs through increasing expression of histone deacetylase 7 (HDAC7), which is a member of the class II HDAC family (7). HDACs are part of the transcriptional corepressor complexes and are key regulators in the differentiation of stem cells toward a specific cell lineage (15, 16). Four different classes of human HDACs have been defined based on their homology to HDACs found in Saccharomyces cerevisiae (17, 18). The class II HDACs (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) trafficking between the cytoplasm and nucleus contain both nuclear localization and export signals (19). Class II HDACs are reported to be important for cell differentiation in tissues such as muscles (20–25). There is also evidence that demonstrates the direct link between HDAC7 expression and SMC differentiation. For example, a recent study showed that HDAC7 knock-out mice had fewer smooth muscle actin (SMA)-positive cells in the vessel wall of embryos and that SMA staining in the dorsal aorta was less prominent. Moreover, fewer than normal SMCs were observed surrounding the dorsal aorta in the knock-out embryos at E11, indicating that HDAC7 has an essential role in SMCs (26).
Despite the fact that we have identified HDAC7, which plays an important role in SMC differentiation from stem cells, the molecular mechanisms of increased HDAC7 expression in SMC differentiation are currently unknown. In this study, we have identified a binding site for the transcription factor, specificity protein 1 (Sp1), in the Hdac7 promoter, which is crucial for the regulation of Hdac7 gene expression in SMC differentiation from stem cells.
MATERIALS AND METHODS
Cell Culture and PDGF-BB Treatment
Mouse ES cells (ES-D3 cell line, CRL-1943) were obtained from American Type Culture Collection (Manassas, VA) and grown on gelatin-coated flasks. Cell passages 5–20 were used for experiments. To maintain the ES cells in an undifferentiated state, leukemia inhibitory factor (1,000 units/ml) was added to the culture medium (DMEM, ATCC) supplemented with 10% FBS (Invitrogen, lot 3095073K), 2 mm l-glutamine, 100 mg/liter gentamicin, and 10−4 m 2-mercaptoethanol.
To induce SMC differentiation, ES cells were seeded on mouse collagen IV (5 μg/ml)-coated flasks or plates in differentiation medium (DMEM supplemented with 10% FBS, 0.05 mm 2-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin) for 1–9 days before further treatment as described previously (5–8). The medium was refreshed every other day. For PDGF treatment, the differentiating ES cells were cultured in serum-free α-minimal essential medium supplemented with 1% bovine serum albumin (BSA), 10 ng/ml insulin (Sigma), 0.05 mm 2-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin for 1 h, followed by the addition of PDGF-BB (Sigma) with the indicated concentrations and further incubation for 12 h. In some experiments, the RNA synthesis inhibitor, actinomycin D (1 μg/ml), was included for 6 h.
Plasmid Construction
Reporter vector harboring sequences of the mouse Hdac7 promoters were created using genomic DNA from 3-day pre-differentiated ES cells. The 5′-flanking region (−1134/−53 bp) of the Hdac7 gene was amplified by PCR with primers shown in Table 1 and cloned into the pGL3-basic vector, designated as pGL3-Luc-HD7 (7). A serial deletion of Hdac7 gene promoter reporter vector was generated from pGL3-Luc-HD7 with specific primers shown in Table 1, designated as pGL3-Luc-HD7-d1 to pGL3-Luc-HD7-d6. Sp1-binding site mutation was introduced into pGL3-Luc-HD7-d4 by PCR-based mutagenesis, designated as pGL3-Luc-HD7-d4-Sp1mut. Sp1 consensus binding site was introduced into pGL3-Luc-HD7-d4 in replacement of the Sp1 native-binding site by using QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies), designated as pGL3-Luc-HD7-d4-Sp1con. The expression vectors pShuttle2-HDAC7 and pShuttle2-Sp1 encoding the mouse HDAC7, Sp1 proteins, and reporter vector pGL3-Luc-SM22α were generated in our group as reported previously (7, 27). All vectors were verified by DNA sequencing.
TABLE 1.
Primers and cloning sites used for generation of full-length HDAC7 promoter, deletions, and mutation
| Vector | Primers | Cloning sites |
|---|---|---|
| pGL3-Luc-HD7 | F, atccctgaggacagtctgtggctgc | −1096/−54 bp |
| R, acagagagagggagcagg | ||
| pGL3-Luc-HD7-d2 | F, cccatcggagatcactgtag | −391/−54 bp |
| R, acagagagagggagcagg | ||
| pGL3-Luc-HD7-d4 | F, tccagggaggagcatgtttag | −343/−54 bp |
| R, gctcggtacctatcgatagag | ||
| pGL3-Luc-HD7-d5 | F, tcgttcaaacaccaccagtg | −292/−54 bp |
| R, gctcggtacctatcgatagag | ||
| pGL3-Luc-HD7-d6 | F, tctgtgcacagttcatgtgac | −262/−54 bp |
| R, gctcggtacctatcgatagag | ||
| pGL3-Luc-HD7-d4-Sp1 mut | F, tccagttagtagcatgtttag | Sp1-binding motif mutations at −339/−338/−335 bp |
| R, gctcggtacctatcgatagag | ||
| pGL3-Luc-HD7-d4-Sp1 con | F, cagcccccggcccggggcggggtatgtttagtggagga | Native Sp1-binding sequence replaced by Sp1 consensus binding sequence in pGL3-Luc-HD7-d4 |
| R, tcctccactaaacataccccgccccgggccgggggctg |
Transient Transfection
Differentiating ES cells were cultured on collagen IV and transfected at 80% confluency at day 4 with HDAC7 and Sp1 expression plasmid (pShuttle2-HDAC7/Sp1) or empty vector (pShuttle2) with FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Cells were harvested at 48 or 72 h after transfection, and real time RT-PCR and Western blot analysis were performed to analyze mRNA and protein levels, respectively.
siRNA Knockdown
The Silencer® Select siRNAs for negative control (number 3, AM4615), Sp1 (S74195), and HDAC7 (S80225) were purchased from Ambion. Mouse ESCs were seeded on collagen IV and cultured in differentiation medium. 5 days after plating, 10 μl of 10 μm siRNA were introduced with siIMPORTER transfection reagent (Millipore) according to the manufacturer's instructions. Cells were harvested at 48 or 72 h after transfection, and real time RT-PCR and Western blot analysis were performed to analyze mRNA and protein levels, respectively.
Luciferase Activity Assay
ES cells were cultured on collagen IV-coated 12-well plates for 4 days and then transfected with reporter gene alone or together with expression plasmid as indicated in the figure legends, using FuGENE 6 reagent (Roche Applied Science), according to the manufacturer's instructions. In some experiments, ES cells were cultured on collagen IV-coated 12-well plates for 5 days; HDAC7 siRNA or Sp1 siRNA oligonucleotides were cointroduced into each well using siIMPORTER transfection reagent with reporter plasmids pGL3-SM22-Luc or pGL3-Luc-HD7-d4. The modified pShuttle2 vector and control siRNA were included as mock/negative control, and Renilla luciferase was used as internal control, respectively. Luciferase and Renilla activities were detected 48 h after transfection using a standard protocol. Relative luciferase unit was defined as the ratio of Firefly versus Renilla with that of the control set as 1.0. At least three independent transfections were performed in triplicate.
