Abstract
Group A streptococci (GAS) utilize soluble human complement regulators to evade host complement attack. Here, we characterized the binding of the terminal complement complex inhibitor complement Factor H-related protein 1 (CFHR1) and of the C3 convertase regulator Factor H to the streptococcal collagen-like proteins (Scl). CFHR1 and Factor H, but no other member of the Factor H protein family (CFHR2, CFHR3, or CFHR4A), bound to the two streptococcal proteins Scl1.6 and Scl1.55, which are expressed by GAS serotypes M6 and M55. The two human regulators bound to the Scl1 proteins via their conserved C-terminal attachment region, i.e. CFHR1 short consensus repeats 3–5 (SCR3–5) and Factor H SCR18–20. Binding was affected by ionic strength and by heparin. CFHR1 and the C-terminal attachment region of Factor H did not bind to Scl1.1 and Scl2.28 proteins but did bind to intact M1-type and M28-type GAS, which express Scl1.1 and Scl2.28, respectively, thus arguing for the presence of an additional binding mechanism to CFHR1 and Factor H. Furthermore mutations within the C-terminal heparin-binding region and Factor H mutations that are associated with the acute renal disease atypical hemolytic uremic syndrome blocked the interaction with the two streptococcal proteins. Binding of CFHR1 affected the complement regulatory functions of Factor H on the level of the C3 convertase. Apparently, streptococci utilize two types of complement regulator-acquiring surface proteins; type A proteins, as represented by Scl1.6 and Scl1.55, bind to CFHR1 and Factor H via their conserved C-terminal region and do not bind the Factor H-like protein 1 (FHL-1). On the contrary, type B proteins, represented by M-, M-like, and the fibronectin-binding protein Fba proteins, bind Factor H and FHL-1 via domain SCR7 and do not bind CFHR1. In conclusion, binding of CFHR1 is at the expense of Factor H-mediated regulatory function at the level of C3 convertase and at the gain of a regulator that controls complement at the level of the C5 convertase and formation of the terminal complement complex.
Keywords: Bacteria, Complement, Immunology, Innate Immunity, Multifunctional Protein, CFHR1 Binding, TCC Inhibition, Terminal Pathway, Complement Evasion
Introduction
Streptococcus pyogenes is a specialized Gram-positive β-hemolytic human pathogen. Group A streptococci (GAS)2 infections cause a variety of diseases including superficial infections of the throat and skin, such as pharyngitis and impetigo, and deep soft tissue infections like necrotizing fasciitis and myositis, as well as live threatening septic infections including toxic shock syndrome and puerperal sepsis (1, 2).
Upon infection of a human host, S. pyogenes, similar to other microbes, is immediately confronted and attacked by the human complement system. This central defense system represents an enzymatic cascade which is initiated via three major pathways, the alternative, the classical, and the lectin pathway. The alternative complement pathway (AP) is activated spontaneously, continuously, and independently of antibodies. Upon activation, a C3 convertase is formed on the surface of an invading microbe; this enzyme cleaves the central complement compound C3 into surface-opsonizing C3b and into the chemotactic and antimicrobial anaphylatoxin C3a (3). If activation proceeds, a C5 convertase is formed that generates the potent anaphylactic peptide C5a and further initiates the terminal complement complex (TCC), which forms a pore in the membrane of the microbe and leads to target lysis (4–6). In addition, cytokine-like activity is described for soluble TCC (7, 8).
Host cells are generally protected from the damaging effects of alternative complement attack by surface-expressed as well as surface-acquired complement regulators. Factor H is the major soluble regulator of the AP, and this protein is composed of 20 domains termed short consensus repeats (SCR). The C terminus (SCR18–20) forms the major binding region of the protein and facilitates surface binding (9–12).
The complement regulatory region is located in the N terminus (SCR1–4) of the protein. Factor H-like protein 1 (FHL-1), which is derived from an alternatively spliced transcript of the Factor H gene, consists of the first seven SCRs including the regulatory region. Both regulators act on the level of the C3 convertase, accelerating the decay of the convertase and acting as cofactors for the C3b-cleaving serine protease Factor I (13–15). The complement Factor H-related protein 1 (CFHR1) consists of five SCRs (16). CFHR1 is a complement inhibitor that inhibits the C5 convertase and blocks formation of the terminal complement complex (17). CFHR1 shares the C-terminal surface binding region with Factor H, and the three most C-terminal domains of both proteins display almost 99% identity on the amino acid level. CFHR1 lacks the N-terminal regulatory region of Factor H and consequently lacks regulatory functions on the C3 convertase level. The three regulators, CFHR1, Factor H, and FHL-1, are members of the Factor H protein family that also include four additional Factor H-related proteins (CFHR2–CFHR5). All members of the Factor H protein family share common structural features and are composed exclusively of short consensus repeats, which have high sequence similarities among each other (18).
S. pyogenes expresses several surface proteins that recruit the host complement regulators Factor H, FHL-1, CFHR1, and C4-binding protein (C4BP) to the bacterial surface. This allows the pathogen to control host complement attack, to inactivate complement effector proteins, and consequently to block C3b opsonization and the generation of inflammatory anaphylatoxins (C3a and C5a), as well as TCC formation and surface deposition. The known streptococcal binding proteins are M-protein, M-like proteins, and fibronectin-binding protein (Fba) (19, 20). The anti-phagocytic M-protein, which is expressed by S. pyogenes, shows high sequence variations and is therefore used for epidemiological typing. More than 140 GAS M-protein variants are identified on the basis of emm gene typing (19, 21–27).
Streptococcal collagen-like (Scl) proteins are expressed by all S. pyogenes M-types and have a conserved domain structure: a C-terminal cell wall membrane-spanning region, with an LPXTG cell wall-binding motif followed by an extracellular collagen-like domain with the repetitive GXY amino acid motif, and an adjacent N-terminal variable domain. The predicted structure indicates a “lollipop-like” domain organization, where the collagen-like domain forms a triple helical stalk and the variable domain folds into a globular head. S. pyogenes expresses two distinct Scl proteins, Scl1 and Scl2. The two bacterial proteins have a rather similar domain organization, but Scl1 includes an additional linker region between the cell wall membrane-spanning region and the collagen-like domain (28–33).
Recently the Scl1 of serotypes M6 and M55 (Scl1.6 and Scl1.55, respectively) were identified as CFHR1 and Factor H-binding surface protein (34). Binding was observed exclusively for Scl1 of the M6 and M55 serotypes, whereas Scl1 and Scl2 proteins of other serotypes, e.g. Scl1 of M1-type bacteria (Scl1.1), Scl1 and Scl2 of M28-type bacteria (Scl1.28 and Scl2.28), and others (34), did not bind. CFHR1 and Factor H bind to the variable domain of both Scl1.6 and Scl1.55 (34), which show 85% sequence identity. In contrast, the variable domain of nonbinding Scl proteins Scl1.1 and Scl2.28 have a lower level of identity of 35 and 40%, respectively, when compared with the binding Scl proteins. Scl proteins bind CFHR1 and Factor H and also additional human proteins, including α2β1 integrin, low density lipoprotein (LDL), fibronectin, laminin, and the thrombin-activatable fibrinolysis inhibitor (34–39).
GAS express Factor H-binding proteins, e.g. M-protein and Fba, that bind Factor H and FHL-1 via SCR7. Scl1, derived from S. pyogenes serotype M6 and M55 (Scl1.6 and Scl1.55), bind CFHR1 and Factor H, but the binding region within the two host proteins is thus far unknown. In the present study we localized the binding regions in the human CFHR1 and Factor H proteins for Scl1 interaction with the C termini of the two complement regulators by using recombinant deletion fragments. CFHR1 and Factor H compete for binding to both Scl1.6 and Scl1.55, indicating that the two human regulators bind the same epitope(s) within the streptococcal proteins. By peptide spot assays a linear Scl binding sequence with a length of five amino acid residues was identified in SCR4 of CFHR1. This sequence is also conserved in SCR19 of Factor H. In addition, several residues located within SCR20 of Factor H (or SCR5 CFHR1) are also relevant for binding to Scl1.6 and Scl1.55. Based on this unique binding profile for the CFHR1 and Factor H C-terminal regions, Scl1.6 and Scl1.55 proteins represent a new class of streptococcal CFHR1/Factor H-binding proteins. They bind a different region of the Factor H protein as compared with the previously identified streptococcal Factor H-binding proteins M-, M-like, and Fba protein. This difference in recognition and binding is relevant for GAS immune evasion as complement is blocked at multiple levels.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Scl Proteins
Production of recombinant Scl proteins has been reported (30, 41). Briefly, Escherichia coli strain DH5α was used in cloning experiments, and E. coli BL21 was used for protein expression. E. coli strains harboring plasmid constructs were routinely grown in Luria-Bertani (LB) liquid medium (BD Biosciences) supplemented with ampicillin (100 μg/ml). Recombinant Scl proteins were produced in the E. coli periplasm using the Strep-Tag II expression and purification system (IBA GmbH, Goettingen, Germany).
Expression of Recombinant Factor H Family Proteins and Fragments
Factor H fragments SCR8–11, SCR11–15, SCR8–20, SCR15–18, SCR15–19, SCR15–20, and SCR19–20, as well as Factor H SCR8–20-based mutant proteins (V1197A, W1183V, R1210C, and R1215G), Factor H SCR15–20-based mutant HepG (R1203E, R1206E, R1210S, K1230S, and R1231A), and Factor H family proteins CFHR2, CFHR3, CFHR4A, and FHL-1, were expressed using the baculovirus expression system described previously (42, 43). Briefly, following cultivation of insect cells of Spodoptera frugiperda (Sf9) at 28 °C in protein-free Grace medium (BioWhittaker), cells were infected with recombinant baculovirus. After 9 days the culture supernatant was harvested, and the recombinant proteins were purified by affinity chromatography using nickel-nitrilotriacetic acid-agarose (Qiagen).