Western Blot Analysis
The procedure used was similar to that described previously (28). Antibody against smooth muscle myosin heavy chain was from AbD Serotec (rabbit, AHP1117). Antibodies against α-tubulin and monoclonal anti-α-smooth muscle actin (SMαA) (clone 1A4, A5228) were from Sigma. Antibodies against Sp1 and HDAC7 were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phosphoserine/phosphothreonine/phosphotyrosine monoclonal antibodies were purchased from Millipore. All secondary antibodies were from Dako (Denmark). Specific antibody-antigen complexes were detected by using the ECL Western blot detection kit (Amersham Biosciences).
Coimmunoprecipitation
The procedure was performed as described previously (7). In brief, cell samples were lysed by rotation for 1 h at 4 °C. 1 mg of whole lysate was subjected to a standard coimmunoprecipitation procedure. Lysates were pre-cleared with normal IgG, then incubated with antibodies for 2 h at 4 °C, and precipitated by incubation for a further 2 h with protein-G-Sepharose beads. Precipitated proteins were resolved by SDS-gel electrophoresis and subsequently immunoblotted with antibodies.
RNA Extraction and RT-PCR
Total RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer's protocol. Two μg of RNA were reverse-transcribed into cDNA with random primer by Moloney murine leukemia virus reverse transcriptase (RT) (Promega). Fifty ng of cDNA (relative to RNA amount) was amplified by standard PCR with Taq DNA polymerase (Invitrogen) and primers.
Real Time PCR
Primers were designed using Primer Express software (Applied Biosystems) and a published sequence for the mouse genes as follows: SMA, TCCTGACGCTGAAGTATCCGAT/GGCCACACGAAGCTCGTTATAG (X13297); Sm-mhc, AAGCAGCCAGCATCAAGGAG/AGCTCTGCCATGTCCTCCAC (NM_013607); Hdac7, CCCAGTGTGCTCTACATTTCCC/CACGTTGACATTGAAGCCCTC (AF207749); and Sp1, ACAGGGTGCCAATGGCTGGC/GCCCACCAGAGACTGTGCGG (NM_013672). For each gene, SYBR Green® (Applied Biosystems) was used in place of a labeled probe. 18 S ribosomal RNA was used as the internal control for each gene, and target gene was amplified in duplex in PCR mixtures (25 μl final volume) containing 12.5 μl of SYBR Green® PCR master mix (Applied Biosystems), cDNA template (20 ng), and optimized primers. PCR thermal cycle parameters were as follows: 2 min at 50 °C, 10 min at 95 °C and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Reactions were performed, and fluorescence was monitored in an ABI Prism 7000 sequence detector system (Applied Biosystems). Relative mRNA expression level was defined as the ratio of target gene expression level to 18 S rRNA expression level with that of the control sample set as 1.0.
Chromatin Immunoprecipitation (ChIP)
The ChIP assays were performed as described previously (8). In brief, ES cells were treated with 1% (v/v) formaldehyde at room temperature for 10 min and then quenched with glycine at room temperature. The medium was removed, and cells were harvested for sonication. The sheared samples were diluted into 1 ml of immunoprecipitation buffer containing 25 mm Tris-HCl, pH 7.2, 0.1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, and immunoprecipitation was conducted with antibody raised against Sp1, together with single strand salmon sperm DNA saturated with protein G-Sepharose beads. Normal IgG was used as a control. The immunoprecipitates were eluted from the beads using 100 μl of elution buffer (50 mm NaHCO3, 1% SDS). A total of 200 μl of proteinase K solution was added to a total elution volume of 300 μl and incubated at 60 °C overnight. DNA was extracted and purified and then used to amplify target sequences by PCR. The primers used to amplify the promoter regions were as follows: GAGATCACTGTAGCCCTCAGC/TCCTCACTTAATCCTGTCGTG (−372/−173 bp). Aliquots of chromatin were also analyzed before immunoprecipitation and served as an input control. The data obtained from four independent experiments were quantified, using Photoshop. Fold of relative binding was defined as the ratio of band intensity in ChIP samples to that in the input sample with that of vehicle group set as 1.0.
Statistical Analysis
Data were expressed as the means ± S.E. The statistical analysis was performed with a two-tailed Student's t test. For multiple treatment groups, a one-way analysis of variance was applied. A value of p < 0.05 was considered to be statistically significant.
RESULTS
PDGF-BB Induced SMC Differentiation from ES Cells through HDAC7 Activation
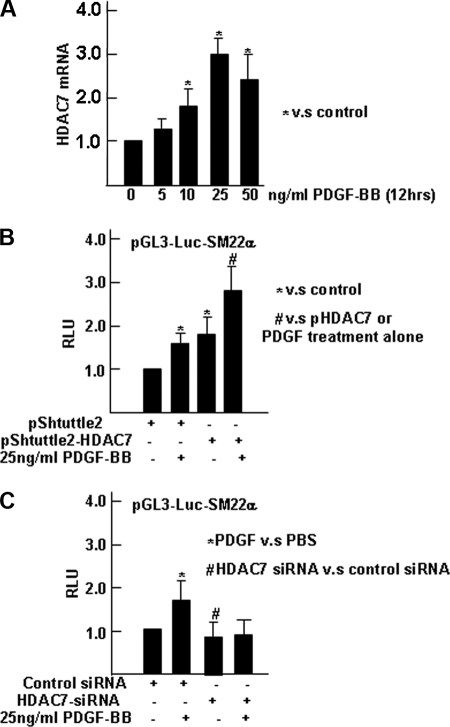
Previous studies from our laboratory showed that PDGF-BB enhanced the expression of the Hdac7 reporter gene in differentiating ES cells, indicating that PDGF-BB induces Hdac7 transcription. We extended these studies to assess the effect of PDGF-BB on levels of Hdac7 mRNA. A dose-dependent manner was detected whereby Hdac7 mRNA levels were visibly increased after treatment with PDGF-BB at 10 ng/ml for 12 h. At 25 ng/ml PDGF-BB, 12 h of treatment was sufficient to render Hdac7 mRNA levels as the most significant expression (Fig. 1A). Further experiments were performed to detect the effect of HDAC7 activation on PDGF-BB-induced change of SMC-specific differentiation gene SM22α in differentiating ES cells. As shown in Fig. 1B, enforced expression of HDAC7 significantly increased PDGF-BB-induced SM22α-Luc reporter gene expression in differentiating ES cells, whereas HDAC7 siRNA dramatically attenuated it (Fig. 1C). These data provide compelling evidence that HDAC7 plays an important role in mediating PDGF-BB-induced SMC differentiation from ES cells.
FIGURE 1.
PDGF-BB induced SMC differentiation from ES cells through activation of HDAC7. A, PDGF-BB induced HDAC7 mRNA level in a dose-dependent manner. ES cells were pre-differentiated for 5 days, followed by serum starvation for 6 h and treatment with the indicated concentration of PDGF-BB in serum-free medium for an additional 12 h. HDAC7 mRNA level was quantified by real time RT-PCR analysis. B, overexpression of HDAC7 enhanced PDGF-BB-induced SM22α transcription. Four days of differentiating ES cells were cotransfected with pGL3-Luc-SM22α (0.33 μg/5·104 cells) and pShuttle2 or pShuttle2-HDAC7 (0.5 μg/5·104 cells), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to luciferase activity analysis. C, knockdown of HDAC7 abolished PDGF-BB-induced SM22α transcription. Five days of differentiating ES cells were cotransfected with pGL3-Luc-SM22α (0.33 μg/5·104 cells) and control siRNA or HDAC7 siRNA (10 nm), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to luciferase activity analysis. pRenilla (0.1 μg/5·104cells) was included as luciferase plasmid control. Luciferase and Renilla activity assays were detected with standard kit 48 h after transfection. Relative luciferase units (RLU) or fold of induction was defined as the ratio of luciferase activity to Renilla activity. The data presented are from three independent experiments (n = 3). *, #, p < 0.05.
PDGF-BB Regulates HDAC7 Expression through Transcriptional Activation
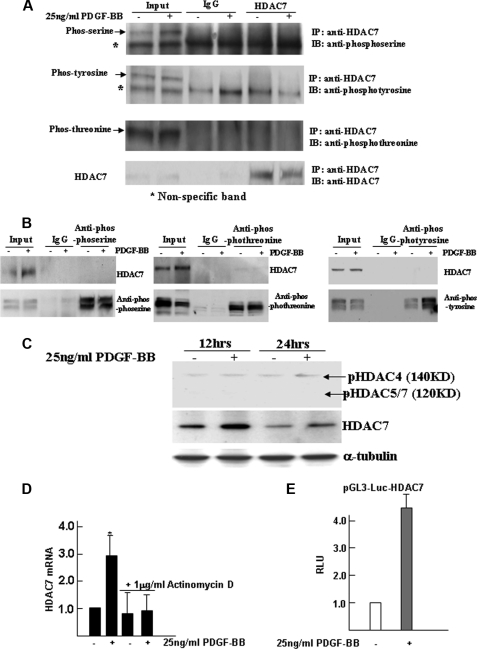
It is a general belief that growth factors can regulate target genes through phosphorylation. We wondered if this is the case for PDGF-BB regulation of HDAC7 expression. To reach this aim, 5 days of differentiating ES cells were subjected to serum starvation for 6 h and then incubated with 25 ng/ml PDGF-BB for an additional 12 h. Cells were harvested and subjected to immunoprecipitation experiments with antibodies against HDAC7 and phosphoserine, phosphotyrosine, and phosphothreonine, respectively. Phosphor bands for serine, tyrosine, and threonine (Fig. 2A, arrow) were detected in the input samples but not in the samples immunoprecipitated with HDAC7 antibody. Similarly, HDAC7 could be clearly detected in the input samples but not in the immunoprecipitated samples with each phosphor antibody (Fig. 2B). These data strongly suggested that PDGF-BB regulation of HDAC7 activation is not via phosphorylation of HDAC7, which is further confirmed by using phospho-HDAC4 (Ser-246)/HDAC5 (Ser-259)/HDAC7 (Ser-155) (D27B5) antibody recently developed by Cell Signaling Technology (catalog no. 3443) (Fig. 2C). To further elucidate the possibility that PDGF-BB regulates HDAC7 expression at the transcription level, 5 days of differentiating ES cells were treated with or without actinomycin D (1 μg/ml) for 6 h in the absence or presence of 25 ng/ml PDGF-BB. As indicated in Fig. 2D, treatment with actinomycin D completely abrogated Hdac7 mRNA induction by PDGF-BB. Luciferase gene reporter assays were performed to further determine the effect of PDGF-BB on Hdac7 transcription regulation. Our data revealed that PDGF-BB significantly increased Hdac7 promoter activity in differentiated ES cells (Fig. 2E), indicating that PDGF-BB directly activates HDAC7 expression at the transcriptional level.
FIGURE 2.
PDGF-BB regulates HDAC7 expression not via phosphorylation but via transcriptional activation. A–C, PDGF-BB regulates HDAC7 expression not via phosphorylation. Five days of differentiating ES cells were subjected to serum starvation for 6 h and then incubated with 25 ng/ml PDGF-BB for an additional 12 h. Cell lysates were harvested and subjected to immunoprecipitation (IP) experiments with antibodies against HDAC7 (A) and phosphoserine, -tyrosine, and -threonine (B), respectively. Arrows in A indicate the phosphor (phos) bands for serine, tyrosine, and threonine. Part of cell lysates were subjected to normal Western blot analysis with phospho-HDAC4 (Ser-246)/HDAC5 (Ser-259)/HDAC7 (Ser-155) (D27B5) antibody (C). Note: positive signal for HDAC4 (140 kDa) can be observed but no such positive signal for HDAC5/7. D, PDGF-BB regulates Hdac7 gene expression through transcriptional activation. Five days of differentiating ES cells were treated with or without actinomycin D (1 μg/ml) for 6 h in the absence or presence of 25 ng/ml PDGF-BB, and RNA samples were subjected to quantitative real time RT-PCR analysis to examine Hdac7 gene expression. E, luciferase reporter assay. ES cells were cultured on collagen IV and transfected at day 4 with reporter genes pGL3-Luc-HDAC7 (0.33 μg/5·104 cells), followed by PDGF-BB treatment. The data presented here are representative (A–C) or an average of three independent experiments (D and E); *, p < 0.05.
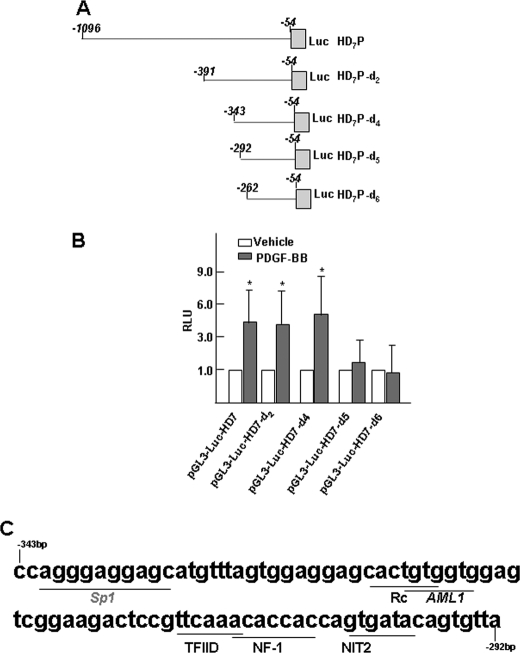
PDGF-BB-induced Increases in Hdac7 Promoter Activity Required Regulatory Elements between −343 and −292 bp of the 5′ Region of the Hdac7 Promoter
To determine the region of the Hdac7 promoter required for increased Hdac7 expression in response to PDGF-BB treatment of differentiating ES cells, we used a series of promoter-reporter plasmids containing various lengths of the Hdac7 promoter (depicted in Fig. 3A). Differentiating ES cells were transfected with different Hdac7 promoter-reporter plasmids and treated with either vehicle or PDGF-BB. Promoter activity was analyzed using luciferase assay. The results showed that PDGF-BB increased the promoter activities of pGL3-Luc-HD7, pGL3-Luc-HD7-d2, and pGL3-Luc-HD7-d4 but was unable to increase the promoter activities of both pGL3-Luc-HD7-d5 and pGL3-Luc-HD7-d6 (Fig. 3B), suggesting that the PDGF-BB-responsive region is located between −343 and −292 bp in the 5′-flanking region of Hdac7 gene. To determine potential transcription factor-binding sites within the promoter region mediating the transcriptional activation of Hdac7 in response to PDGF-BB treatment, we analyzed the “positively acting” Hdac7 promoter sequence between −343 and −292 bp using the transcription element search system (available on line). As depicted in Fig. 3C, there is one potential transcription factor specificity protein 1 (Sp1)-binding site, and there are several other potential binding sites for factors of the AML1, NF-1, and NIT2 proteins. It has been reported previously that transcription factor Sp1 modulates the expression of HDAC4 (29), which is a member of class II HDACs as HDAC7. Therefore, we hypothesized that Sp1 may regulate HDAC7 expression in differentiating ES cells upon PDGF-BB treatment.