The recombinant CFHR1 protein was expressed by using the Pichia pastoris expression system (Invitrogen). The CFHR1-encoding gene was cloned into the pPicZαB vector and subsequently introduced into P. pastoris X33. Expression was induced with 1% methanol. The recombinant protein-containing supernatant was harvested after 4 days of expression and purified by using the FPLC Äkta purifier (GE Healthcare) together with 1 ml nickel-chelating columns (HiTrap HP, GE Healthcare). The purified proteins were concentrated (Amicon Ultra-15), and buffer was changed to 1× Dulbecco's phosphate-buffered saline (DPBS, Lonza).
Enzyme-linked Immunosorbent Assay (ELISA)
The recombinant Scl proteins Scl1.1, Scl1.6, Scl1.55, and Scl2.28 (10 μg/ml in DPBS) were immobilized overnight at 4 °C onto a microtiter plate (Maxisorb, Nunc). All samples were probed in triplicate. Following washing, plates were blocked with 1× PBS containing 2% bovine serum albumin (BSA, Applichem) and 1× Roti-Block (Roth) for 1.5 h at 37 °C. Plates were washed three times, and the ligand protein, Factor H (Comptech) or CFHR1, respectively, was added with increasing amounts in a range of 0 to 20 μg/ml in DPBS. Following incubation for 1 h at room temperature, plates were washed extensively with 1× PBS containing 0.05% Tween 20 (Sigma). Bound proteins were detected with polyclonal Factor H antiserum (1:1000, Comptech) and a secondary horseradish peroxidase (HRP)-coupled anti-goat antibody (1:1000, Dako), each diluted in blocking buffer. Serum-derived CFHR1 was detected with monoclonal CFHR1 antibody JHD10 (1:1000, Prof. Dr. Wallich, Heidelberg University) and a secondary HRP-coupled anti-mouse antibody (1:1000, Dako) (17). The reaction was developed with the chromogen substrate 1,2-phenylenediamine dihydrochloride (OPD, Dako), and absorbency was measured at 490 nm.
For the reverse setting, Factor H, CFHRs, or Factor H fragments (SCR1–7, SCR8–11, SCR11–15, SCR15–20, SCR15–18, and SCR19–20) were immobilized (10 μg/ml), and Scl were used as ligand (15 μg/ml). Bound Scl proteins were detected with anti-Strep-Tag II-HRP.
Ionic Strength
Dependence was measured by using 1× PBS buffer (2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4) with increasing NaCl molarities ranging from 0 to 560 mm for protein diluents, blocking solution, and washing buffer. Factor H and CFHR1 were immobilized (10 μg/ml), and Scl (15 μg/ml) was added.
Heparin Blocking
Experiments were performed as described above. Upon immobilization of Scl (10 μg/ml), Factor H or CFHR1 (5 μg/ml) were added to the wells, together with increasing amounts of heparin sodium salt (Fluka) ranging from 0 to 512 μg/ml.
Catch ELISA
A catch ELISA was performed to measure the interaction of Factor H SCR8–20-based mutant proteins with Scl1.6, Scl1.55, and Scl2.28. The mutations were located at positions V1197A, W1183V, R1210C, and R1215G. In addition, a Factor H SCR15–20 fragment (a non-disease-associated protein) was added. This mutant construct harbors amino acid exchanges of five heparin-binding residues, i.e. R1203E, R1206E, R1210S, K1230S, and R1231A, that form a positively charged patch in SCR20. All mutants used in this assay showed impaired heparin and C3b binding.
Heparin binding was assayed as described above, with the following modifications. First, a Factor H SCR15–18-specific antiserum was immobilized (1:300 in DPBS) overnight at 4 °C. Following blocking, baculovirus supernatant containing the various mutant proteins was added and incubated for 2 h at room temperature. Two plates were prepared in parallel. The first plate was used to verify equal binding of mutant proteins by adding a polyclonal Factor H antiserum (1:1000, Comptech) and a secondary anti-goat-HRP antibody (1:1000, Dako). The second plate was used to analyze the Scl interaction. Then Scl was added (10 μg/ml) and detected with anti-Strep-Tag II-HRP.
Whole-cell ELISA
This was performed as follows. 25 ml of brain-heart infusion (BHI, Fluka) medium was inoculated with 0.5 ml of an overnight culture of GAS types M1, M6, M28, and M55. The cultures were grown at 37 °C for 4 h until mid-log phase. GAS at 5 × 106 colony-forming units in DPBS were added to each well and incubated overnight at 4 °C. Equal attachment was verified by staining bound bacteria with crystal violet in a parallel approach. The ligand proteins were added at 15 μg/ml, and the assay was developed as described above. The M-type-dependent antibody background level was measured and subtracted to compare ligand binding among the serotypes.
Competition ELISA
This was performed by adding different molar ratios of Factor H:CFHR1 (1:0, 1:0.5, 1:1, 1:2, 1:4, and 0:1) to immobilized Scl proteins (10 μg/ml). The ELISA was performed as described above. Bound Factor H was detected with a polyclonal rabbit Factor H SCR1–4 antiserum (1:1000) and a secondary anti-rabbit-HRP antiserum (1:1000, Dako).
Surface Plasmon Resonance (SPR) Studies
Protein-protein interactions were analyzed by the SPR technique using a Biacore 3000 instrument (Biacore AB) as described (44) with the following modifications. The recombinant Scl proteins (i.e. Scl1.1, Scl1.6, Scl2.28, and Scl1.55) were immobilized on the sensorchip by Strep-Tactin (IBA), which was fixed by standard amine coupling to the surface of a CM5 chip (3000 relative light units). All SPR experiments were performed in DPBS.
Peptide Spot Analysis
Forty peptides representing CFHR1 SCR4 and SCR5 (residues 202–330) with a length of 13 amino acids and an overlap of 10 amino acids were synthesized and coupled to a cellulose membrane (JPT, Berlin, Germany). The membranes were treated as described (44). Then 1 μg/ml Scl1.6 or Scl2.28 protein in DPBS was used. Bound protein was detected with 1:1000 anti-Strep-Tag II-HRP antibody.
SDS-PAGE and Western Blot Analysis
Samples were separated by SDS-PAGE using 12% gels. After transfer of the proteins onto nitrocellulose membranes (Protran, Whatman) by semidry blotting, the membranes were blocked with 4% (w/v) dried milk, 1% BSA, and 0.1% Tween 20 in DPBS (Lonza) overnight at 4 °C. Blots were incubated with the indicated primary antibodies for 1 h at room temperature followed by the corresponding secondary antisera coupled with horseradish peroxidase. Antibodies were diluted in blocking buffer. ECL plus solution (GE Healthcare) was used for protein detection, and images were taken using Bio-Imaging Systems MF-ChemiBIS 3.2 (Biostep).
Flow Cytometry
GAS types M1, M6, M28, and M55 were grown overnight at 37 °C in 10 ml of brain-heart infusion. Bacteria were washed twice with DPBS, and 1 × 108 GAS colony-forming units/sample were incubated with Factor H SCR15–20 (15 μg/ml), Factor H SCR15–18, CFHR1, or buffer without protein for 30 min at 37 °C. Next, bacteria were washed twice, a polyclonal antiserum (1:100, Comptech) that also detects CFHR1 and Factor H was added, and the mixture was incubated for 20 min at room temperature. Bacteria were washed again, a secondary anti-goat F′ab2 antibody coupled with Alexa 488 (1:100, Invitrogen) was added, and the mixture was incubated for 20 min at 4 °C. Bacteria were washed with DPBS and examined by flow cytometry (LSR II, BD Biosciences). Forward and sideward scatter were used to identify bacteria, and 10,000 events were counted.
Complement Cofactor Assay
The functional activity of Scl-bound Factor H was analyzed using a cofactor assay. First, Scl1.6, Scl1.55, and Scl2.28 protein (each at 10 μg/ml) was immobilized overnight at 4 °C. After washing (1× PBS and 0.05% Tween 20) and blocking (1× PBS, 2% BSA, and 1× Roti-Block), Factor H was added (15 μg/ml in DPBS), and the mixture was incubated for 1 h at room temperature. Wells without Factor H were prepared in parallel to verify Factor H specificity. Next, the plate was washed, and C3b (10 μg/ml in DPBS) together with Factor I (0.7 μg/ml in DPBS) was added and incubated for 30 min at 37 °C. Samples were collected, separated by SDS-PAGE, and analyzed by Western blotting. C3b cleavage was visualized using a C3-specific antiserum (1:1000, Calbiochem) and a secondary anti-goat-HRP antibody (1:2000, Dako).
In addition a cofactor assay was performed to determine the functional relevance of the Factor H and CFHR1 competition upon Scl binding. Various molar ratios of Factor H:CFHR1 (1:0, 1:0.5, 1:1, 1:2, 1:4, 1:8, and 0:1) were added to immobilized Scl proteins (10 μg/ml). C3b cleavage was assayed as described. In addition, competition of CFHR1 and Factor H was followed by ELISA (see “Competition ELISA” above).