FIGURE 3.
PDGF-BB-responsive elements in the promoter of HDAC7. A, schematic illustration of the cloned HDAC7 promoter deletions. B, luciferase (Luc) reporter analysis showing the different activity of HDAC7 promoter deletions. Four days of differentiating ES cells were transfected with reporter genes pGL3-Luc-HDAC7 or their derivates/deletions (0.33 μg/5·104 cells), followed by PDGF-BB treatment. pRenilla (0.1 μg/5·104cells) was included as luciferase plasmid control. Luciferase and Renilla activity assays were detected 48 h after transfection. The data presented here are the averages of three independent experiments. *, p < 0.05. C, potential binding sites and sequence for transcription factors within PDGF-BB-response element in the HDAC7 promoter.
PDGF-BB-induced Increase in Hdac7 Promoter Activity Is Dependent on the Sp1 Site in the Hdac7 Promoter
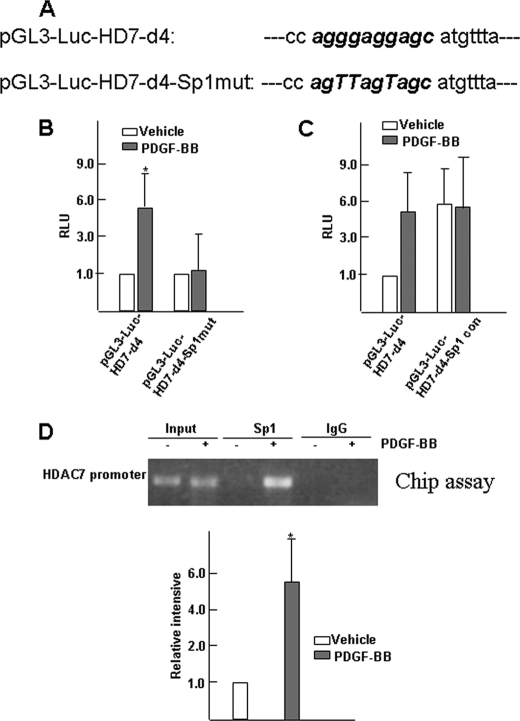
The functional significance of the potential Sp1-binding site in PDGF-BB-induced increase of Hdac7 expression was investigated by site-specific mutagenesis. Within the context of the pGL3-Luc-HDAC7-d4 vector that has maximal promoter activity in response to PDGF-BB treatment, the sequence of the potential Sp1-binding site, 5′-agggaggagc-3′, was mutated to a sequence 5′-agTTagTagc-3′ (as depicted in Fig. 4A) which will not bind to Sp1 factor (29). To test the effect of this mutation, ES cells were transfected with either the wild-type or Sp1 mutant pGL3-Luc-HDAC7-d4-Sp1mut plasmid and treated with vehicle or PDGF-BB. As shown in Fig. 4B, the mutation of the Sp1 site prevented PDGF-BB-induced increase in Hdac7 promoter activity. In contrast, replacing the sequence of the native Sp1 site in the Hdac7 promoter with a consensus Sp1-binding sequence (5′-GgggCggGgc-3′) resulted in a more than 5-fold increase in Hdac7 promoter activity (Fig. 4C) and eliminated the effect of PDGF-BB on Hdac7 promoter activity, suggesting the possibility that the native sequence in the Hdac7 promoter is a weaker Sp1-binding site compared with the consensus Sp1-binding sequence, making the promoter sensitive to change in Sp1 level/activity and responsive to Sp1 up-regulation.
FIGURE 4.
PDGF promotes Sp1 binding to HDAC7 promoter. A, schematic illustration of the triple mutations within HDAC7 promoter deletion 4. B, luciferase reporter analysis showing the Sp1-binding site is necessary for PDGF-BB-induced HDAC7 activation. The data presented here are the average of three independent experiments. *, p < 0.05. C, Sp1 consensus binding site eliminates the effects of PDGF-BB on HDAC7 promoter activity. The data presented here are the average of three independent experiments. D, chromatin immunoprecipitation (CHIP) assays were performed using antibodies against Sp1. Appropriate IgG was used as a negative control. Aliquots of chromatin before immunoprecipitation served as an input control. The data presented here are representative (upper panels) or an average of three independent experiments (lower panels). The data obtained from three independent experiments were quantified, using ImageQuant TL (Amersham Biosciences). Fold of relative binding was defined as the ratio of band intensity in ChIP samples to that in the Input sample with that of vehicle group set as 1.0. *, p < 0.05.
We next sought to determine whether PDGF-BB enhances the binding of Sp1 protein to the endogenous Hdac7 promoter within intact chromatin. ChIP assays were performed on chromatin isolated from differentiated ES cells treated with vehicle or PDGF-BB. As shown in Fig. 4D, Sp1 binding was enhanced more than 5-fold in ES cells treated with PDGF-BB compared with IgG antibody controls. These results demonstrated that PDGF-BB stimulated the binding of Sp1 to the endogenous Hdac7 promoter in differentiating ES cells. Taken together, these data strongly suggested that transcription factor Sp1 up-regulated HDAC7 in differentiating ES cells treated with PDGF-BB by direct binding to the Sp1 site in the Hdac7 promoter.
Transcription Factor Sp1 Plays a Crucial Role in Hdac7 Gene Expression Mediated by PDGF-BB
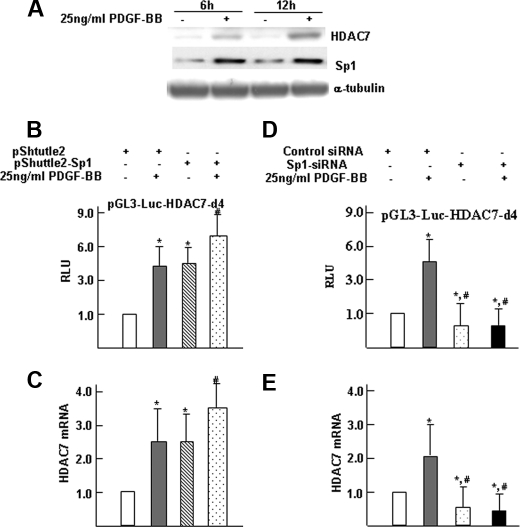
We further investigated the link between altered expression of Sp1 and Hdac7 transcription. First, Western blot analysis demonstrated that PDGF-BB up-regulated Sp1 and HDAC7 in a similar manner (Fig. 5A). To determine whether the enforced expression of Sp1 could induce HDAC7 expression, we next cotransfected differentiating ES cells with the Hdac7 reporter plasmid (pGL3-Luc-HDAC7-d4) and an Sp1 expression plasmid (pShuttle2-Sp1) or control vector. Following overexpression, cells were treated with vehicle or PDGF-BB, and promoter activity was analyzed by luciferase assay. The results showed that the overexpression of Sp1 alone increased the promoter activity of pGL3-Luc-HDAC7-d4 plasmid (Fig. 5B). To ensure that our results were not merely an artifact due to the use of artificial, truncated, episomal promoter-reporter constructs, endogenous HDAC7 expression was measured by quantitative real time RT-PCR. As shown in Fig. 5C, enforced expression of Sp1 significantly increased the Hdac7 mRNA level in differentiating ES cells. To further determine whether Sp1 is required for the PDGF-BB-induced activation of the Hdac7 transcription, siRNA knockdown experiments were performed in differentiating ES cells using control or Sp1-targeted siRNAs. Following knockdown experiments, cells were treated with vehicle or PDGF-BB. Hdac7 reporter gene expression was analyzed by luciferase assay, and mRNA levels were measured by quantitative real time RT-PCR. As shown in Fig. 5, D and E, the knockdown of Sp1 significantly ablated PDGF-BB-induced Hdac7 promoter activity and mRNA expression in differentiating ES cells. These data demonstrated that Sp1 was required for the PDGF-BB-mediated Hdac7 transcription activation.
FIGURE 5.
Transcription factor Sp1 plays a crucial role in Hdac7 gene expression mediated by PDGF-BB. A, HDAC7 up-regulation is parallel with Sp1 upon PDGF-BB treatment. Day 5 differentiating ES cells were treated with PDGF-BB for 6 and 12 h, respectively; cells were harvested and subjected to Western blot analysis to examine the protein levels of Sp1 and HDAC7. B, overexpression Sp1 enhanced PDGF-BB-induced Hdac7 gene transcriptional activation. Four days of differentiating ES cells were cotransfected with pGL3-Luc-HDAC7-d4 (0.33 μg/5·104 cells) and pShuttle2 or pShuttle2-Sp1 (0.5 μg/5·104 cells), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to luciferase activity analysis. pRenilla (0.1 μg/5·104 cells) was included as luciferase plasmid control. C, overexpression Sp1 enhanced PDGF-BB-induced Hdac7 gene expression. Four days of differentiating ES cells were transfected with pShuttle2 or pShuttle2-Sp1 (1.0 μg/1·106 cells), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to real time RT-PCR analysis. D, knockdown of Sp1 abolished PDGF-BB-induced Hdac7 gene transcriptional activation. Five days of differentiating ES cells were cotransfected with pGL3-Luc-HDAC7-d4 (0.33 μg/5·104 cells) and control siRNA or Sp1 siRNA (10 nm), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to luciferase activity analysis. pRenilla (0.1 μg/5·104cells) was included as luciferase plasmid control. E, knockdown of Sp1 abolished PDGF-BB-induced Hdac7 gene expression. Five days of differentiating ES cells were transfected with control siRNA or Sp1 siRNA (10 nm), followed by PDGF-BB treatment. After 48 h of transfection, cells were harvested and subjected to real time RT-PCR analysis. The data presented here are representative (A) or an average of three independent experiments (B–E), #, p < 0.05 (versus control); *, p < 0.05; 4th versus 2nd or 3rd column in B and C or 2nd versus 3rd or 4th columns in D and E.
Transcription Factor Sp1 Is Required for PDGF-BB-induced SMC Differentiation
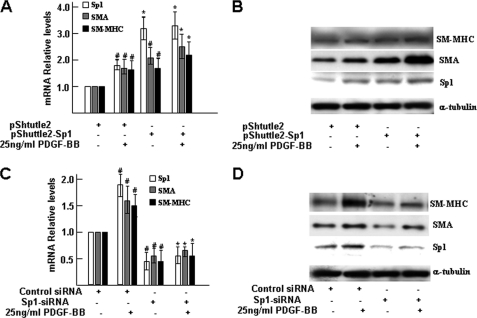
We finally investigated the effects of altered expression of Sp1 on PDGF-BB-induced SMC differentiation. As indicated in Fig. 6, A and B, enforced expression of Sp1 significantly up-regulated SMC-specific marker expression, such as SMA and smooth muscle myosin heavy chain, at both mRNA and protein levels. To further investigate whether Sp1 expression was necessary for smooth muscle marker expression, siRNA knockdown experiments were performed in differentiating ES cells using Sp1 or control siRNA, followed by PDGF-BB treatment. Our data revealed that the PDGF-BB-induced up-regulation of SMC-specific markers was significantly inhibited by Sp1 siRNA at both mRNA and protein levels, indicating that Sp1 expression was essential for PDGF-BB-induced SMC differentiation gene expression (Fig. 6, C and D). Taken together, these results suggested that PDGF might promote ES cell differentiation into SMCs through transcription factor Sp1.
FIGURE 6.
Transcription factor Sp1 is essential for PDGF-BB-induced SMC differentiation. A and B, overexpression Sp1 enhanced PDGF-BB-induced SMC differentiation gene expression. Four days of differentiating ES cells were transfected with pShuttle2 or pShuttle2-Sp1 (1.0 μg/1·106 cells), followed by PDGF-BB treatment. After 48 h of transfection, total RNA and protein were harvested and subjected to real time RT-PCR analysis (A) and Western blot analysis (B), respectively. C and D, knockdown of Sp1 abolished PDGF-BB-induced SMC differentiation gene expression. Five days of differentiating ES cells were transfected with control siRNA or Sp1 siRNA (10 nm), followed by PDGF-BB treatment. After 48 h of transfection, total RNA and protein were harvested and subjected to real time RT-PCR analysis (C) and Western blot analysis (D), respectively. The data presented here are representative (B and D) or an average of three independent experiments (A and C). #, p < 0.05; *, p < 0.01 (versus control) in A; #, p < 0.05 (versus control); *, p < 0.05; 4th versus 2nd columns in C.
HDAC7 Is Required for Sp1-induced SMC Differentiation Gene Expression
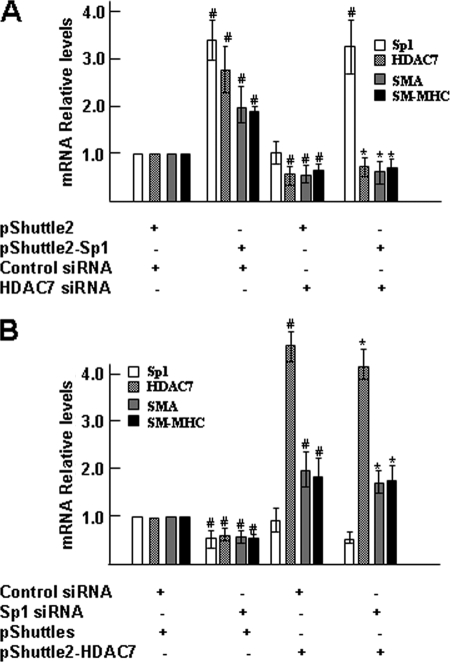
To further clarify whether HDAC7 is required for SP1-mediated SMC differentiation gene expression, Sp1 expression plasmids and HDAC7-specific siRNA were cotransfected into differentiating ES cells. Data shown in Fig. 7A revealed that Sp1 overexpression results in significant induction of Hdac7 as well as SMC differentiation genes, whereas knockdown of HDAC7 significantly down-regulated SMC differentiation gene expression, even in the presence of high levels of Sp1, which strongly suggests that HDAC7 is essential for Sp1 overexpression-induced SMC differentiation gene expression. Furthermore, data from other sets of cotransfection experiments with Sp1 siRNA and HDAC7 expression plasmids displayed the comparable levels of SMC differentiation gene expression between control siRNA and Sp1 siRNA-treated samples in the presence of high levels of HDAC7, indicating that inhibition of SMC differentiation gene expression induced by Sp1 knockdown could be rescued by HDAC7 overexpression. Taken together, these data suggested that Sp1-induced SMC differentiation gene expression is required for HDAC7 activation.