CFHR1 TCC Inhibition Assay
Scl proteins Scl1.6, Scl1.55, and Scl2.28 or human serum albumin (each at 15 μg/ml) was immobilized for 1 h at 37 °C, washed, and blocked, and then CFHR1 and Factor H were added as ligands. CFHR1 and Factor H binding was verified with a polyclonal Factor H antiserum. In parallel, purified TCC compounds (Comptech) diluted in HEPES buffer (20 mm HEPES, 144 mm NaCl, 7 mm MgCl2, and 10 mm EGTA) were added to the wells. C5b6 (3.8 μl, 20 μg/ml) and C7 (5 μl, 20 μg/ml) were adjusted to 40 μl with HEPES buffer and added to the sample. This mixture was incubated for 10 min at room temperature, and then C8 and C9 (both at 5 μl, 20 μg/ml) were added. After incubation for 30 min at 37 °C, the samples were washed, and surface-deposited TCC was detected using a polyclonal SC5b9 (TCC)-reacting rabbit antiserum (1:1000, Comptech) and a secondary anti-rabbit HRP-coupled serum (Dako).
Hemolysis Assay
CFHR1, used at increasing amounts (0–2 μm) was incubated together with increasing amounts of Scl1.6, Scl1.55, or Scl2.28 (0–2 μm) in HEPES buffer. Samples were split; one aliquot was analyzed for CFHR1-Scl complex formation, whereas the other part was used in a hemolytic assay. The level of Scl-bound CFHR1 was identified using the catch ELISA described above, with the following modifications. Anti-Strep-Tag II antibody (1:400) was immobilized to bind Scl protein. Samples diluted 1:100 in DPBS were added to the wells and incubated for 1 h at room temperature, and bound complexes were detected with a polyclonal CFHR1 antiserum (1:1000) and a secondary HRP-coupled anti-goat antiserum (1:1000). Unbound CFHR1 protein was detected upon transfer of the supernatant, which contained free, non-absorbed CFHR1 protein to a new ELISA plate that had the immobilized JHD10 monoclonal antibody, specific for CFHR1 (1:1000).
The second part of the probe was assayed in a hemolysis assay. The sample (10 μl) was incubated together with C5b6 (4.5 μl; 20 μg/ml), C7 (6 μl; 20 μg/ml), and 30 μl of HEPES buffer containing 2 × 108 sheep erythrocytes (Rockland). The volume was adjusted to 48 μl with HEPES buffer. Samples were incubated for 10 min at room temperature, and then C8 and C9 (each 6 μl, 20 μg/ml) were added. The reaction was incubated for 30 min at 37 °C with agitation and then centrifuged for 2 min at 5000 rpm. Erythrocyte lysis was determined by measuring the absorbance at 414 nm.
RESULTS
CFHR1 and Factor H Binding to Scl
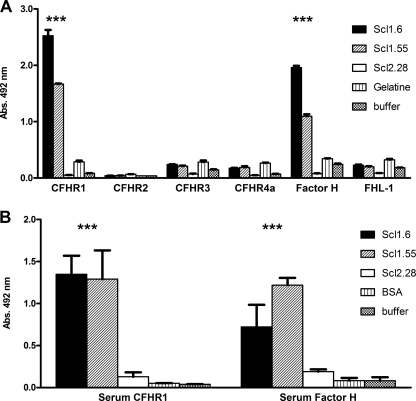
To characterize the role of Scl proteins for immune and complement escape of GAS, we analyzed the binding of recombinant Scl1.6 and Scl1.55 to four immobilized CFHR proteins (i.e. CFHR1, CFHR2, CFHR3, and CFHR4A), as well as to Factor H and FHL-1. Both Scl1.6 and Scl1.55 bound to immobilized CFHR1 and Factor H but did not bind to any other CFHR protein or FHL-1 (Fig. 1A). In this assay Scl1.6 bound the two human complement regulators with stronger intensity as compared with Scl1.55. Streptococcal Scl2.28 was used as a negative control, and this protein did not bind to any tested Factor H family protein. Binding of CFHR1 and Factor H to immobilized Scl proteins was confirmed using 10% human serum as a protein source (Fig. 1B). Bound CFHR1 was detected with the specific mouse monoclonal antibody JHD10 and bound Factor H with Factor H-specific antiserum. Serum-derived CFHR1 and Factor H bound to immobilized streptococcal Scl1.6 and Scl1.55 proteins but not the related Scl2.28 protein.
FIGURE 1.
Binding of Scl proteins to Factor H protein family members. A, CFHR1, CFHR2, CFHR3, and CFHR4A, as well as Factor H and FHL-1, were immobilized, and binding of streptococcal proteins Scl1.6, Scl1.55, and Scl2.28 was followed using a Strep-Tag II-specific HRP-coupled antibody. Streptococcal proteins Scl1.6 and Scl1.55 bound to CFHR1 and Factor H but not to any other Factor H family protein. Scl2.28 bound to none of the tested proteins (***, p < 0.001). B, 10% normal human serum was added to immobilized Scl1.6, Scl1.55, or Scl2.28, and bound CFHR1 and Factor H derived from human plasma were detected by a monoclonal antibody specific for CFHR1 (JHD10) and by a polyclonal antiserum specific for Factor H SCR1–4. The human regulators bound to Scl1.6 and Scl1.55 but not to Scl2.28. Mean values from triplicate wells of a representative experiment ± S.D. are shown (total of three independent experiments; ***, p < 0.001).
Dose-dependent Binding of CFHR1 and Factor H to Scl1.6 and Scl1.55
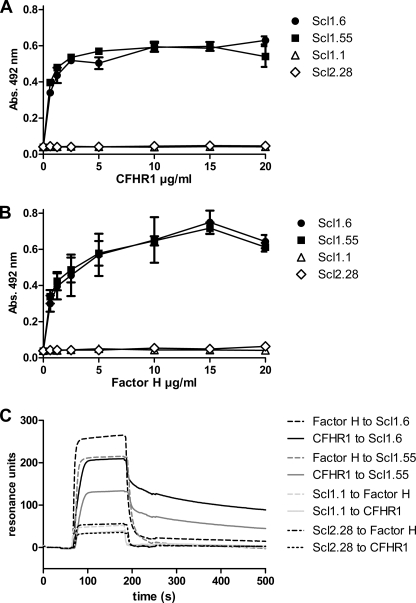
CFHR1, a regulator of the C5 convertase and terminal complement pathway, and Factor H, a regulator of the C3 convertase, bind to streptococcal Scl1.6 and Scl1.55 proteins. The two human complement inhibitors bound to immobilized Scl1.6 as well as to Scl1.55 in a dose-dependent manner (Fig. 2, A and B). CFHR1 binding was saturated at a concentration of ∼5 μg/ml and Factor H binding at ∼10 μg/ml. Given the plasma concentrations of CFHR1 and Factor H of ∼100 and 500 μg/ml, respectively, the binding is relatively efficient. Neither CFHR1 nor Factor H bound to Scl proteins Scl1.1 and Scl2.28, expressed by the streptococcal strains serotypes M1 and M28 (Table 1).
FIGURE 2.
Dose-dependent binding of CFHR1 and Factor H to streptococcal Scl1.6 and Scl1.55. Purified CFHR1 (A) and Factor H (B) bound dose-dependently to immobilized Scl1.6 (black circles) and Scl1.55 (black squares). No binding was detected for Scl1.1 (white triangles) and Scl2.28 (white diamonds). Error bars indicate S.D. C, binding of CFHR1 and Factor H to immobilized Scl1.6 (black), Scl1.55 (gray), Scl1.1 (light gray), and Scl2.28 (black dotted/dashed dotted) was analyzed by surface plasmon resonance SPR. CFHR1 (solid lines) and Factor H (dashed lines) bound to Scl1.6 and Scl1.55 but not to Scl1.1 or to Scl2.28. A representative experiment of three is shown.
TABLE 1.
S. pyogenes strains used in this study
| GAS strain | Scl protein | M-type | Complement regulator binding to intact bacteria | Complement regulator binding to purified Scl proteins |
|---|---|---|---|---|
| MGAS6169 | Scl1.6 | M6 | CFHR1 and Factor H SCRs 15–20 | CFHR1 and Factor H |
| MGAS1863 | Scl1.55 | M55 | CFHR1 and Factor H SCRs 15–20 | |
| MGAS5005 | Scl1.1 | M1 | None | |
| MGAS6143 | Scl1.28, Scl2.28 | M28 | CFHR1 and Factor H SCRs 15–20 |
Binding was further characterized by SPR. CFHR1 and Factor H bound to immobilized Scl1.6 and Scl1.55 but did not bind to either Scl1.1 or Scl2.28 (Fig. 2C). The two human regulators showed different binding profiles, i.e. CFHR1 had a slower association than Factor H but also a slower dissociation profile. The two human complement regulators, CFHR1 and Factor H, bound to the streptococcal proteins Scl1.6 and Scl1.55, with CFHR1 binding appearing the stronger.
CFHR1 and Factor H Bind Scl1.6 and Scl1.55 via Their C-terminal Surface Binding Regions
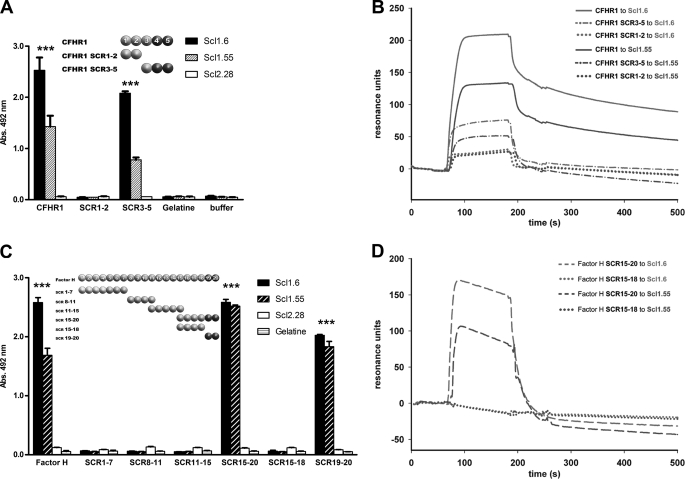
Next we aimed to localize the binding region of both human regulators for the streptococcal proteins. To this end recombinant fragments of both CFHR1 and Factor H were used. The C-terminal CFHR1 fragment, SCR3–5, similar to the intact full-length protein CFHR1, bound to both streptococcal proteins Scl1.6 and Scl1.55 but not to Scl2.28 (Fig. 3A). The N-terminal fragment CFHR1 SCR1–2 did not bind to any Scl protein.
FIGURE 3.
Binding of CFHR1 and Factor H, and of deletion fragments to Scl1.6 and Scl1.55. A, binding of Scl1.6, Scl1.55, and Scl2.28 to CFHR1 and to the two CFHR1 fragments, SCR1–2 and SCR3–5, was assayed. Scl1.6 and Scl1.55, but not Scl2.28, bound to CFHR1 and the C-terminal CFHR1 fragment SCR3–5. No Scl protein bound to the N-terminal CFHR1 fragment SCR1–2. Mean values from triplicate wells of a representative experiment ± S.D. are shown (total of three independent experiments). The insert shows the schematic domain structure of CFHR1 and that of the two fragments (***, p < 0.001). B, binding of CFHR1 as well as CFHR1 fragments to immobilized Scl1.6 (light gray) and Scl1.55 (dark gray) was assayed by SPR. CFHR1 (solid line) and the C-terminal fragment SCR3–5 (dashed-dotted line) bound to Scl1.6 and Scl1.55. The N-terminal CFHR1 SCR1–2 fragment (dotted line) did not bind to Scl1.6 and Scl1.55. C, binding of Scl1.6, Scl1.55, and Scl2.28 to Factor H, and the indicated immobilized Factor H fragments was assayed by ELISA. Scl1.6 and Scl1.55 bound to full-length Factor H as well as the C-terminal Factor H fragments SCR15–20 and SCR19–20. Scl2.28 did not bind to any Factor H fragment tested. Mean values from triplicate wells of a representative experiment ± S.D. are shown (total of three independent experiments). The insert shows the schematic domain structure of Factor H and that of the various fragments (***, p < 0.001). D, binding was also analyzed by SPR. Again, the C-terminal Factor H SCR15–20 (dashed line) bound to immobilized Scl1.6 (light gray) and Scl1.55 (dark gray). The Factor H SCR15–18 fragment (dotted line) did not bind.
Binding was confirmed by SPR analysis. Again, C-terminal SCR3–5 bound to the immobilized streptococcal proteins Scl1.6 and Scl1.55. The N-terminal CFHR1 SCR1–2 fragment did not bind. In this setup, however, the full-length CFHR1 protein bound more strongly than the C-terminal fragment (Fig. 3B). Thus CFHR1 binds both streptococcal proteins, i.e. Scl1.6 and Scl1.55, via the C-terminal surface binding region.
Factor H also bound to the two streptococcal proteins Scl1.6 and Scl1.55. Using several Factor H fragments, the binding region for the two streptococcal proteins was localized to C-terminal SCR19–20. Scl1.6 and Scl1.55 bound to the two C-terminal Factor H constructs SCR15–20 and SCR19–20, whereas Scl proteins did not bind to constructs lacking SCR19–20, i.e. SCR1–7, SCR8–11, SCR11–15, and SCR15–18 (Fig. 3C). Similar results were obtained in the reverse setting with immobilized streptococcal proteins (data not shown). Thus the C-terminal surface binding region of Factor H, i.e. SCR19–20, contacts the two streptococcal proteins.
Binding of the C-terminal Factor H construct SCR15–20 to Scl1.6 and Scl1.55 was also assayed by SPR. In this case, the association profile of the Factor H fragment was stronger for Scl1.6 as compared with Scl1.55. Again, Factor H SCR15–18 fragment did not bind (Fig. 3D). In summary, both CFHR1 and Factor H bound to the two streptococcal proteins, Scl1.6 and Scl1.55, via their C-terminal surface binding regions. Binding was detected in both orientations either with immobilized CFHR1/Factor H or with immobilized streptococcal proteins (data not shown), and in this assay both human regulators bound more strongly to Scl1.6 than to Scl1.55.
Identification of a Linear Scl1-binding Motif within SCR4 of CFHR1
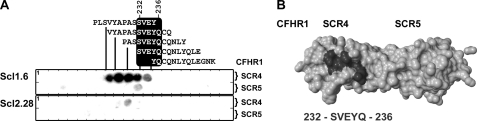
Having localized the Scl1-binding region within the C-terminal surface binding region of both CFHR1 and Factor H, we aimed to identify linear binding sequences within this region. To this end, peptides with a length of 13 amino acids and an overlap of 10 residues that cover the entire sequence of CFHR1 SCR4 and SCR5 (residues 202–330) were spotted onto a membrane, and binding of Scl1.6 was assayed. Scl1.6 bound to five consecutive peptides that represent the sequence 223PLSVYAPASSVEYQCQNLYQLEGNK247 (core residues are shown in bold) (Fig. 4A, upper panel). GAS protein Scl2.28 did not bind to any peptide, thus confirming the specificity of the Scl1.6-CFHR1 interaction (Fig. 4A, lower panel). The core residues SVEYQ, which represent the major linear Scl1-binding motif in SCR4 of CFHR1, are conserved in the corresponding SCR19 of Factor H. The positions of the residues that form this linear Scl1.6-binding motif were localized in a structural model of CFHR1 SCR4–5 (Fig. 4B). The five binding residues are surface-exposed and thus accessible for ligand interaction.
FIGURE 4.
Identification of a linear Scl1.6-binding motif in SCR4 of CFHR1. A, linear peptides representing SCR4–5 of CFHR1 (residues 202–330), each with a length of 13 amino acids and an overlap of 10 residues, were spotted onto a membrane, and binding of Scl1.6 (upper panel) and Scl2.28 (lower panel) was followed. Scl1.6 bound to five spots representing the core motif, 232SVEYQ236 (upper panel). The nonbinding Scl2.28 did not bind to any peptide (lower panel). SCR4 of CFHR1 and SCR19 of the Factor H protein share sequence identity, and thus the binding motif is also contained in SCR19 (1133SVEYQ1137) of Factor H. B, structure of SCR4 and SCR5 of CFHR1 (corresponding to SCR19 and SCR20 of Factor H). The residues that form the linear binding motif are shown in black and are surface-exposed (Protein Data Bank code: 2G7I).
CFHR1 and Factor H Binding to Scl1.6 and Scl1.55 Is Influenced by Ionic Strength
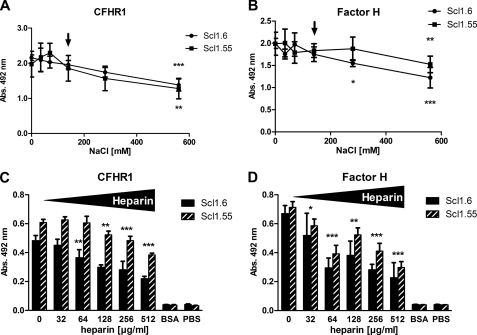
To characterize the nature of the CFHR1-Factor H interaction with the two streptococcal virulence factors the effect of ionic strength (NaCl) was assayed. First, the influence of different salt concentrations for CFHR1 or Factor H binding to immobilized Scl1.6 and Scl1.55 was tested. Both complement regulators bound to the streptococcal proteins in the absence of NaCl (Fig. 5, A and B). Increasing NaCl concentrations reduced the binding of both complement regulators, and the effect was dose-dependent. At physiological NaCl concentrations (140 mm; Fig. 5A, marked by an arrow), the binding of CFHR1 to both Scl1.6 and Scl1.55 was reduced by ∼8–10%, and at 560 mm binding was reduced by ∼35% (Fig. 5A).
FIGURE 5.
Ionic strength (NaCl) and heparin affect CFHR1 and Factor H binding to Scl1.6 and Scl1.55. A, the influence of ionic strength on CFHR1 binding to immobilized Scl1.6 and Scl1.55 was analyzed by increasing the NaCl concentration. CFHR1 bound to immobilized Scl proteins in the absence of NaCl, and NaCl reduced binding in a dose-dependent manner. B, similarly, NaCl affected the interaction of Factor H with Scl1.6 and Scl1.55. The arrows (A and B) indicate the physiological NaCl concentration. C, increasing concentrations of heparin dose-dependently affected the binding of CFHR1 to immobilized Scl1.6 and Scl1.55. D, similarly, heparin affected the binding of Factor H to immobilized streptococcal Scl1.6 and Scl1.55 proteins. Again, the effect was dose-dependent. The mean values derived from at least three separate experiments ± S.D. are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001; versus 0 mm NaCl or 0 μg/ml heparin.
Similarly, NaCl affected Factor H binding to the streptococcal proteins. At physiological concentrations binding was reduced by 12 and 8%, and at higher NaCl concentrations by 39 and 23% (Fig. 5B). Thus NaCl affected the binding of both CFHR1 and Factor H to Scl1.6 and Scl1.55 at comparable levels, suggesting a related interaction. The modest effect at the physiological concentration (140 mm) demonstrates that CFHR1·Scl and Factor H·Scl complexes form under physiological conditions.