FIGURE 7.
HDAC7 is required for Sp1-induced SMC differentiation gene expression. A, knockdown of HDAC7 abolished Sp1 overexpression-induced SMC differentiation gene expression. Four days of differentiating ES cells were cotransfected with pShuttle2 or pShuttle2-Sp1 (1.0 μg/1·106 cells) and control siRNA or HDAC7 siRNA (10 nm). After 48 h of transfection, total RNA were harvested and subjected to real time RT-PCR. B, HDAC7 overexpression rescued the inhibitory effects of Sp1 knockdown on SMC differentiation gene expression. Five days of differentiating ES cells were cotransfected with control siRNA or Sp1 siRNA (10 nm) and pShuttle2 or pShuttle2-HDAC7 (1.0 μg/1·106 cells. After 48 h of transfection, total RNA were harvested and subjected to real time RT-PCR analysis. The data presented here are an average of three independent experiments; #, p < 0.05 (versus control); *, p < 0.05 (4th versus 2nd columns).
DISCUSSION
The major conclusion in this study is that transcription factor Sp1 plays an important role in the regulation of Hdac7 gene expression in SMC differentiation from ES cells, which is supported by several lines of evidence. (a) Hdac7 transcription activation was required for PDGF-BB-induced ES cell differentiation toward SMCs. (b) The promoter region (between −343 and −292 bp) of the Hdac7 gene contains an Sp1-binding site that confers the responsiveness to PDGF-BB. (c) Stimulation with PDGF-BB enhanced the binding of Sp1 to the Hdac7 promoter in vivo. (d) RNA interference knockdown of Sp1 abolished PDGF-BB-induced Hdac7 promoter activity and mRNA expression. (e) Sp1-dependent activation of HDAC7 was required for PDGF-BB-induced expression of SMC-specific markers in differentiating ES cells. These findings enhance our knowledge of SMC differentiation and provide useful insights in the mechanisms of vessel development.
Acetylation of chromatin proteins and transcription factors is part of a complex signaling system that is largely involved in the control of gene expression (30, 31). Generally, it is believed that acetylation of lysine in the histone tail by histone acetyltransferases can relax the nucleosome at promoters in advance of initiation, which is crucial for gene transcription (32), whereas deacetylation carried out by HDACs, which remove acetyl groups from lysine residues in histones and subsequently cause a particular region of chromatin to be condensed, results in the repression of certain gene expression. HDACs are classified into four groups according to sequence identity (33–35). Class II HDACs (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) contain an N-terminal extension important for protein-protein interactions and are highly expressed in skeletal muscle, heart, and brain (36, 37). The restricted expression of class II HDACs suggests an important role for their activity in these tissues. Moreover, class II HDACs have also been reported to act as signal-responsive repressors of cardiac hypertrophy, skeletal muscle differentiation, and bone development (24, 38, 39) and play a key role in the differentiation of stem cells toward a specific cell lineage (15, 16, 20). In particular, HDAC7 has been initially implicated in the regulation of vascular integrity as shown in a study that HDAC7-deficient mice have a small number of vascular SMCs, which died during embryonic stage because of vascular leakage (26). Further evidence suggests that HDAC7 plays a crucial role in vascular endothelial cell functions through modulation of matrix metalloproteinase 10 (26) and β-catenin (40). Recent evidence, however, also indicate that HDAC7 is a key mediator in SMC differentiation from ES cells. Our recent study revealed for the first time that PDGF-BB promoted the differentiation of ESCs toward the SMC lineage via the up-regulation of Hdac7 transcription and its alternative splicing (7). Despite our data depicting that HDAC7 plays an important role in PDGF-BB-induced ES cell differentiation into SMCs, the molecular mechanisms of increased HDAC7 expression in SMC differentiation are largely unknown. In this study, we further confirmed a clear correlation between HDAC7 expression and PDGF-BB-induced SMC differentiation from ES cells. Up-regulation of HDAC7 expression increased the expression of SMC markers, whereas its down-regulation significantly decreased SMC marker expression, indicating a crucial role for HDAC7 in PDGF-BB-induced SMC differentiation. In addition, we have demonstrated here that PDGF-BB regulates HDAC7 expression not through phosphorylation but through transcription activation.
Understanding the molecular mechanisms that underlie the transcriptional regulation of HDAC7 is essential, in view of the importance of alterations of HDAC7 mRNA expression in its activation and determining PDGF-BB-induced ES cell differentiation toward an SMC lineage. The results from this study showed that PDGF-BB-induced expression of HDAC7 was dependent on an Sp1-binding site in the Hdac7 promoter. Our initial studies were done using a truncated Hdac7 promoter fragment; therefore, it was possible that there were additional unidentified regulatory regions (in addition to Sp1) that may also play a role in PDGF-BB-induced HDAC7 expression. However, we confirmed that Sp1 did bind to the region of the endogenous Hdac7 promoter containing an Sp1-binding site within intact chromatin in response to PDGF-BB treatment of the differentiated ES cells and that knockdown of Sp1 prevented PDGF-BB-induced increase in endogenous HDAC7 expression. These data demonstrated that the effect of Sp1 was not an artifact due to the use of truncated, episomal Hdac7 promoter-reporter plasmid but represented a functionally relevant signaling pathway. In addition, we further reported that Sp1 played a critical role in the PDGF-BB-induced up-regulation of SMC marker gene expression. Enforced expression of Sp1 significantly up-regulated HDAC7 activity and SMC-specific marker expression, such as SMA and smooth muscle myosin heavy chain, both at mRNA and protein levels, whereas siRNA-mediated suppression of Sp1 could ablate PDGF-BB-induced HDAC7-Luc reporter gene expression and SMC-specific differentiation gene expression in differentiating ES cells.