Heparin Influences Factor H and CFHR1 Binding to Scl1.6 and Scl1.55
The C terminus of CFHR1 and Factor H includes several residues that are relevant for heparin binding (17, 45). Therefore we assayed the effect of heparin on CFHR1 and Factor H binding to immobilized Scl1.6 and Scl1.55. Both human regulators bound to the streptococcal proteins in the absence of heparin (Fig. 5, C and D). Heparin decreased the binding of CFHR1 to Scl1.6 and Scl1.55 in a dose-dependent manner (Fig. 5C). At 512 μg/ml, heparin inhibited CFHR1 binding to Scl1.6 by 55% and to Scl1.55 by 37%. Similarly, heparin affected Factor H binding to both streptococcal proteins (Fig. 5D). At 512 μg/ml, heparin inhibited Factor H binding to Scl1.6 by 75% and to Scl1.55 by 58%. Thus heparin affects the interaction of both human complement regulators with each of the streptococcal proteins.
Disease-associated Factor H Variants Do Not Bind to Scl1.6 and Scl1.55
Factor H bound to the two streptococcal proteins Scl1.6 and Scl1.55 with the C-terminal surface binding region; this binding was affected by both NaCl and by heparin.
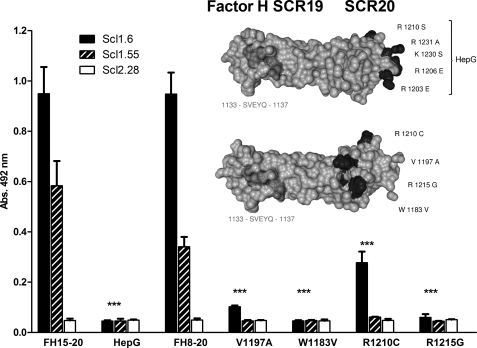
First, binding of the C-terminal Factor H SCR15–20 mutant “HepG” was assayed; it has five heparin-interacting residues in SCR20 of Factor H mutated (i.e. R1203E, R1206E, R1210S, K1230S, and R1231A) and does not bind heparin. This Factor H mutant construct was attached via an immobilized antibody to allow proper orientation and accessibility of the protein. Scl1.6 and Scl1.55 were applied to the fluid phase, and bound proteins were detected with a specific antibody. None of the Scl1 proteins bound to the immobilized HepG mutant, but they did bind the wild-type SCR15–20 construct (Fig. 6).
FIGURE 6.
Mutations in SCR20 and also disease-associated sequence variants affect Factor H binding to Scl1.6 and Scl1.55. Factor H SCR8–20, as well as four fragments that include aHUS-associated mutations (R1210C, V1197A, R1215G, and W1183V), and Factor H SCR15–20, which has heparin-binding residues exchanged (HepG), were attached via an immobilized monoclonal antibody (equal immobilization was verified in a parallel approach). Binding of Scl1.6, Scl1.55, and Scl2.28 applied to the fluid phase was detected using a Strep-Tag II-specific HRP-coupled antibody. Scl1.6 and Scl1.55, but not Scl2.28, bound to the unmodified fragment Factor H SCR8–20 and to Factor H SCR15–20, but the streptococcal proteins did not bind to any of the mutant proteins. Scl1.6 bound weakly to the Factor H SCR8–20 R1210C mutant construct. The mean values from triplicate wells of a representative experiment ± S.D. are shown (total of three independent experiments, ***, p < 0.001 versus no mutated proteins). The insert shows the structural model of SCR19–20 of Factor H (Protein Data Bank code: 2G7I). The positions of the five exchanged amino acids are indicated for the HepG mutant (upper panel). The disease-associated amino acids that were exchanged individually in four different aHUS cases are shown in black (lower panel). The linear Scl-binding motif is marked in dark gray.
Atypical hemolytic uremic syndrome (aHUS)-associated mutations that cluster within the C-terminal recognition region of Factor H affect heparin, C3b, and human cell binding (45, 46). Therefore, we investigated whether mutations in the C terminus of Factor H affect binding to Scl1.6 and Scl1.55. Binding of four aHUS-associated C-terminal Factor H SCR8–20 mutant fragments, which harbor the disease-associated mutations V1197A, W1183V, R1210C, and R1215G, was assayed. Again, none of the Scl1 variants bound to aHUS-associated mutants V1197A, W1183V, and R1215G. As expected, both Scl1 variants bound to wild-type construct SCR8–20 (Fig. 6). Scl1.6 bound weakly to mutant Factor H SCR8–20 with the R1210C exchange, but binding was of relatively weak intensity (∼30%). Thus, mutations of positively charged residues in SCR20 of Factor H, as well as aHUS-associated single amino acid substitutions, affected and blocked Scl-Factor H interaction.
CFHR1 and Factor H Bind to Intact Streptococci
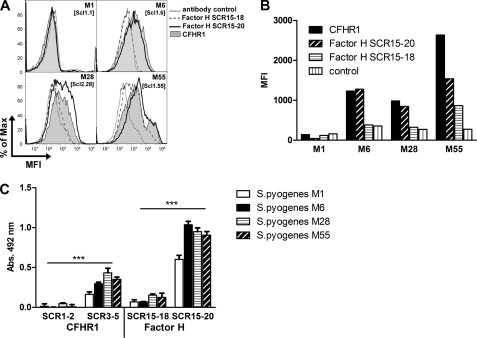
CFHR1 and Factor H bind to Scl1.6 and Scl1.55 via their C-terminal surface binding regions. We therefore asked whether the two host regulators use the same C-terminal region to bind to intact bacteria. Binding of CFHR1 and Factor H C-terminal constructs was compared for the streptococcal serotypes M6 and M55, which express the CFHR1- and Factor H-binding Scl1.6 and Scl1.55 proteins. In addition, binding was also assayed for GAS serotypes M1 and M28 that express the non-CFHR1- and non-Factor H-binding Scl proteins Scl1.1 and Scl2.28, respectively. Binding of CFHR1 and of the C-terminal Factor H fragments, SCR15–20 and SCR15–18, to intact streptococci was assayed by flow cytometry (Fig. 7, A and B) using a polyclonal antiserum that detects CFHR1 and Factor H. Both CFHR1 and Factor H SCR15–20 bound strongly to serotypes M6 and M55 (CFHR1, mean fluorescence intensity (MFI) 1278 and 1541; SCR15–20, MFI 1231 and 2638) (Fig. 7B) and with lower intensity to the M28-type strain (CFHR1, MFI 847; SCR15–20, MFI 984). No binding was observed for serotype M1, which expresses the Scl1.1 protein. Factor H SCR15–18, which lacks the C-terminal Scl1-binding region, SCR19–20, did not bind to bacteria of serotypes M1, M6, and M28 and bound weakly to M55-type bacteria. Thus CFHR1, as well as the C-terminal surface binding region of Factor H, binds to intact bacteria. We hypothesized that binding of the two human regulators to GAS strain serotypes M6 and M55 is mediated by Scl1.6 and Scl1.55 proteins. Binding to the M28-type strain suggests the presence additional binding proteins.
FIGURE 7.
CFHR1 and Factor H bind to various S. pyogenes serotypes. A and B, binding of CFHR1, Factor H SCR15–20, or Factor H SCR15–18 to S. pyogenes M1-, M6-, M28- and M55-type, which express Scl1.1, Scl1.6, Scl2.28, and Scl1.55, respectively, was assayed by flow cytometry (A), and the MFI are shown (B). CFHR1 and Factor H SCR15–20 bound to the Scl1.55-expressing M55-type and to the Scl1.6-expressing M6-type bacteria. Both proteins also bound with weaker intensity to the M28-type streptococci, which express the non-CFHR1/C-terminal Factor H-binding Scl1.28 and Scl2.28 surface proteins. The two human proteins did not bind to M1-type streptococci. Factor H SCR15–18 did not bind to any of the tested streptococcal serotypes. One representative experiment of five independent experiments is shown. C, binding of CFHR1 and of Factor H fragments to immobilized S. pyogenes serotypes M1, M6, M28, and M55 was analyzed by whole-cell ELISA. Bound proteins were identified with a polyclonal antiserum that detects CFHR1 and Factor H. The C-terminal CFHR1 SCR3–5, but not the N-terminal CFHR1 SCR1–2 construct, bound to all four streptococcal serotypes, however with different intensities. Factor H construct SCR15–20 also bound, whereas construct SCR15–18 did not bind. The background intensities were subtracted to allow direct comparison of the four serotypes. The mean values derived from at least three separate experiments ± S.D. are shown (***, p < 0.001).
The role of the C-terminal surface binding region of CFHR1 and Factor H was confirmed using a whole-cell ELISA. Streptococci were attached to a microtiter plate, and binding of CFHR1 and Factor H fragments was analyzed (Fig. 7C). In this assay, the C-terminal CFHR1 construct, i.e. SCR3–5 but not the N-terminal construct CFHR1 SCR1–2, bound to all four streptococcal serotypes, i.e. M1, M6, M28, and M55. Similarly, the C-terminal construct of Factor H SCR15–20, but not construct SCR15–18, bound to all four serotypes. The discrepancy observed for Factor H SCR15–18 in binding to S. pyogenes serotype M55 may be due to different methods applied. Flow cytometry revealed weak binding (Fig. 7B), whereas a whole-cell ELISA showed no binding of the same fragment to strain M55 (Fig. 7C). The two C-terminal constructs bound strongly to M6-, M28-, and M55-type GAS and weakly to the M1-type bacteria. This pattern reflects the results obtained by flow cytometry, where residual binding to M1-type bacteria was also observed.
Depending on the assay, each of the four GAS M-types, i.e. M1, M6, M28, and M55, bind CFHR1 as well as the C terminus of Factor H. Scl1.6 and Scl1.55 expressed by S. pyogenes serotypes M6 and M55 bind the two regulators via their C-terminal domains, but the Scl1.1 and Scl2.28 surface proteins expressed by serotype M1 and M28 do not bind to the two human complement regulators. Binding of the CFHR1 and the C-terminal fragment of Factor H at least to M28-type bacteria argues for the existence of additional binding proteins expressed on the surface GAS.
CFHR1 and Factor H Compete for Binding to Scl1.6 and Scl1.55 and Modulate Complement Function
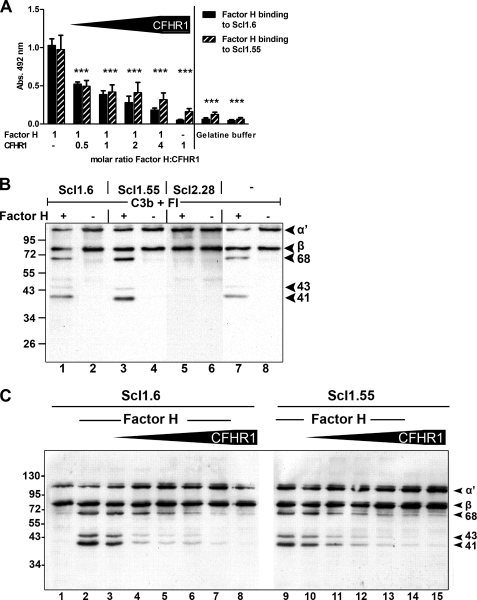
CFHR1 and Factor H, which are both present in human plasma, bind Scl1.6 and Scl1.55 proteins with their conserved C-terminal surface attachment regions. We therefore asked whether the two human proteins compete for binding to streptococcal Scl1 proteins. To this end, the effect of increasing amounts of CFHR1 on Factor H binding to Scl1 proteins was assayed (Fig. 8A). Factor H binding to both Scl1.6 and Scl1.55 was reduced upon increasing the concentration of CFHR1. Thus, CFHR1 and Factor H compete for binding to both streptococcal proteins. Interestingly, at the physiological CFHR1:Factor H molar ratio of ∼1:2, Factor H binding was reduced by ∼50%, suggesting that binding of equal amounts of both regulators bind to these streptococcal proteins.
FIGURE 8.
Factor H binding to Scl proteins is influenced by CFHR1, CFHR1 competes with Factor H for binding, and CFHR1 affects cofactor activity of Scl-bound Factor H. A, Factor H bound to Scl1.6 and Scl1.55 (columns 1 and 2). CFHR1 competes with Factor H for binding, and increasing amounts of CFHR1 affected Factor H binding in a dose-dependent manner (columns 3–10). Bound Factor H was detected with a Factor H SCR1–4-specific antiserum. Mean values from triplicate wells of a representative experiment ± S.D. are shown (total of three independent experiments). ***, p < 0.001 versus column 1 and 2. B, Factor H bound to Scl1.6 and Scl1.55 retains complement regulatory activity. Factor H bound to the indicated Scl proteins; then C3b and Factor I was added, and after incubation the mixture was separated by SDS-PAGE and transferred to a membrane. Cofactor activity of bound Factor H was analyzed by assaying C3b cleavage products by Western blotting. C3b cleavage results in the appearance of the α′ 68-, α′ 43-, and α′ 41-kDa fragments for Factor H bound to Scl1.6 (lane 1) or Scl1.55 (lane 3). A control reaction with Factor H in the fluid phase was assayed in lane 7. A representative experiment of three is shown. C, CFHR1 affects Factor H-mediated complement control at the level of C3 convertase. Factor H, used at constant amounts together with increasing concentrations of CFHR1, bound to immobilized Scl1.6 or Scl1.55 at molar ratios of Factor H:CFHR1 1:0.5, 1:1, 1:2, 1:4, and 1:8 (lanes 3–7 and lanes 10–14). Following washing, C3b and Factor I were added, and after incubation the mixture was separated by SDS-PAGE and transferred to a membrane. Cofactor activity of bound Factor H was analyzed by assaying the appearance of C3b cleavage products by Western blotting using a polyclonal C3b antiserum. C3b cleavage is identified by the appearance of the α′ 68-, α′ 43-, and α′ 41-kDa fragments. CFHR1, by displacing Factor H, affects Factor H cofactor activity on the level of the C3 convertase. A representative experiment of three is shown.
Factor H bound to Scl1.6 and Scl1.55 is functionally active and acts as cofactor for Factor I in C3b cleavage, as revealed by the appearance of specific C3b cleavage products, i.e. α′ 68 kDa, α′ 43 kDa, and α′ 41 kDa (Fig. 8B, lanes 1 and 3) (34). As expected, no cofactor activity was observed in samples containing the Scl2.28 protein, which does not bind Factor H (Fig. 8B, lane 5).
CFHR1 lacks cofactor activity at the level of C3 convertase but instead acts as a regulator of the C5 convertase and of TCC formation. On the basis of the competitive binding to Scl1 proteins, we investigated whether CFHR1 binding to Scl1 affects complement regulatory activity of Factor H at the level of C3 convertase. Therefore, the effect of increasing amounts of CFHR1 for cofactor activity of Scl1-bound Factor H was assayed. Factor H bound to Scl1.6 and Scl1.55 in the presence of increasing concentrations of CFHR1. After addition of a constant amount of C3b and Factor I and upon incubation, samples were separated by SDS-PAGE and then transferred to a membrane, and C3b degradation fragments were identified by Western blotting. Increasing concentrations of CFHR1 reduced Factor H-mediated cofactor activity, as evidenced by the decreased intensity of the specific C3b cleavage products, i.e. α′ 68 kDa, α′ 43 kDa, and α′ 41 kDa fragments (Fig. 8C). This effect was dose-dependent. At the physiological molar ratio of CFHR1:Factor H of 0.5:1, C3b cleavage was reduced by ∼30% (Fig. 8C, lanes 3 and 10), as determined by densitometry of the C3b α′ 43 kDa band. The effect was even more pronounced at higher CFHR1:Factor H ratios (Fig. 8C, lanes 4–7 and 11–14). At the highest ratio of 8:1, cofactor activity was reduced by almost 90% (Fig. 8C, lanes 7 and 14).
CFHR1 Bound to Scl1.6 and Scl1.55 Inhibits TCC Deposition
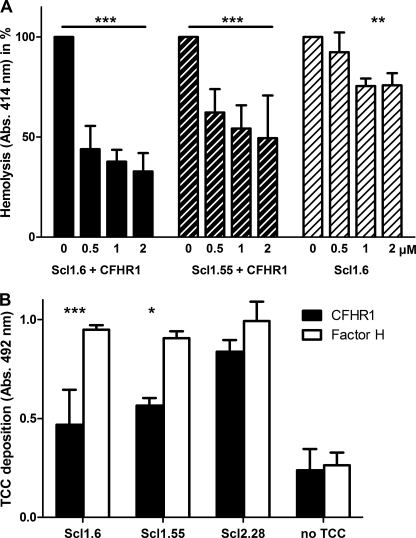
CFHR1 inhibits TCC formation by the terminal complement pathway (17). First, complex formation of CFHR1 and Scl proteins was analyzed, and also the level of free, uncomplexed CFHR1 was assayed. The other part of the sample was tested in a hemolytic assay with sheep erythrocytes to evaluate the regulatory activity of CFHR1 complexed with Scl1.6 or Scl1.55. CFHR1 formed complexes with Scl1.6 and Scl1.55 in fluid phase (supplemental Fig. 1A). Complex formation was dose-dependent, and the level of free, uncomplexed CFHR1 was rather low. Samples were used for hemolysis assay. Increasing amounts of CFHR1·Scl complexes or Scl1.6 alone were first added to purified terminal complement compounds C5b6, C7, C8, and C9 and then incubated with sheep erythrocytes. After incubation TCC-mediated erythrocyte lysis was recorded. In the absence of CFHR1, TCC was formed, and sheep erythrocytes were lysed (Fig. 9A, 0 μm). Increasing amounts of CFHR1·Scl1.6 and CFHR1·Scl1.55 complexes inhibited TCC-mediated erythrocyte lysis. This effect was dose-dependent, and at 2 μm CFHR1 lysis was reduced by 70 and 50%, respectively. In the absence of CFHR1, Scl1.6 alone affected lysis to a much smaller extend. CFHR1 did not form complexes with Scl2.28 (supplemental Fig. 1B).
FIGURE 9.
CFHR1 bound to Scl1.6 and Scl1.55 inhibits TCC formation. A, the functional relevance of CFHR1·Scl1.6 and CFHR1·Scl1.55 complexes was analyzed in a hemolysis assay with sheep erythrocytes. TCC formation on sheep erythrocytes was induced with C5b6, C7, C8, and C9 and assayed by following erythrocyte lysis. Lysis was inhibited by increasing amounts of CFHR1·Scl1.6 and CFHR1·Scl1.55 complexes. Scl1.6 alone had a minor effect on hemolysis that was not amplified by increasing amounts. The mean values ± S.D. of three independent experiments are shown. B, TCC deposition was analyzed by the addition of TCC compounds C5b6, C7, C8, and C9 to CFHR1 bound to immobilized Scl1.6 and Scl1.55. For the negative control sample (no TCC), no C9 was added. TCC was detected using a TCC-specific antiserum. CFHR1 bound to Scl1.6 and Scl1.55 significantly reduced TCC deposition, whereas bound Factor H did not affect the TCC. Scl2.28, which bound to neither CFHR1 nor Factor H, did not influence TCC deposition. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Additionally, the role of CFHR1 bound to Scl proteins on C5b9 (TCC) deposition was assayed. CFHR1 bound to Scl1.6 or to Scl1.55 (supplemental Fig. 1C) was functionally active and inhibited TCC formation by 45 and 33%, respectively (Fig. 9B). Factor H, which lacks TCC regulatory activity, also bound to Scl1.6 and Scl1.55 and did not block TCC formation. As CFHR1 does not bind to Scl2.28, no TCC inhibition was detectable when this streptococcal protein was immobilized. Thus, CFHR1 bound to Scl1.6 and Scl1.55 is functionally active and acts as a regulator of the terminal complement pathway.