The transcription factor Sp1 is ubiquitously expressed in many tissues and cell lines and possesses three C2H2-type zinc fingers as a DNA-binding domain, which binds to GC-boxes with the consensus sequence 5′-(G/T)GGGCGG(G/A)(G/A)(C/T)-3′ or 5′-(G/T)(G/A)GGCG(G/T)(G/A)(G/A)(C/T)-3′. Sp1 also binds to CT-boxes and GT-boxes, but with significantly lower affinities. For example, the CT-box 5′-GGGGAGGGGC-3′ and the GT-box 5′-GGGTGGGGC-3′ are bound 3- or 6-fold more weakly, respectively, than the GC-box (5′-GGGGCGGGGC-3′) (reviewed in Ref. 41). Sp1 also has two glutamine-rich transactivation domains, namely the domains A and B, each interacting directly with both TATA-binding protein and TAF4 (TATA-binding protein-associated factor 4) to coordinate the regulation of gene expression (41). It is suggested that there are over 12,000 Sp1-binding sites in the full human genome (42). Initially, the ubiquitous transcription factor Sp1 was considered as a constitutive activator of housekeeping genes and other TATA-less genes. However, accumulating evidence also strongly suggest that, similar to many tissue-specific transcription factors or mediators, Sp1 also has many divergent functional roles in regulating tissue- and/or cell type-specific gene expression, for instance, synergistic transactivation with erythroid cell-specific transcription factor GATA-1 to regulate their downstream genes in erythroid cells such as α/γ-globins and erythropoietin receptor (43, 44). In this study, we showed that Sp1 is required for Hdac7 transcriptional activation and SMC differentiation gene expression because knockdown of Sp1 itself significantly down-regulated Hdac7 transcriptional activation. However, it remains uncertain whether this effect of Sp1 on Hdac7 transcription is because Sp1 is important for Hdac7 basal transcription or because it is responsible for HDAC7 induction by PDGF-BB and whether the modest up-regulation of Sp1 in PDGF-BB-treated cells is indeed an important regulator of HDAC7 expression in response to PDGF-BB. In addition, it is unclear whether the relatively small (3-fold) increase in HDAC7 alone would be sufficient for PDGF-BB-induced SMC differentiation from ES cells. These are difficult questions to answer conclusively and represent the limitations of this study. Pertinent to the question about Sp1, our study showed that when the weak Sp1 site in the native Hdac7 promoter was converted to a strong Sp1 site, the basal transcription of the promoter increased, and the promoter was no longer responsive to PDGF-BB. This result provides some support for the hypothesis that the weak Sp1 site in the native promoter may make the promoter sensitive to Sp1 up-regulation, although it is also possible that the strong Sp1 site artificially introduced to the Hdac7 promoter may have an overriding effect on other mechanisms that are involved in transcriptional induction.
Our unpublished data5 suggest that Sp1 knockdown down-regulates SMC differentiation gene expression without affecting other cell lineage genes, including genes reported to be changed during SMC differentiation from stem cells (5) and reported SMC/endothelial cell-specific differentiation regulators, such as NADPH oxidase 4 (Nox4) (5), nuclear factor erythroid 2-related factor (NRF) 3 (8), and HDAC3 (3, 9). However, it is unknown whether it is because these genes are not Sp1 targets or because the Sp1 knockdown is incomplete, and these genes are less sensitive to changes in Sp1 concentration.
In the past decade, Sp1 has emerged as a crucial mediator in many important cellular physiological/pathological functions and signal transductions through modulating its respective target genes, such as cell proliferation related-genes (CCN D1/D2/D3/E and cdk2 in G1/S transition, CCNB1 and CDC25c in G2/M transition, and CCNB1 and survivin in cell mitosis), cell survival (IGF-II and IGF-IR), cell growth (c-myc and n-myc), cell invasion and metastasis (MMP2, MMP9, and uPA), and angiogenesis (VEGF, VEGF-R1/R2, and FGF-R1) (41). Importantly, the functional role of Sp1 in embryonic development and cell growth/differentiation was further highlighted in Sp1-deficient mice (Sp1−/−). Sp1−/− embryos survive until day 9.5 (E9.5) of gestation but display a severely retarded growth and show a broad range of phenotypic abnormalities, which suggests that Sp1 is essential for early normal embryonic development and plays an important role in the maintenance of differentiated cells through the regulation of genes like MeCP2 (45).
Based on previous findings and the data presented here, we postulate that the mechanisms of mediating the promoter activity of Hdac7 by Sp1 are reminiscent of that previously described for Hdac4, another member of the class II HDAC family. Activation of Hdac4 expression by Sp1 was shown to be dependent on specific sequences in the Hdac4 promoter, which was increased by histone deacetylase inhibitor TSA (29). We similarly found that Sp1 regulated Hdac7 promoter activity, mRNA, and protein expression also through its binding to the Hdac7 promoter. Notably, some potential binding sites for other transcription factors such as AML1, NF-1, and NIT2 also exist in the PDGF-BB-responsive minimal element within the Hdac7 gene promoter, which might also play some part in Hdac7 gene regulation by PDGF-BB during SMC differentiation from ES cells. The potential functions of these transcription factors in regulation of Hdac7 activation and SMC differentiation are currently under further investigation in our group.
Apart from transcriptional regulation, HDAC activity is also believed to be mediated through protein-protein interactions, such as protein complex of silencing mediator of retinoid and thyroid receptor (SMRT) and nuclear receptor corepressor (N-CoR). Both SMRT and N-CoR are nuclear receptor corepressors that bind and enhance the HDAC activity of HDAC3 but not the class II HDACs (HDAC4, -5, -7, and -9) (46–48), which suggests that the class II HDACs may recruit enzymatically active HDAC3-SMRT/N-coR complexes for their functional effects (49). However, whether it is the case for HDAC7 activity regulation by PDGF-BB remains to be elucidated.
In summary, we provide evidence that Sp1 plays an important role in the regulation of Hdac7 gene expression in SMC differentiation from ES cells. Further studies are needed to determine whether the Sp1-dependent induction of HDAC7 expression is a universal mechanism involved in both physiological maintenance of the cardiovascular system during embryonic development and in the pathophysiology of vascular diseases, such as atherosclerosis in adults, and whether other transcription factors also play some roles in Hdac7 gene expression and SMC differentiation.
This work was supported in part by the British Heart Foundation, National Natural Science Foundation of China Grant 30900571, Qiangjiang Talent Project of Science and Technology Department of Zhejiang Province (2010R1066), and Natural Science Foundation of Zhejiang Province Grant Y2090411. This work forms part of the research themes contributing to the translational research portfolio of Barts and the London Cardiovascular Biomedical Research Unit, which is supported and funded by the National Institute of Health Research.
L. Zhang, M. Jin, A. Margariti, G. Wang, Z. Luo, A. Zampetaki, L. Zeng, S. Ye, J. Zhu, and Q. Xiao, unpublished data.
- ES
- embryonic stem
- SMC
- smooth muscle cell
- PDGF-BB
- platelet-derived growth factor-BB
- HDAC
- histone deacetylase
- SMA
- smooth muscle actin.