DISCUSSION
Group A streptococci utilize soluble human complement regulators to evade host complement attack. Here, we have characterized the binding of the TCC inhibitor CFHR1 and of the C3 convertase regulator Factor H to streptococcal collagen-like proteins Scl1.6 and Scl1.55, which are expressed by GAS serotypes M6 and M55. CFHR1 and Factor H bind to the two streptococcal proteins, and the two human regulators bind via their conserved C-terminal attachment region. In contrast, no additional member of the Factor H protein family, neither CFHR2, CFHR3, CFHR4A, nor FHL-1, bound to these streptococcal proteins. Scl1.6 and Scl1.55 are the first streptococcal proteins found to recruit host regulators via their conserved C-terminal surface recognition regions. All previously reported complement regulator-binding GAS proteins, i.e. M-, M-like, and Fba, bind Factor H and FHL-1 via domain SCR7 and do not bind CFHR1 (19, 21, 22).
Purified Factor H and CFHR1 bound more strongly to Scl1.6 followed by Scl1.55. On the contrary, when normal human serum was used, both regulators bound more strongly to Scl1.55 than to Scl1.6. CFHR1 and Factor H are both present in human serum, and as both proteins bind simultaneously they also compete for Scl binding. Furthermore, additional Factor H- and/or CFHR1-binding proteins such as C3 may influence the interaction of the human complement regulators with the bacterial proteins, which likely explains the discrepancy in binding.
The three most C-terminal SCRs, i.e. SCR3–5 of CFHR1 and SCR18–20 of Factor H, represent the conserved surface attachment region that displays 99% sequence identity. Within SCR4 of CFHR1, a linear Scl1.6-binding motif was identified with the core sequence 232SVEYQ236. This motif, which is also present in SCR19 of Factor H, is surface-exposed and thus accessible for ligand binding. In general PepSpot assays are useful for identifying linear binding regions. However, this approach does not identify a more complex three-dimensional folding as occurs for SCR domains. Thus, the linear binding motif does not exclusively mediate interaction. Single amino acid residues within the most C-terminal domain of Factor H also affect the interaction with Scl1.6 and Scl1.55. Scl1.6 and Scl1.55 did not bind to a heparin mutant (HepG) of Factor H, which has five heparin-binding residues exchanged. In addition four proteins carrying aHUS-associated single amino acid mutations within SCR20 of Factor H (V1197A, W1183V, R1210C, and R1215G) bound to neither Scl1.6 nor Scl1.55. This lack of binding shows that several amino acid residues within the most C-terminal domain are relevant for this interaction. Importantly, our data suggest that certain genetic abnormalities in Factor H affect binding to GAS surfaces.
NaCl and heparin influence the binding of both human regulators to the streptococcal Scl1.6 and Scl1.55 proteins. At physiological levels, NaCl (140 mm) reduced the CFHR1/Factor H·Scl interaction by about 10%, and heparin (512 μg/ml) reduced the interaction by ∼50%. The C-terminal surface binding regions of both CFHR1 and Factor H include several heparin-interacting residues, which are positioned in the direct vicinity of the linear Scl-binding residues. This suggests that heparin may occlude the Scl-binding region in the C terminus of Factor H. Thus, the interaction of CFHR1 and Factor H with the streptococcal proteins is complex, most likely includes multiple contact points and sites, and is influenced by charged residues.
Streptococcal Scl1.6 and Scl1.55 proteins bind CFHR1 and Factor H but do not bind FHL-1. In contrast, each of the three previously identified streptococcal complement regulator-binding proteins, M-protein, M-like proteins, and Fba, bind Factor H and FHL-1 but do not bind CFHR1 (21, 47–49) (Table 2). On the basis of these different binding features, we propose that S. pyogenes expresses two distinct types of surface proteins that recruit complement regulators by separate regions. Type A proteins, as identified here for Scl1.6 and Scl1.55, recruit CFHR1 and Factor H via their conserved C-terminal regions and do not bind FHL-1. Type B proteins, as represented by streptococcal M-Protein, M-like proteins, and Fba, bind Factor H and FHL-1 via SCR domain 7 and do not bind CFHR1.
TABLE 2.
Two distinct types of complement regulator recruitment
| Genus and species | Pathogen-encoded host complement regulator-recruiting proteins |
|
|---|---|---|
| Type Aa | Type Bb | |
| S. pyogenes | Scl1.6 and Scl1.55 | M-protein, emm-like protein, and Fba |
| S. aureus | Sbi | |
| B. burgdorferi | CRASP-3, CRASP-4 and CRASP-5 | CRASP-1 and CRASP-2 |
| P. aeruginosa | Tuf | |
| C. albicans | CaCRASP1 and CaCRASP2 | |
a CFHR1- and C-terminal Factor H-binding protein.
b FHL-1- and N-terminal Factor H-binding protein.
Such distinct and selective recruitment of host immune regulators is also used by other pathogens. For example, the Gram-negative bacterium Borrelia burgdorferi expresses three type A proteins, i.e. CRASP3, CRASP4, and CRASP5, that bind CFHR1 and Factor H via their C-terminal surface binding regions (50) and two borrelial type B proteins, i.e. CRASP1 and CRASP2, that bind Factor H and FHL-1 via SCR 7 (50–52). Additional examples of type A proteins include the Sbi protein of Staphylococcus aureus (53), Tuf of Pseudomonas aeruginosa (54), and CaCRASP-1 and CaCRASP-2 of Candida albicans (40, 55) (Table 2). Thus, GAS, similar to other pathogens, recruit different members of the Factor H protein family to the bacterial surface. It will be of interest to determine during which phase of infection the different sets of regulators are relevant.
The attachment of functionally distinct human complement regulators allows the pathogen to control and inhibit the complement cascade at multiple levels, i.e. AP C3 and C5 convertase and TCC formation. Both Factor H and CFHR1 are present in the plasma of healthy individuals and bind simultaneously to the streptococcal Scl proteins Scl1.6 and Scl1.55. Streptococcal type A proteins recruit Factor H, a regulator of the AP C3 convertase C3bBb, and also CFHR1, an inhibitor of the AP C5 convertase (C3bBbC3b) and of TCC formation. Thus, streptococci, by recruiting two different complement regulators, control host complement attack at distinct levels. Acquired Factor H blocks complement activation at the level of C3 and thereby inhibits C3b deposition and release of the anaphylatoxin C3a. In addition, acquired CFHR1 inhibits complement activation at the level of C5 convertase and blocks release of the anaphylatoxin C5a as well as TCC formation. In the presence of CFHR1, Factor H is recruited less efficiently, which at a physiological CFHR1-Factor H ratio of 1:2 reduced Factor H-mediated C3b degradation by ∼30%. Thus, simultaneous binding of CFHR1 and Factor H to Scl1 provides a unique and novel mechanism of complement evasion by GAS.
CFHR1 and Factor H bind to Scl1.6 and Scl1.55 but do not bind to Scl1.1 or to Scl2.28. However, M28-type streptococci, which express the nonbinding Scl2.28 protein, do bind CFHR1 and Factor H via their C termini. The binding appears to be at a lower intensity as compared with the Scl1.6- and Scl1.55-expressing GAS serotypes M6 and M55. This binding suggests that these strains express additional CFHR1- and Factor H-binding type A proteins. The identification of these proteins will be the subject of further studies.
In summary, the two human complement inhibitors CFHR1 and Factor H bind the streptococcal proteins Scl1.6 and Scl1.55. Scl1.6 and Scl1.55 are the first streptococcal proteins found to bind the two human complement regulators via their conserved C-terminal surface attachment regions. The interaction is complex and is likely mediated by several contact points, including a linear binding motif of five amino acids (SVEYQ) in SCR4 of CFHR1 and SCR19 of Factor H, as well as additional residues located in the adjacent domain (i.e. SCR5 of CFHR1 and SCR20 of Factor H).
Both CFHR1 and Factor H are present in human plasma, and both human complement regulators compete for binding to Scl1.6 and Scl1.55. At physiological levels, CFHR1 binding reduces Factor H binding, resulting in decreased regulatory activity of Factor H on AP C3 convertase. However, this effect is at the gain of C5 convertase activity and control of TCC formation. This novel mechanism involving the single surface protein Scl1 of GAS type M6 and M55 and two human ligands, CFHR1 and Factor H, protects GAS from complement attack by inhibiting the complement cascade at multiple levels.
Supplementary Material
Acknowledgment
We thank Steffi Hälbich for excellent assistance with surface plasmon resonance assays.
This work was supported, in whole or in part, by National Institutes of Health Grant AI083683 (to S. L.). This work was also supported by a grant from the “Grundlagenfond” of the Leibniz Institute for Natural Product Research and Infection Biology (to P. F. Z.) and an internal grant from the Office of Research and Graduate Education, West Virginia University Health Sciences Center (to S. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- GAS
- group A streptococci
- AP
- alternative pathway
- TCC
- terminal complement complex
- SCR
- short consensus repeat
- CFHR
- complement Factor H-related (protein)
- Scl
- streptococcal collagen-like (protein)
- DPBS
- Dulbecco's phosphate-buffered saline
- aHUS
- atypical hemolytic uremic syndrome
- MFI
- mean fluorescence intensity.
REFERENCES
- 1.Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005) Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 2.Pichichero M. E. (1998) Pediatr. Rev. 19, 291–302 [DOI] [PubMed] [Google Scholar]
- 3.Zipfel P. F., Reuter M. (2009) Int. J. Pept. Res. Ther. 15, 87–95 [Google Scholar]
- 4.Taylor P., Botto M., Walport M. (1998) Curr. Biol. 8, R259–R261 [DOI] [PubMed] [Google Scholar]
- 5.Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 6.Walport M. J. (2001) N. Engl. J. Med. 344, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 7.Tedesco F., Pausa M., Nardon E., Introna M., Mantovani A., Dobrina A. (1997) J. Exp. Med. 185, 1619–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrina A., Pausa M., Fischetti F., Bulla R., Vecile E., Ferrero E., Mantovani A., Tedesco F. (2002) Blood 99, 185–192 [DOI] [PubMed] [Google Scholar]
- 9.Józsi M., Oppermann M., Lambris J. D., Zipfel P. F. (2007) Mol. Immunol. 44, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zipfel P. F., Hellwage J., Friese M. A., Hegasy G., Jokiranta S. T., Meri S. (1999) Mol. Immunol 36, 241–248 [DOI] [PubMed] [Google Scholar]
- 11.Zipfel P. F., Skerka C., Hellwage J., Jokiranta S. T., Meri S., Brade V., Kraiczy P., Noris M., Remuzzi G. (2002) Biochem. Soc. Trans. 30, 971–978 [DOI] [PubMed] [Google Scholar]
- 12.Oppermann M., Manuelian T., Józsi M., Brandt E., Jokiranta T. S., Heinen S., Meri S., Skerka C., Götze O., Zipfel P. F. (2006) Clin. Exp. Immunol. 144, 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellwage J., Kühn S., Zipfel P. F. (1997) Biochem. J. 326, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J., Wiita P., Kawaguchi R., Honda J., Jorgensen A., Zhang K., Fischetti V. A., Sun H. (2007) Biochemistry 46, 8451–8461 [DOI] [PubMed] [Google Scholar]
- 15.Zipfel P. F., Skerka C. (2009) Nat. Rev. Immunol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 16.Skerka C., Horstmann R. D., Zipfel P. F. (1991) J. Biol. Chem. 266, 12015–12020 [PubMed] [Google Scholar]
- 17.Heinen S., Hartmann A., Lauer N., Wiehl U., Dahse H. M., Schirmer S., Gropp K., Enghardt T., Wallich R., Halbich S., Mihlan M., Schlotzer-Schrehardt U., Zipfel P. F., Skerka C. (2009) Blood 114, 2439–2447 [DOI] [PubMed] [Google Scholar]
- 18.Józsi M., Zipfel P. F. (2008) Trends Immunol. 29, 380–387 [DOI] [PubMed] [Google Scholar]
- 19.Jarva H., Jokiranta T. S., Würzner R., Meri S. (2003) Mol. Immunol. 40, 95–107 [DOI] [PubMed] [Google Scholar]
- 20.Zipfel P. F., Würzner R., Skerka C. (2007) Mol. Immunol. 44, 3850–3857 [DOI] [PubMed] [Google Scholar]
- 21.Kihlberg B. M., Collin M., Olsén A., Björck L. (1999) Infect. Immun. 67, 1708–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotarsky H., Hellwage J., Johnsson E., Skerka C., Svensson H. G., Lindahl G., Sjöbring U., Zipfel P. F. (1998) J. Immunol. 160, 3349–3354 [PubMed] [Google Scholar]
- 23.Akerström B., Lindqvist A., Maelen C. V., Grubb A., Lindahl G., Vaerman J. P. (1994) Mol. Immunol 31, 393–400 [DOI] [PubMed] [Google Scholar]
- 24.Stenberg L., O'Toole P. W., Mestecky J., Lindahl G. (1994) J. Biol. Chem. 269, 13458–13464 [PubMed] [Google Scholar]
- 25.Husmann L. K., Scott J. R., Lindahl G., Stenberg L. (1995) Infect. Immun. 63, 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thern A., Stenberg L., Dahlbäck B., Lindahl G. (1995) J. Immunol. 154, 375–386 [PubMed] [Google Scholar]
- 27.Johnsson E., Thern A., Dahlbäck B., Hedén L. O., Wikström M., Lindahl G. (1996) J. Immunol. 157, 3021–3029 [PubMed] [Google Scholar]
- 28.Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Ireland R. M., Reid S. D., Adams G. G., Musser J. M. (2000) Infect. Immun. 68, 6542–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen M., Edén A., Björck L. (2000) Infect. Immun. 68, 6370–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002) J. Biol. Chem. 277, 27312–27318 [DOI] [PubMed] [Google Scholar]
- 31.Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Shelvin B. J., Graviss E. A., Musser J. M. (2001) Infect. Immun. 69, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whatmore A. M. (2001) Microbiology 147, 419–429 [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen M., Björck L. (2001) Mol. Microbiol. 40, 1427–1438 [DOI] [PubMed] [Google Scholar]
- 34.Caswell C. C., Han R., Hovis K. M., Ciborowski P., Keene D. R., Marconi R. T., Lukomski S. (2008) Mol. Microbiol. 67, 584–596 [DOI] [PubMed] [Google Scholar]
- 35.Caswell C. C., Barczyk M., Keene D. R., Lukomska E., Gullberg D. E., Lukomski S. (2008) J. Biol. Chem. 283, 36168–36175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han R., Caswell C. C., Lukomska E., Keene D. R., Pawlowski M., Bujnicki J. M., Kim J. K., Lukomski S. (2006) Mol. Microbiol. 61, 351–367 [DOI] [PubMed] [Google Scholar]
- 37.Humtsoe J. O., Kim J. K., Xu Y., Keene D. R., Höök M., Lukomski S., Wary K. K. (2005) J. Biol. Chem. 280, 13848–13857 [DOI] [PubMed] [Google Scholar]
- 38.Påhlman L. I., Marx P. F., Mörgelin M., Lukomski S., Meijers J. C., Herwald H. (2007) J. Biol. Chem. 282, 24873–24881 [DOI] [PubMed] [Google Scholar]
- 39.Caswell C. C., Oliver-Kozup H., Han R., Lukomska E., Lukomski S. (2010) FEMS Microbiol. Lett. 303, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poltermann S., Kunert A., von der Heide M., Eck R., Hartmann A., Zipfel P. F. (2007) J. Biol. Chem. 282, 37537–37544 [DOI] [PubMed] [Google Scholar]
- 41.Han R., Zwiefka A., Caswell C. C., Xu Y., Keene D. R., Lukomska E., Zhao Z., Höök M., Lukomski S. (2006) Appl. Microbiol. Biotechnol. 72, 109–115 [DOI] [PubMed] [Google Scholar]
- 42.Kühn S., Skerka C., Zipfel P. F. (1995) J. Immunol. 155, 5663–5670 [PubMed] [Google Scholar]
- 43.Skerka C., Hellwage J., Weber W., Tilkorn A., Buck F., Marti T., Kampen E., Beisiegel U., Zipfel P. F. (1997) J. Biol. Chem. 272, 5627–5634 [DOI] [PubMed] [Google Scholar]
- 44.Wieland G. D., Nehmann N., Müller D., Eibel H., Siebenlist U., Sühnel J., Zipfel P. F., Skerka C. (2005) J. Cell Sci. 118, 3203–3212 [DOI] [PubMed] [Google Scholar]
- 45.Hellwage J., Jokiranta T. S., Friese M. A., Wolk T. U., Kampen E., Zipfel P. F., Meri S. (2002) J. Immunol. 169, 6935–6944 [DOI] [PubMed] [Google Scholar]
- 46.Józsi M., Heinen S., Hartmann A., Ostrowicz C. W., Hälbich S., Richter H., Kunert A., Licht C., Saunders R. E., Perkins S. J., Zipfel P. F., Skerka C. (2006) J Am. Soc. Nephrol. 17, 170–177 [DOI] [PubMed] [Google Scholar]
- 47.Blackmore T. K., Fischetti V. A., Sadlon T. A., Ward H. M., Gordon D. L. (1998) Infect. Immun. 66, 1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischetti V. A. (1989) Clin. Microbiol. Rev. 2, 285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandiripally V., Wei L., Skerka C., Zipfel P. F., Cue D. (2003) Infect. Immun. 71, 7119–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haupt K., Kraiczy P., Wallich R., Brade V., Skerka C., Zipfel P. F. (2007) J. Infect. Dis. 196, 124–133 [DOI] [PubMed] [Google Scholar]
- 51.Kraiczy P., Hellwage J., Skerka C., Kirschfink M., Brade V., Zipfel P. F., Wallich R. (2003) Eur. J. Immunol. 33, 697–707 [DOI] [PubMed] [Google Scholar]
- 52.Wallich R., Pattathu J., Kitiratschky V., Brenner C., Zipfel P. F., Brade V., Simon M. M., Kraiczy P. (2005) Infect. Immun. 73, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haupt K., Reuter M., van den Elsen J., Burman J., Hälbich S., Richter J., Skerka C., Zipfel P. F. (2008) PLoS Pathog. 4, e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunert A., Losse J., Gruszin C., Hühn M., Kaendler K., Mikkat S., Volke D., Hoffmann R., Jokiranta T. S., Seeberger H., Moellmann U., Hellwage J., Zipfel P. F. (2007) J. Immunol. 179, 2979–2988 [DOI] [PubMed] [Google Scholar]
- 55.Meri T., Blom A. M., Hartmann A., Lenk D., Meri S., Zipfel P. F. (2004) Infect. Immun. 72, 6633–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.