REFERENCES
- 1.Hu Y., Zhang Z., Torsney E., Afzal A. R., Davison F., Metzler B., Xu Q. (2004) J. Clin. Invest. 113, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rotmans J. I., Heyligers J. M., Verhagen H. J., Velema E., Nagtegaal M. M., de Kleijn D. P., de Groot F. G., Stroes E. S., Pasterkamp G. (2005) Circulation 112, 12–18 [DOI] [PubMed] [Google Scholar]
- 3.Xiao Q., Zeng L., Zhang Z., Margariti A., Ali Z. A., Channon K. M., Xu Q., Hu Y. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 4.Wang C. H., Cherng W. J., Yang N. I., Kuo L. T., Hsu C. M., Yeh H. I., Lan Y. J., Yeh C. H., Stanford W. L. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 54–60 [DOI] [PubMed] [Google Scholar]
- 5.Xiao Q., Luo Z., Pepe A. E., Margariti A., Zeng L., Xu Q. (2009) Am. J. Physiol. Cell Physiol. 296, C711–C723 [DOI] [PubMed] [Google Scholar]
- 6.Xiao Q., Zeng L., Zhang Z., Hu Y., Xu Q. (2007) Am. J. Physiol. Cell Physiol. 292, C342–C352 [DOI] [PubMed] [Google Scholar]
- 7.Margariti A., Xiao Q., Zampetaki A., Zhang Z., Li H., Martin D., Hu Y., Zeng L., Xu Q. (2009) J. Cell Sci. 122, 460–470 [DOI] [PubMed] [Google Scholar]
- 8.Pepe A. E., Xiao Q., Zampetaki A., Zhang Z., Kobayashi A., Hu Y., Xu Q. (2010) Circ. Res. 106, 870–879 [DOI] [PubMed] [Google Scholar]
- 9.Zeng L., Xiao Q., Margariti A., Zhang Z., Zampetaki A., Patel S., Capogrossi M. C., Hu Y., Xu Q. (2006) J. Cell Biol. 174, 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishishita T., Lin P. C. (2004) J. Cell. Biochem. 91, 584–593 [DOI] [PubMed] [Google Scholar]
- 11.Miyata T., Iizasa H., Sai Y., Fujii J., Terasaki T., Nakashima E. (2005) J. Cell. Physiol. 204, 948–955 [DOI] [PubMed] [Google Scholar]
- 12.Holycross B. J., Blank R. S., Thompson M. M., Peach M. J., Owens G. K. (1992) Circ. Res. 71, 1525–1532 [DOI] [PubMed] [Google Scholar]
- 13.Blank R. S., Owens G. K. (1990) J. Cell. Physiol. 142, 635–642 [DOI] [PubMed] [Google Scholar]
- 14.Yin X., Mayr M., Xiao Q., Wang W., Xu Q. (2006) Proteomics 6, 6437–6446 [DOI] [PubMed] [Google Scholar]
- 15.Kato H., Tamamizu-Kato S., Shibasaki F. (2004) J. Biol. Chem. 279, 41966–41974 [DOI] [PubMed] [Google Scholar]
- 16.Sterner D. E., Berger S. L. (2000) Microbiol. Mol. Biol. Rev. 64, 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taunton J., Hassig C. A., Schreiber S. L. (1996) Science 272, 408–411 [DOI] [PubMed] [Google Scholar]
- 18.Verdin E., Dequiedt F., Kasler H. G. (2003) Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 19.Wang A. H., Yang X. J. (2001) Mol. Cell. Biol. 21, 5992–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dressel U., Bailey P. J., Wang S. C., Downes M., Evans R. M., Muscat G. E. (2001) J. Biol. Chem. 276, 17007–17013 [DOI] [PubMed] [Google Scholar]
- 21.Deng X., Ewton D. Z., Mercer S. E., Friedman E. (2005) J. Biol. Chem. 280, 4894–4905 [DOI] [PubMed] [Google Scholar]
- 22.Lu J., McKinsey T. A., Zhang C. L., Olson E. N. (2000) Mol. Cell 6, 233–244 [DOI] [PubMed] [Google Scholar]
- 23.Zhang C. L., McKinsey T. A., Olson E. N. (2002) Mol. Cell. Biol. 22, 7302–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinsey T. A., Zhang C. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karamboulas C., Swedani A., Ward C., Al-Madhoun A. S., Wilton S., Boisvenue S., Ridgeway A. G., Skerjanc I. S. (2006) J. Cell Sci. 119, 4305–4314 [DOI] [PubMed] [Google Scholar]
- 26.Chang S., Young B. D., Li S., Qi X., Richardson J. A., Olson E. N. (2006) Cell 126, 321–334 [DOI] [PubMed] [Google Scholar]
- 27.Zampetaki A., Zeng L., Xiao Q., Margariti A., Hu Y., Xu Q. (2007) Am. J. Physiol. Cell Physiol. 293, C1226–C1238 [DOI] [PubMed] [Google Scholar]
- 28.Zampetaki A., Zhang Z., Hu Y., Xu Q. (2005) Am. J. Physiol. Heart Circ. Physiol. 288, H2946–H2954 [DOI] [PubMed] [Google Scholar]
- 29.Liu F., Pore N., Kim M., Voong K. R., Dowling M., Maity A., Kao G. D. (2006) Mol. Biol. Cell 17, 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinsey T. A., Olson E. N. (2005) J. Clin. Invest. 115, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Backs J., Olson E. N. (2006) Circ. Res. 98, 15–24 [DOI] [PubMed] [Google Scholar]
- 32.Mellor J. (2005) Mol. Cell 19, 147–157 [DOI] [PubMed] [Google Scholar]
- 33.de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregoretti I. V., Lee Y. M., Goodson H. V. (2004) J. Mol. Biol. 338, 17–31 [DOI] [PubMed] [Google Scholar]
- 35.Witt O., Deubzer H. E., Milde T., Oehme I. (2009) Cancer Lett. 277, 8–21 [DOI] [PubMed] [Google Scholar]
- 36.Fischle W., Kiermer V., Dequiedt F., Verdin E. (2001) Biochem. Cell. Biol. 79, 337–348 [PubMed] [Google Scholar]
- 37.Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 38.Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 39.Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margariti A., Zampetaki A., Xiao Q., Zhou B., Karamariti E., Martin D., Yin X., Mayr M., Li H., Zhang Z., De Falco E., Hu Y., Cockerill G., Xu Q., Zeng L. (2010) Circ. Res. 106, 1202–1211 [DOI] [PubMed] [Google Scholar]
- 41.Wierstra I. (2008) Biochem. Biophys. Res. Commun. 372, 1–13 [DOI] [PubMed] [Google Scholar]
- 42.Cawley S., Bekiranov S., Ng H. H., Kapranov P., Sekinger E. A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A. J., Wheeler R., Wong B., Drenkow J., Yamanaka M., Patel S., Brubaker S., Tammana H., Helt G., Struhl K., Gingeras T. R. (2004) Cell 116, 499–509 [DOI] [PubMed] [Google Scholar]
- 43.Fischer K. D., Haese A., Nowock J. (1993) J. Biol. Chem. 268, 23915–23923 [PubMed] [Google Scholar]
- 44.Merika M., Orkin S. H. (1995) Mol. Cell. Biol. 15, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marin M., Karis A., Visser P., Grosveld F., Philipsen S. (1997) Cell 89, 619–628 [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., Kalkum M., Chait B. T., Roeder R. G. (2002) Mol. Cell 9, 611–623 [DOI] [PubMed] [Google Scholar]
- 47.Wen Y. D., Perissi V., Staszewski L. M., Yang W. M., Krones A., Glass C. K., Rosenfeld M. G., Seto E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7202–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guenther M. G., Barak O., Lazar M. A. (2001) Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. (2002) Mol. Cell 9, 45–57 [DOI] [PubMed] [Google Scholar]